Independent Effects of a Herbivore’s Bacterial Symbionts on Its Performance and Induced Plant Defences
Abstract
:1. Introduction
2. Results
2.1. Bacterial Communities in Antibiotics-Treated and Untreated Mite Lines of Tetranychus urticae DeLier-1 and Santpoort-2
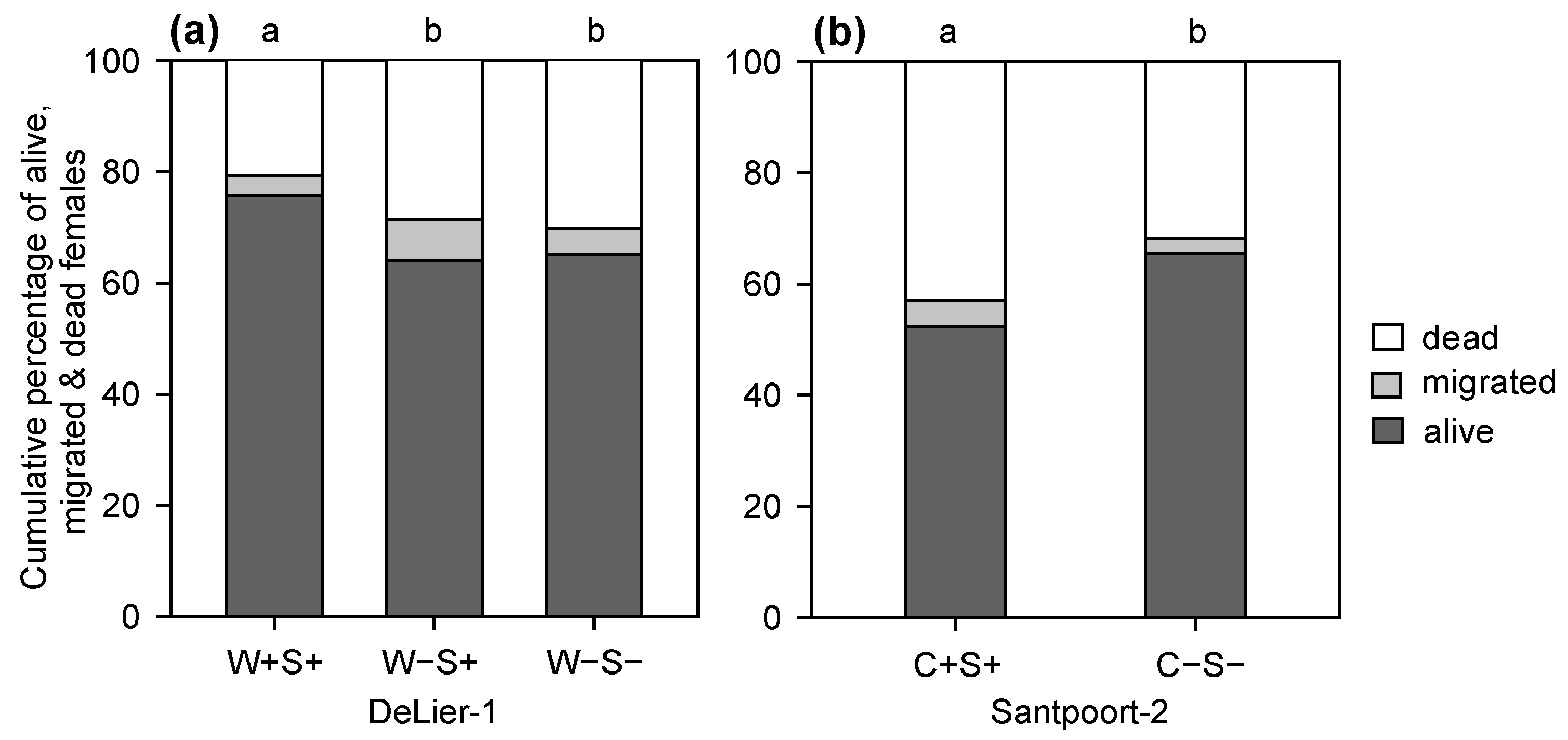
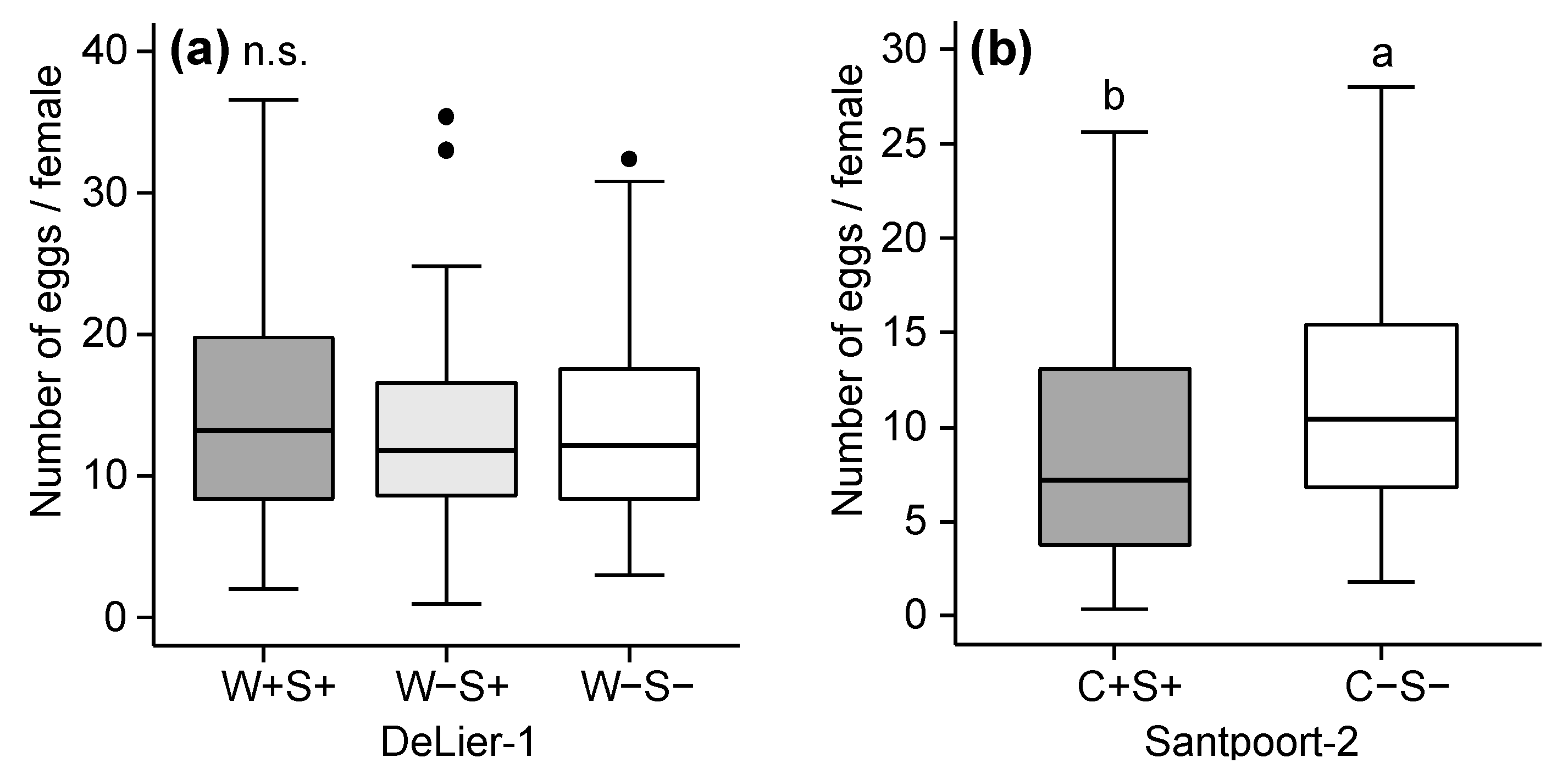
2.2. Effects of Wolbachia, Cardinium and Spiroplasma on Spider Mite Performance
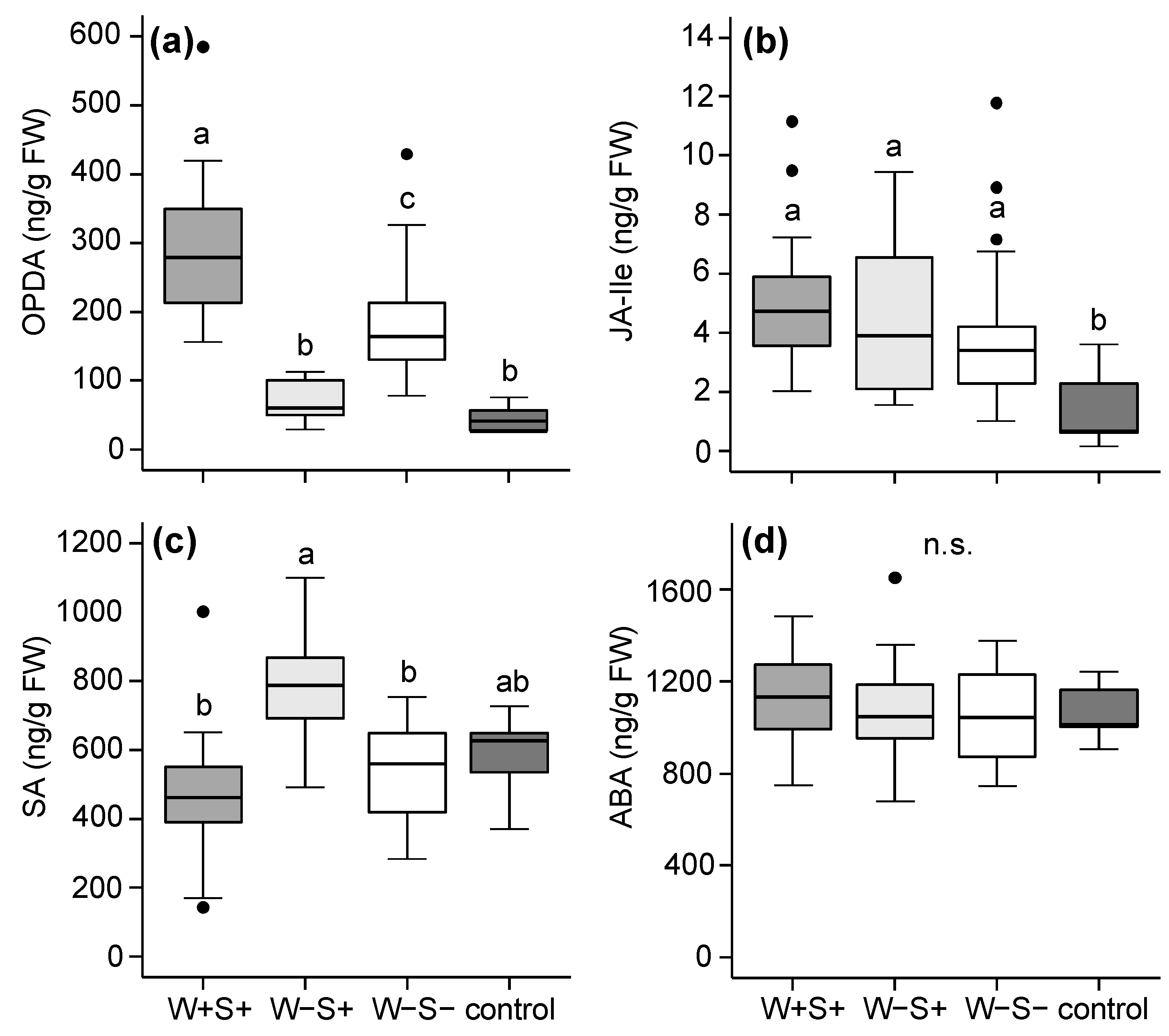
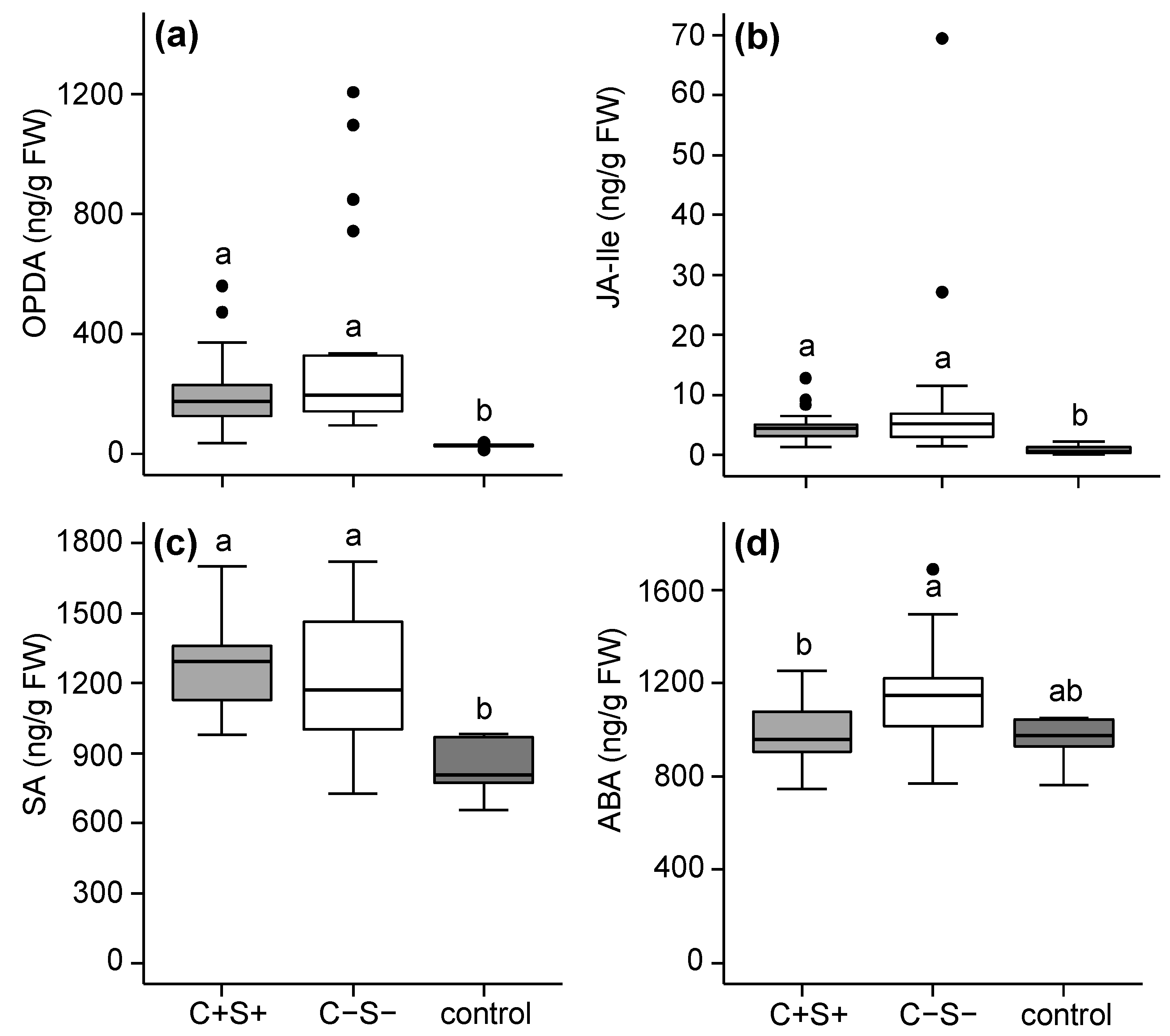
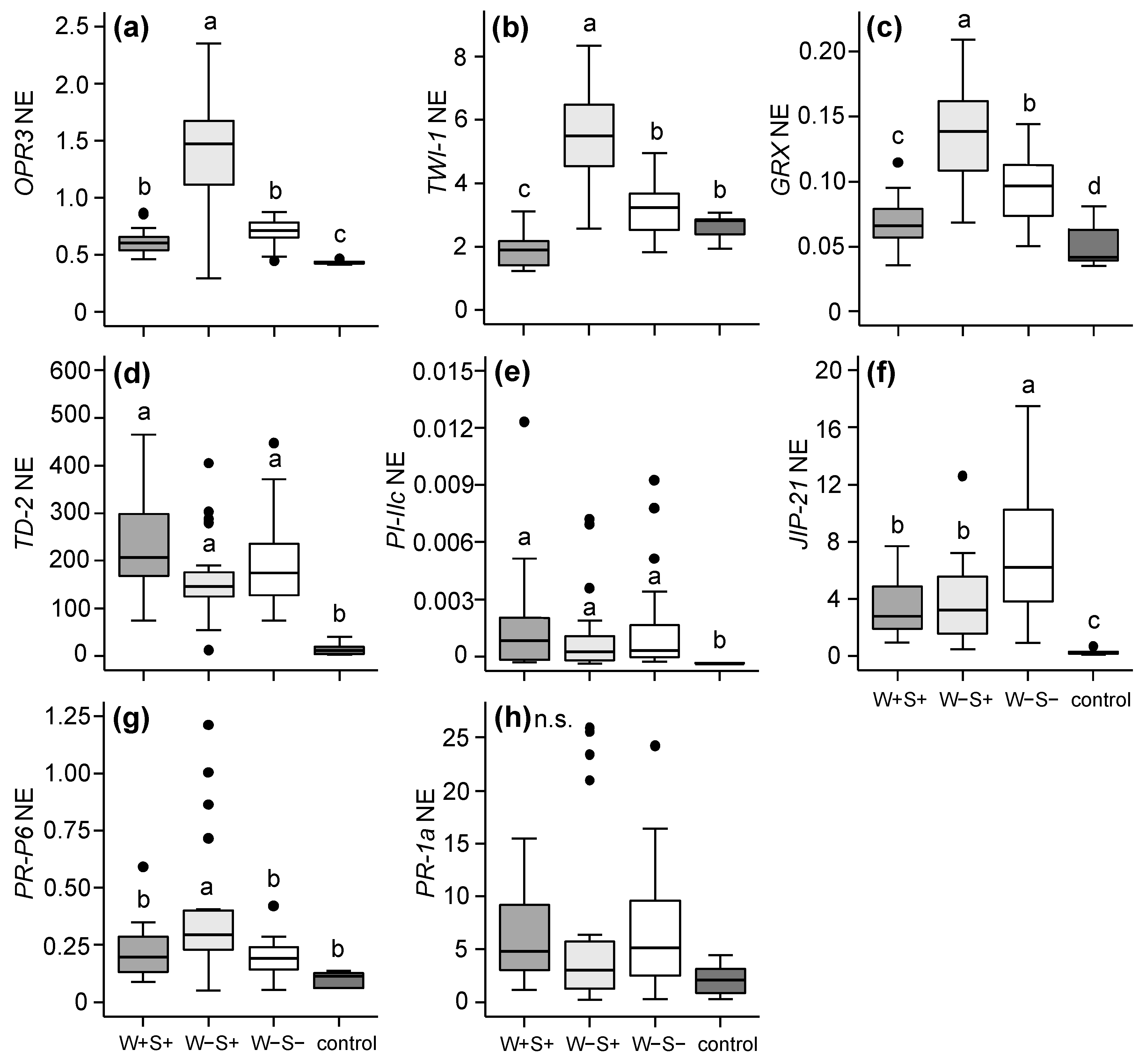
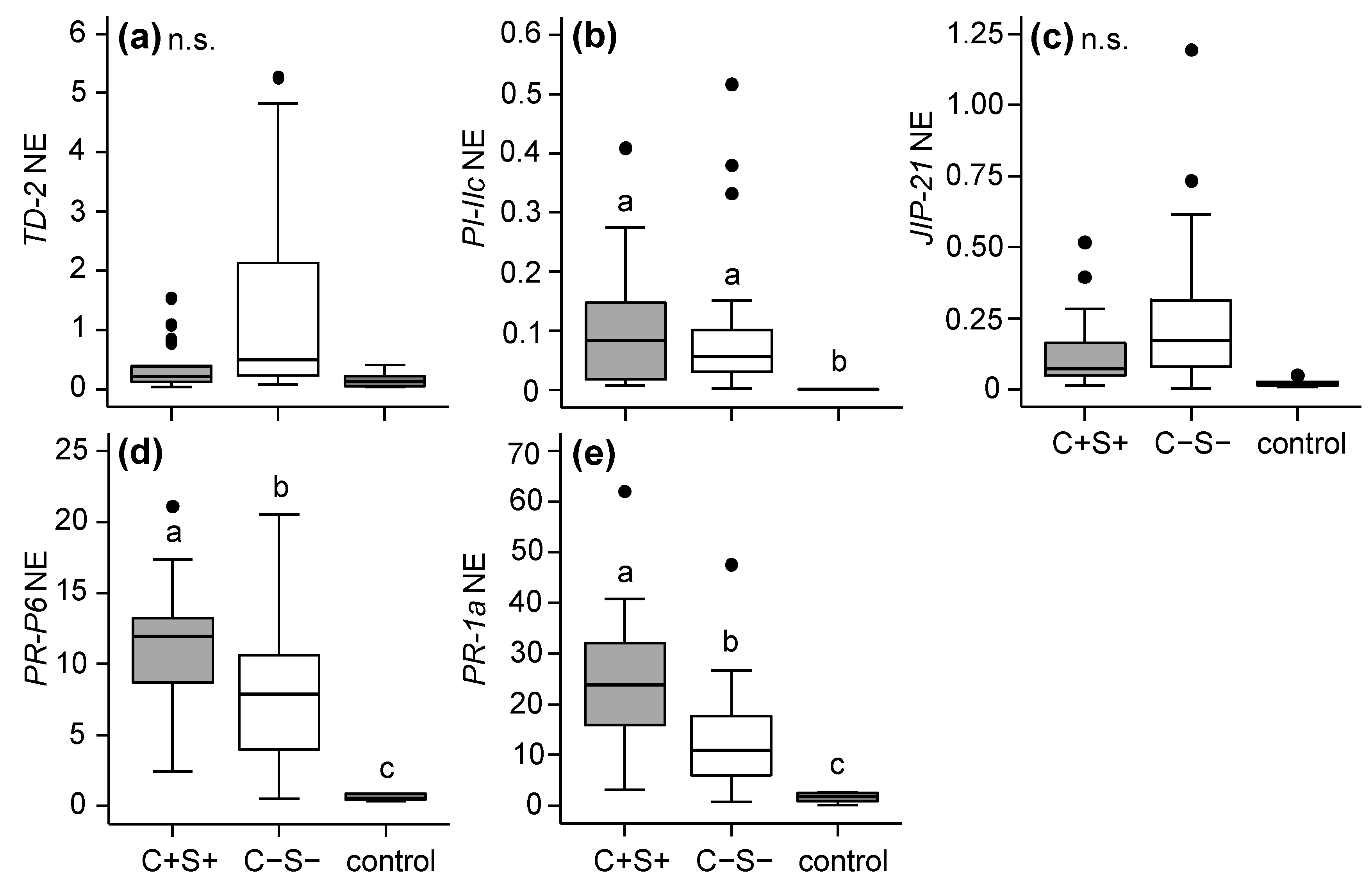
2.2.1. Effects of Mite-Associated Wolbachia, Cardinium and Spiroplasma on Tomato Induced Responses
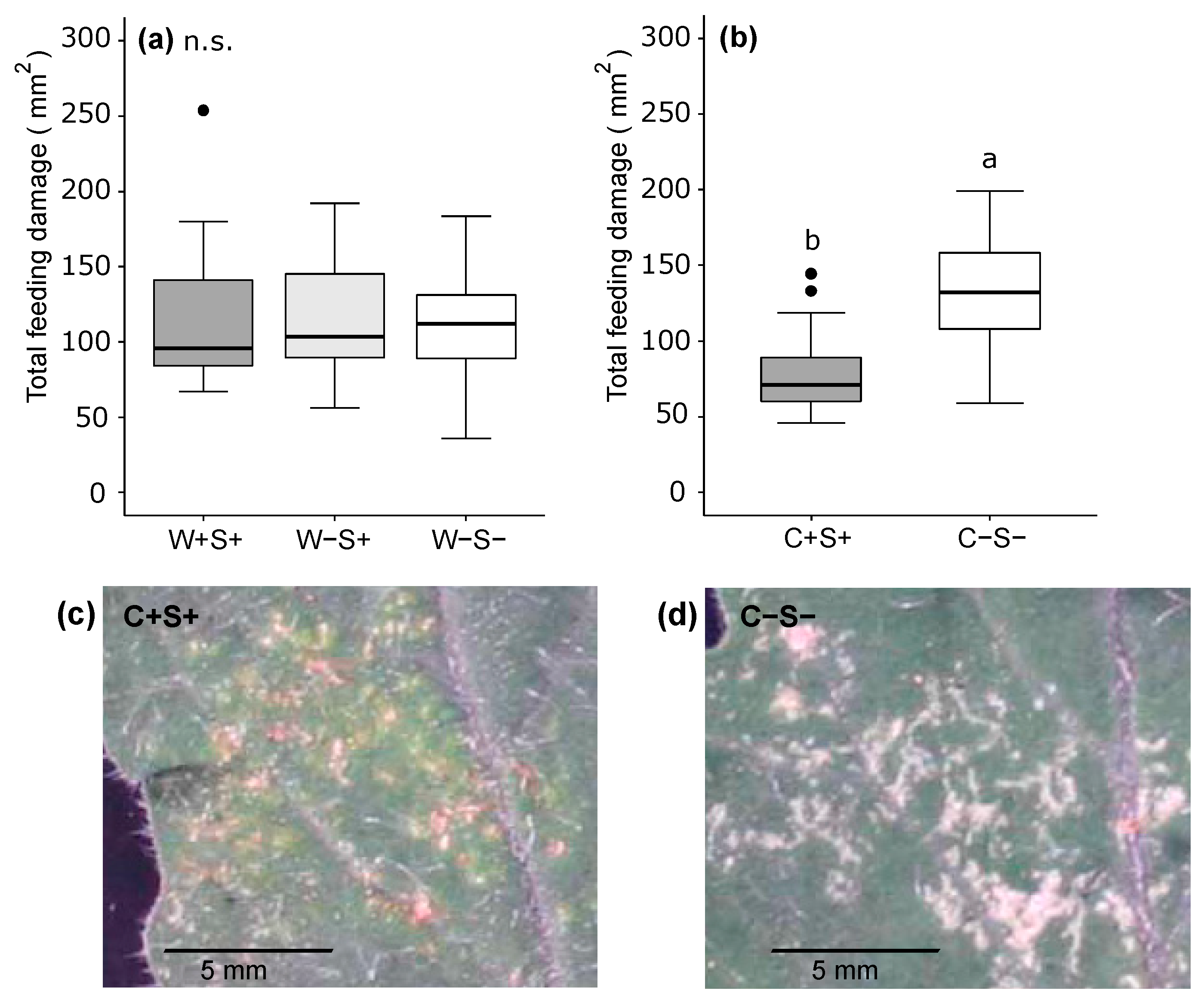
2.2.2. Effects of Wolbachia, Cardinium and Spiroplasma on the Amount of Feeding Damage Inflicted by Spider Mites
3. Discussion
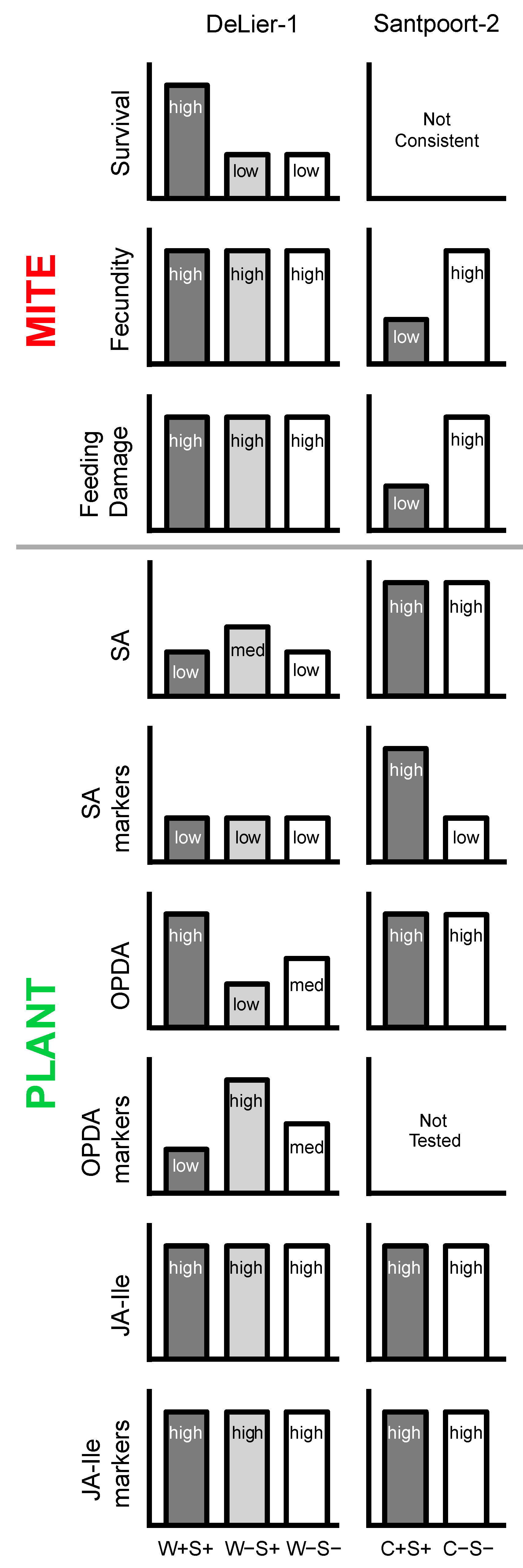
3.1. Effects of Wolbachia, Spiroplasma and Cardinium on Spider Mite Performance
3.2. Effects of Wolbachia, Cardinium and Spiroplasma on Tomato Induced Responses
3.3. The Combined Presence of Wolbachia and Spiroplasma Bacteria Has Consequences for Induced Plant Responses
4. Material and Methods
4.1. Plants
4.2. Spider Mites
4.2.1. Bacterial Communities in Antibiotics-Treated and Untreated Mite Lines of Tetranychus urticae DeLier-1 and Santpoort-2
4.2.2. Antibiotics Treatments and Nomenclature of Mite Lines
4.2.3. Illumina Sequencing
4.2.4. Diagnostic PCRs on Mites
4.3. Effects of Wolbachia, Cardinium and Spiroplasma on Spider Mite Performance
4.3.1. Spider Mite Performance Assay
4.3.2. Statistical Analysis of Spider Mite Performance Assay
4.4. Effects of SpiderMite-Associated Wolbachia, Cardinium and Spiroplasma on Induced Plant Responses
4.4.1. Plant Infestation Assay
4.4.2. Isolation of Phytohormones and Analysis by Means of Liquid Chromatography Tandem Mass Spectrometry
4.4.3. Gene Expression Analysis by Quantitative Reverse-Transcription PCR
4.4.4. Statistical Analysis of the Plant Infestation Assay Data—Phytohormones, Quantitative Reverse-Transcription PCR, Feeding Damage
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Schoonhoven, L.M.; Van Loon, J.J.; Dicke, M. Insect-Plant Biology, 2nd ed.; Oxford University Press: Oxford, UK, 2005; p. 440. [Google Scholar]
- Karban, R.; Baldwin, I.T. Induced Responses to Herbivory; University of Chicago Press: Chicago, IL, USA, 2007; p. 330. [Google Scholar]
- Mithöfer, A.; Boland, W. Plant defense against herbivores: Chemical aspects. Annu. Rev. Plant Biol. 2012, 63, 431–450. [Google Scholar] [CrossRef] [PubMed]
- De Bary, A. Die Erscheinung der Symbiose: Vortrag Gehalten auf der Versammlung Deutscher Naturforscher und Aerzte zu Cassel; Karl, J., Ed.; Trubner: Strasbourg, Germany, 1879. [Google Scholar]
- Douglas, A.E. The microbial dimension in insect nutritional ecology. Funct. Ecol. 2009, 23, 38–47. [Google Scholar] [CrossRef]
- Feldhaar, H. Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol. Entomol. 2011, 36, 533–543. [Google Scholar] [CrossRef]
- Engel, P.; Moran, N.A. The gut microbiota of insects–diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef] [PubMed]
- Hansen, A.K.; Moran, N.A. The impact of microbial symbionts on host plant utilization by herbivorous insects. Mol. Ecol. 2014, 23, 1473–1496. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, P.; Krischik, V.A.; Jones, C.G. Microbial Mediation of Plant-Herbivore Interactions; John Wiley & Sons: New York, NY, USA, 1991. [Google Scholar]
- Frago, E.; Dicke, M.; Godfray, H.C. Insect symbionts as hidden players in insect-plant interactions. Trends Ecol. Evol. 2012, 27, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Casteel, C.L.; Hansen, A.K. Evaluating insect-microbiomes at the plant-insect interface. J. Chem. Ecol. 2014, 40, 836–847. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.H.; Rosa, C.; Scully, E.D.; Peiffer, M.; Tooker, J.F.; Hoover, K.; Luthe, D.S.; Felton, G.W. Herbivore exploits orally secreted bacteria to suppress plant defenses. Proc. Natl. Acad. Sci. USA 2013, 110, 15728–15733. [Google Scholar] [CrossRef] [PubMed]
- Duron, O.; Bouchon, D.; Boutin, S.; Bellamy, L.; Zhou, L.; Engelstadter, J.; Hurst, G.D. The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol. 2008, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Werren, J.H. Biology of Wolbachia. Annu. Rev. Entomol. 1997, 42, 587–609. [Google Scholar] [CrossRef] [PubMed]
- Werren, J.H.; Baldo, L.; Clark, M.E. Wolbachia: Master manipulators of invertebrate biology. Nat. Rev. Microbiol. 2008, 6, 741–751. [Google Scholar] [CrossRef] [PubMed]
- Engelstädter, J.; Hurst, G.D.D. The Ecology and Evolution of Microbes that Manipulate Host Reproduction. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 127–149. [Google Scholar] [CrossRef]
- Zug, R.; Hammerstein, P. Still a host of hosts for Wolbachia: Analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS ONE 2012, 7, e38544. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.A.; Hercus, M.; Dagher, H. Population dynamics of the Wolbachia infection causing cytoplasmic incompatibility in Drosophila melanogaster. Genetics 1998, 148, 221–231. [Google Scholar] [PubMed]
- Fry, A.J.; Palmer, M.R.; Rand, D.M. Variable fitness effects of Wolbachia infection in Drosophila melanogaster. Heredity 2004, 93, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Sugio, A.; Dubreuil, G.; Giron, D.; Simon, J.C. Plant-insect interactions under bacterial influence: Ecological implications and underlying mechanisms. J. Exp. Bot. 2015, 66, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Zug, R.; Hammerstein, P. Bad guys turned nice? A critical assessment of Wolbachia mutualisms in arthropod hosts. Biol. Rev. Camb. Philos. Soc. 2015, 90, 89–111. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Vilchez, I.; Mateos, M. Spiroplasma bacteria enhance survival of Drosophila hydei attacked by the parasitic wasp Leptopilina heterotoma. PLoS ONE 2010, 5, e12149. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Butler, S.; Sanchez, G.; Mateos, M. Male killing Spiroplasma protects Drosophila melanogaster against two parasitoid wasps. Heredity 2014, 112, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Jaenike, J.; Unckless, R.; Cockburn, S.N.; Boelio, L.M.; Perlman, S.J. Adaptation via symbiosis: Recent spread of a Drosophila defensive symbiont. Science 2010, 329, 212–215. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, P.T.; Leong, J.S.; Koop, B.F.; Perlman, S.J. Transcriptional responses in a Drosophila defensive symbiosis. Mol. Ecol. 2014, 23, 1558–1570. [Google Scholar] [CrossRef] [PubMed]
- Moreira, L.A.; Iturbe-Ormaetxe, I.; Jeffery, J.A.; Lu, G.; Pyke, A.T.; Hedges, L.M.; Rocha, B.C.; Hall-Mendelin, S.; Day, A.; Riegler, M.; et al. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 2009, 139, 1268–1278. [Google Scholar] [CrossRef] [PubMed]
- Walker, T.; Johnson, P.H.; Moreira, L.A.; Iturbe-Ormaetxe, I.; Frentiu, F.D.; McMeniman, C.J.; Leong, Y.S.; Dong, Y.; Axford, J.; Kriesner, P.; et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 2011, 476, 450–453. [Google Scholar] [CrossRef] [PubMed]
- Hedges, L.M.; Brownlie, J.C.; O’Neill, S.L.; Johnson, K.N. Wolbachia and virus protection in insects. Science 2008, 322, 702. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, L.; Ferreira, A.; Ashburner, M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008, 6, e2. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, P.T.; Peng, F.; Boulanger, M.J.; Perlman, S.J. A ribosome-inactivating protein in a Drosophila defensive symbiont. Proc. Natl. Acad. Sci. USA 2016, 113, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Foster, J.; Ganatra, M.; Kamal, I.; Ware, J.; Makarova, K.; Ivanova, N.; Bhattacharyya, A.; Kapatral, V.; Kumar, S.; Posfai, J.; et al. The Wolbachia genome of Brugia malayi: Endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 2005, 3, e121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brownlie, J.C.; Cass, B.N.; Riegler, M.; Witsenburg, J.J.; Iturbe-Ormaetxe, I.; McGraw, E.A.; O’Neill, S.L. Evidence for metabolic provisioning by a common invertebrate endosymbiont, Wolbachia pipientis, during periods of nutritional stress. PLoS Pathog. 2009, 5, e1000368. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, T.; Koga, R.; Kikuchi, Y.; Meng, X.Y.; Fukatsu, T. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc. Natl. Acad. Sci. USA 2010, 107, 769–774. [Google Scholar] [CrossRef] [PubMed]
- Unckless, R.L.; Jaenike, J. Maintenance of a male-killing Wolbachia in Drosophila innubila by male-killing dependent and male-killing independent mechanisms. Evolution 2012, 66, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, W.; Huguet, E.; Casas, J.; Commin, C.; Giron, D. Plant green-island phenotype induced by leaf-miners is mediated by bacterial symbionts. Proc. Biol. Sci. R. Soc. 2010, 277, 2311–2319. [Google Scholar] [CrossRef] [PubMed]
- Body, M.; Kaiser, W.; Dubreuil, G.; Casas, J.; Giron, D. Leaf-miners co-opt microorganisms to enhance their nutritional environment. J. Chem. Ecol. 2013, 39, 969–977. [Google Scholar] [CrossRef] [PubMed]
- Barr, K.L.; Hearne, L.B.; Briesacher, S.; Clark, T.L.; Davis, G.E. Microbial symbionts in insects influence down-regulation of defense genes in maize. PLoS ONE 2010, 5, e11339. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.A.; Frank, D.L.; Leach, K.A.; Turlings, T.C.; Hibbard, B.E.; Erb, M. Direct and indirect plant defenses are not suppressed by endosymbionts of a specialist root herbivore. J. Chem. Ecol. 2013, 39, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Breeuwer, J.; Jacobs, G. Wolbachia: Intracellular manipulators of mite reproduction. Exp. Appl. Acarol. 1996, 20, 421–434. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, T.; Noda, H.; Hong, X. Wolbachia distribution and cytoplasmic incompatibility based on a survey of 42 spider mite species (Acari: Tetranychidae) in Japan. Heredity 2003, 91, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Enigl, M.; Schausberger, P. Incidence of the endosymbionts Wolbachia, Cardinium and Spiroplasma in phytoseiid mites and associated prey. Exp. Appl. Acarol. 2007, 42, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, T.; Noda, H.; Ito, S. Cardinium symbionts cause cytoplasmic incompatibility in spider mites. Heredity 2007, 98, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Bolland, H.R.; Gutierrez, J.; Flechtmann, C.H. World Catalogue of the Spider Mite Family (Acari: Tetranychidae); Brill: Amsterdam, The Netherlands, 1998. [Google Scholar]
- Grbić, M.; Van Leeuwen, T.; Clark, R.M.; Rombauts, S.; Rouzé, P.; Grbić, V.; Osborne, E.J.; Dermauw, W.; Ngoc, P.C.T.; Ortego, F.; et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 2011, 479, 487–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Migeon, A.; Nouguier, E.; Dorkeld, F. Spider Mites Web: A Comprehensive Database for the Tetranychidae; Trends in Acarology: Amsterdam, The Netherlands; Springer: Amsterdam, The Netherlands, 2010; pp. 557–560. [Google Scholar]
- Erb, M.; Meldau, S.; Howe, G.A. Role of phytohormones in insect-specific plant reactions. Trends Plant Sci. 2012, 17, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef] [PubMed]
- Kessler, A.; Halitschke, R.; Baldwin, I.T. Silencing the jasmonate cascade: Induced plant defenses and insect populations. Science 2004, 305, 665–668. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef] [PubMed]
- Kant, M.R.; Ament, K.; Sabelis, M.W.; Haring, M.A.; Schuurink, R.C. Differential timing of spider mite-induced direct and indirect defenses in tomato plants. Plant Physiol. 2004, 135, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Zhurov, V.; Navarro, M.; Bruinsma, K.A.; Arbona, V.; Santamaria, M.E.; Cazaux, M.; Wybouw, N.; Osborne, E.J.; Ens, C.; Rioja, C.; et al. Reciprocal responses in the interaction between Arabidopsis and the cell-content-feeding chelicerate herbivore spider mite. Plant Physiol. 2014, 164, 384–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alba, J.M.; Schimmel, B.C.J.; Glas, J.J.; Ataide, L.M.; Pappas, M.L.; Villarroel, C.A.; Schuurink, R.C.; Sabelis, M.W.; Kant, M.R. Spider mites suppress tomato defenses downstream of jasmonate and salicylate independently of hormonal crosstalk. New Phytol. 2015, 205, 828–840. [Google Scholar] [CrossRef] [PubMed]
- Martel, C.; Zhurov, V.; Navarro, M.; Martinez, M.; Cazaux, M.; Auger, P.; Migeon, A.; Santamaria, M.E.; Wybouw, N.; Diaz, I.; et al. Tomato Whole Genome Transcriptional Response to Tetranychus urticae Identifies Divergence of Spider Mite-Induced Responses between Tomato and Arabidopsis. Mol. Plant Microbe Interact. 2015, 28, 343–361. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Williams, M.M.; Loh, Y.T.; Lee, G.I.; Howe, G.A. Resistance of cultivated tomato to cell content-feeding herbivores is regulated by the octadecanoid-signaling pathway. Plant Physiol. 2002, 130, 494–503. [Google Scholar] [CrossRef] [PubMed]
- Ament, K.; Kant, M.R.; Sabelis, M.W.; Haring, M.A.; Schuurink, R.C. Jasmonic acid is a key regulator of spider mite-induced volatile terpenoid and methyl salicylate emission in tomato. Plant Physiol. 2004, 135, 2025–2037. [Google Scholar] [CrossRef] [PubMed]
- Kant, M.R.; Sabelis, M.W.; Haring, M.A.; Schuurink, R.C. Intraspecific variation in a generalist herbivore accounts for differential induction and impact of host plant defences. Proc. Biol. Sci. R. Soc. 2008, 275, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Ament, K.; Krasikov, V.; Allmann, S.; Rep, M.; Takken, F.L.; Schuurink, R.C. Methyl salicylate production in tomato affects biotic interactions. Plant J. 2010, 62, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Ataide, L.M.; Pappas, M.L.; Schimmel, B.C.; Lopez-Orenes, A.; Alba, J.M.; Duarte, M.V.; Pallini, A.; Schuurink, R.C.; Kant, M.R. Induced plant-defenses suppress herbivore reproduction but also constrain predation of their offspring. Plant Sci. 2016, 252, 300–310. [Google Scholar] [CrossRef] [PubMed]
- Villarroel, C.A.; Jonckheere, W.; Alba, J.M.; Glas, J.J.; Dermauw, W.; Haring, M.A.; van Leeuwen, T.; Schuurink, R.C.; Kant, M.R. Salivary proteins of spider mites suppress defenses in Nicotiana benthamiana and promote mite reproduction. Plant J. 2016, 86, 119–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yano, S.; Kanaya, M.; Takafuji, A. Genetic basis of color variation in leaf scars induced by the Kanzawa spider mite. Entomol. Exp. Appl. 2003, 106, 37–44. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Bordenstein, S.R.; Werren, J.H. Do Wolbachia influence fecundity in Nasonia vitripennis? Heredity 2000, 84, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Ebbert, M.A. The interaction phenotype in the Drosophila willistoni-Spiroplasma symbiosis. Evolution 1991, 45, 971–988. [Google Scholar] [CrossRef]
- Hoffmann, A.A.; Clancy, D.J.; Merton, E. Cytoplasmic incompatibility in Australian populations of Drosophila melanogaster. Genetics 1994, 136, 993–999. [Google Scholar] [PubMed]
- Vala, F.; Breeuwer, J.A.; Sabelis, M.W. Wolbachia-induced ‘hybrid breakdown’ in the two-spotted spider mite Tetranychus urticae Koch. Proc. R. Soc. Lond. B Biol. Sci. 2000, 267, 1931–1937. [Google Scholar] [CrossRef] [PubMed]
- Fukatsu, T.; Tsuchida, T.; Nikoh, N.; Koga, R. Spiroplasma symbiont of the pea aphid, Acyrthosiphon pisum (Insecta: Homoptera). Appl. Environ. Microbiol. 2001, 67, 1284–1291. [Google Scholar] [CrossRef] [PubMed]
- Fry, A.J.; Rand, D.M. Wolbachia interactions that determine Drosophila melanogaster survival. Evolution 2002, 56, 1976–1981. [Google Scholar] [CrossRef] [PubMed]
- Perrot-Minnot, M.J.; Cheval, B.; Migeon, A.; Navajas, M. Contrasting effects of Wolbachia on cytoplasmic incompatibility and fecundity in the haplodiploid mite Tetranychus urticae. J. Evol. Biol. 2002, 15, 808–817. [Google Scholar] [CrossRef]
- Weeks, A.R.; Stouthamer, R. Increased fecundity associated with infection by a cytophaga-like intracellular bacterium in the predatory mite, Metaseiulus occidentalis. Proc. Biol. Sci. R. Soc. 2004, 271 (Suppl. S4), S193–S195. [Google Scholar] [CrossRef] [PubMed]
- Montenegro, H.; Petherwick, A.; Hurst, G.; Klaczko, L. Fitness effects of Wolbachia and Spiroplasma in Drosophila melanogaster. Genetica 2006, 127, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Ros, V.I.; Breeuwer, J.A. The effects of, and interactions between, Cardinium and Wolbachia in the doubly infected spider mite Bryobia sarothamni. Heredity 2009, 102, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Anbutsu, H.; Fukatsu, T. Spiroplasma as a model insect endosymbiont. Environ. Microbiol. Rep. 2011, 3, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.-R.; Chen, X.-L.; Hong, X.-Y. Variable fitness and reproductive effects of Wolbachia infection in populations of the two-spotted spider mite Tetranychus urticae Koch in China. Appl. Entomol. Zool. 2011, 46, 95–102. [Google Scholar] [CrossRef]
- Stefanini, A.; Duron, O. Exploring the effect of the Cardinium endosymbiont on spiders. J. Evol. Biol. 2012, 25, 1521–1530. [Google Scholar] [CrossRef] [PubMed]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, S.; Chini, A.; Hamberg, M.; Adie, B.; Porzel, A.; Kramell, R.; Miersch, O.; Wasternack, C.; Solano, R. (+)-7-iso-Jasmonoyl-l-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 2009, 5, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Stintzi, A.; Weber, H.; Reymond, P.; Browse, J.; Farmer, E.E. Plant defense in the absence of jasmonic acid: The role of cyclopentenones. Proc. Natl. Acad. Sci. USA 2001, 98, 12837–12842. [Google Scholar] [CrossRef] [PubMed]
- Taki, N.; Sasaki-Sekimoto, Y.; Obayashi, T.; Kikuta, A.; Kobayashi, K.; Ainai, T.; Yagi, K.; Sakurai, N.; Suzuki, H.; Masuda, T.; et al. 12-oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol. 2005, 139, 1268–1283. [Google Scholar] [CrossRef] [PubMed]
- Dabrowska, P.; Freitak, D.; Vogel, H.; Heckel, D.G.; Boland, W. The phytohormone precursor OPDA is isomerized in the insect gut by a single, specific glutathione transferase. Proc. Natl. Acad. Sci. USA 2009, 106, 16304–16309. [Google Scholar] [CrossRef] [PubMed]
- Stotz, H.U.; Jikumaru, Y.; Shimada, Y.; Sasaki, E.; Stingl, N.; Mueller, M.J.; Kamiya, Y. Jasmonate-dependent and COI1-independent defense responses against Sclerotinia sclerotiorum in Arabidopsis thaliana: Auxin is part of COI1-independent defense signaling. Plant Cell Physiol. 2011, 52, 1941–1956. [Google Scholar] [CrossRef] [PubMed]
- Farmer, E.E.; Mueller, M.J. ROS-mediated lipid peroxidation and RES-activated signaling. Annu. Rev. Plant Biol. 2013, 64, 429–450. [Google Scholar] [CrossRef] [PubMed]
- Park, S.W.; Li, W.; Viehhauser, A.; He, B.; Kim, S.; Nilsson, A.K.; Andersson, M.X.; Kittle, J.D.; Ambavaram, M.M.; Luan, S.; et al. Cyclophilin 20-3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proc. Natl. Acad. Sci. USA 2013, 110, 9559–9564. [Google Scholar] [CrossRef] [PubMed]
- Bosch, M.; Wright, L.P.; Gershenzon, J.; Wasternack, C.; Hause, B.; Schaller, A.; Stintzi, A. Jasmonic acid and its precursor 12-oxophytodienoic acid control different aspects of constitutive and induced herbivore defenses in tomato. Plant Physiol. 2014, 166, 396–410. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.M.; Li, H.C.; Zhou, S.R.; Xue, H.W.; Miao, X.X. Cis-12-oxo-phytodienoic acid stimulates rice defense response to a piercing-sucking insect. Mol. Plant 2014, 7, 1683–1692. [Google Scholar] [CrossRef] [PubMed]
- Shabab, M.; Khan, S.A.; Vogel, H.; Heckel, D.G.; Boland, W. OPDA isomerase GST16 is involved in phytohormone detoxification and insect development. FEBS J. 2014, 281, 2769–2783. [Google Scholar] [CrossRef] [PubMed]
- Scalschi, L.; Sanmartin, M.; Camanes, G.; Troncho, P.; Sanchez-Serrano, J.J.; Garcia-Agustin, P.; Vicedo, B. Silencing of OPR3 in tomato reveals the role of OPDA in callose deposition during the activation of defense responses against Botrytis cinerea. Plant J. 2015, 81, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Mueller, S.; Hilbert, B.; Dueckershoff, K.; Roitsch, T.; Krischke, M.; Mueller, M.J.; Berger, S. General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 2008, 20, 768–785. [Google Scholar] [CrossRef] [PubMed]
- Uquillas, C.; Letelier, I.; Blanco, F.; Jordana, X.; Holuigue, L. NPR1-independent activation of immediate early salicylic acid-responsive genes in Arabidopsis. Mol. Plant Microbe Interact. 2004, 17, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Langlois-Meurinne, M.; Gachon, C.M.; Saindrenan, P. Pathogen-responsive expression of glycosyltransferase genes UGT73B3 and UGT73B5 is necessary for resistance to Pseudomonas syringae pv tomato in Arabidopsis. Plant Physiol. 2005, 139, 1890–1901. [Google Scholar] [CrossRef] [PubMed]
- Ndamukong, I.; Abdallat, A.A.; Thurow, C.; Fode, B.; Zander, M.; Weigel, R.; Gatz, C. SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 2007, 50, 128–139. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, P.J.; Truesdale, M.R.; Calvert, C.M.; Dorans, A.; Roberts, M.R.; Bowles, D.J. A novel tomato gene that rapidly responds to wound-and pathogen-related signals. Plant J. 1998, 14, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Herman, M.; Davidson, J.; Smart, C. Induction of plant defense gene expression by plant activators and Pseudomonas syringae pv. tomato in greenhouse-grown tomatoes. Phytopathology 2008, 98, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, Y.; Hase, S.; Takenaka, S.; Kanayama, Y.; Yoshioka, H.; Kamoun, S.; Takahashi, H. INF1 Elicitin Activates Jasmonic Acid-and Ethylene-mediated Signalling Pathways and Induces Resistance to Bacterial Wilt Disease in Tomato. J. Phytopathol. 2009, 157, 287–297. [Google Scholar] [CrossRef]
- Tornero, P.; Gadea, J.; Conejero, V.; Vera, P. Two PR-1 genes from tomato are differentially regulated and reveal a novel mode of expression for a pathogenesis-related gene during the hypersensitive response and development. Mol. Plant Microbe Interact. 1997, 10, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Fidantsef, A.; Stout, M.; Thaler, J.; Duffey, S.; Bostock, R. Signal interactions in pathogen and insect attack: Expression of lipoxygenase, proteinase inhibitor II, and pathogenesis-related protein P4 in the tomato, Lycopersicon esculentum. Physiol. Mol. Plant Pathol. 1999, 54, 97–114. [Google Scholar] [CrossRef]
- Su, Q.; Oliver, K.M.; Xie, W.; Wu, Q.; Wang, S.; Zhang, Y.; Biere, A. The whitefly-associated facultative symbiont Hamiltonella defensa suppresses induced plant defences in tomato. Funct. Ecol. 2015, 29, 1007–1018. [Google Scholar] [CrossRef]
- De Torres Zabala, M.; Bennett, M.H.; Truman, W.H.; Grant, M.R. Antagonism between salicylic and abscisic acid reflects early host-pathogen conflict and moulds plant defence responses. Plant J. 2009, 59, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Dommel, M.; Mou, Z. Abscisic Acid Promotes Proteasome-Mediated Degradation of the Transcription Coactivator NPR1 in Arabidopsis thaliana. Plant J. 2016, 86, 20–34. [Google Scholar] [CrossRef] [PubMed]
- Audenaert, K.; De Meyer, G.B.; Höfte, M.M. Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 2002, 128, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, R.; Ozawa, R.; Uefune, M.; Gotoh, T.; Takabayashi, J. Intraspecies variation in the Kanzawa spider mite differentially affects induced defensive response in lima bean plants. J. Chem. Ecol. 2006, 32, 2501–2512. [Google Scholar] [CrossRef] [PubMed]
- Hughes, G.L.; Dodson, B.L.; Johnson, R.M.; Murdock, C.C.; Tsujimoto, H.; Suzuki, Y.; Patt, A.A.; Cui, L.; Nossa, C.W.; Barry, R.M. Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. Proc. Natl. Acad. Sci. USA 2014, 111, 12498–12503. [Google Scholar] [CrossRef] [PubMed]
- Rossi, P.; Ricci, I.; Cappelli, A.; Damiani, C.; Ulissi, U.; Mancini, M.V.; Valzano, M.; Capone, A.; Epis, S.; Crotti, E.; et al. Mutual exclusion of Asaia and Wolbachia in the reproductive organs of mosquito vectors. Parasites Vectors 2015, 8, 278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rousset, F.; Braig, H.R.; O’Neill, S.L. A stable triple Wolbachia infection in Drosophila with nearly additive incompatibility effects. Heredity 1999, 82, 620–627. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.-X.; Zhang, X.-F.; Chen, D.-S.; Zhang, Y.-K.; Hong, X.-Y. Wolbachia-host interactions: Host mating patterns affect Wolbachia density dynamics. PLoS ONE 2013, 8, e66373. [Google Scholar] [CrossRef] [PubMed]
- Goto, S.; Anbutsu, H.; Fukatsu, T. Asymmetrical interactions between Wolbachia and Spiroplasma endosymbionts coexisting in the same insect host. Appl. Environ. Microbiol. 2006, 72, 4805–4810. [Google Scholar] [CrossRef] [PubMed]
- Oliver, K.M.; Moran, N.A.; Hunter, M.S. Costs and benefits of a superinfection of facultative symbionts in aphids. Proc. R. Soc. Lond. B Biol. Sci. 2006, 273, 1273–1280. [Google Scholar] [CrossRef] [PubMed]
- Unckless, R.L.; Boelio, L.M.; Herren, J.K.; Jaenike, J. Wolbachia as populations within individual insects: Causes and consequences of density variation in natural populations. Proc. Biol. Sci. R. Soc. 2009, 276, 2805–2811. [Google Scholar] [CrossRef] [PubMed]
- Schmid-Hempel, P. Evolutionary Parasitology: The Integrated Study of Infections, Immunology, Ecology, and Genetics; Oxford University Press: Oxford, UK, 2011. [Google Scholar]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Bittinger, K.; Bushman, F.D.; DeSantis, T.Z.; Andersen, G.L.; Knight, R. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 2010, 26, 266–267. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Staudacher, H.; Sabelis, M.W. Why do males choose heterospecific females in the red spider mite? Exp. Appl. Acarol. 2016, 68, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Storms, J. Some physiological effects of spider mite infestation on bean plants. Neth. J. Plant Pathol. 1971, 77, 154–167. [Google Scholar] [CrossRef]
- Bates, D.; Maechler, M.; Bolker, B.; Walker, S. Lme4: Linear Mixed-Effects Models Using Eigen and S4. R Package Version 1.7. 2014. Available online: http://cran.r-project.org/package=lme4 (accessed on 14 May 2015).
- Hothorn, T.; Bretz, F.; Westfall, P. Simultaneous inference in general parametric models. Biometr. J. 2008, 50, 346–363. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing, version 3.0.2; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Verwoerd, T.C.; Dekker, B.; Hoekema, A. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 1989, 17, 2362. [Google Scholar] [CrossRef] [PubMed]
- Strassner, J.; Schaller, F.; Frick, U.B.; Howe, G.A.; Weiler, E.W.; Amrhein, N.; Macheroux, P.; Schaller, A. Characterization and cDNA-microarray expression analysis of 12-oxophytodienoate reductases reveals differential roles for octadecanoid biosynthesis in the local versus the systemic wound response. Plant J. 2002, 32, 585–601. [Google Scholar] [CrossRef] [PubMed]
- Lisón, P.; Rodrigo, I.; Conejero, V. A novel function for the cathepsin D inhibitor in tomato. Plant Physiol. 2006, 142, 1329–1339. [Google Scholar] [CrossRef] [PubMed]
- Gonzales-Vigil, E.; Bianchetti, C.M.; Phillips, G.N., Jr.; Howe, G.A. Adaptive evolution of threonine deaminase in plant defense against insect herbivores. Proc. Natl. Acad. Sci. USA 2011, 108, 5897–5902. [Google Scholar] [CrossRef] [PubMed]
- Gadea, J.; Mayda, M.E.; Conejero, V.; Vera, P. Characterization of defense-related genes ectopically expressed in viroid-infected tomato plants. Mol. Plant Microbe Interact. 1996, 9, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Van Kan, J.A.; Joosten, M.H.; Wagemakers, C.A.; van den Berg-Velthuis, G.C.; de Wit, P.J. Differential accumulation of mRNAs encoding extracellular and intracellular PR proteins in tomato induced by virulent and avirulent races of Cladosporium fulvum. Plant Mol. Biol. 1992, 20, 513–527. [Google Scholar] [CrossRef] [PubMed]
- Truesdale, M.R.; Doherty, H.M.; Loake, G.J.; McPherson, M.J.; Roberts, M.; Bowles, D.J. Molecular cloning of a novel wound-induced gene from tomato: Twi1. Plant Physiol. 1996, 112, 446. [Google Scholar]
- Harrell, F.E.; Dupont, C. Hmisc: Harrell Miscellaneous. R Package Version 3.15-0. 2015. Available online: http://biostat.mc.vanderbilt.edu/wiki/Main/Hmisc (accessed on 14 May 2015).
| Mite Strain | Group | Line | Total | Wolbachia | Spiroplasma | Cardinium |
|---|---|---|---|---|---|---|
| DeLier-1 | W+S+ | 1 | 3164 | 1272 | 141 | - |
| 2 | 14,303 | 2131 | 350 | 1 | ||
| 3 | 34,118 | 7940 | 1605 | 2 | ||
| 4 | 19,747 | 8546 | 890 | - | ||
| W−S+ | 1 | 4371 | 1 | 440 | - | |
| 2 | 16,637 | - | 1120 | 1 | ||
| 3 | 27,939 | 6 | 2422 | 2 | ||
| 4 | 8812 | 1 | 567 | - | ||
| W−S− | 1 | 4975 | 3 | - | - | |
| 2 | 20,054 | 4 | - | 1 | ||
| 3 | 10,241 | 2 | - | - | ||
| 4 | 8906 | 1 | - | - | ||
| Santpoort-2 | C+S+ | 5 | 26,384 | 7 | 1159 | 7443 |
| 6 | 18,367 | 5 | 165 | 3643 | ||
| 7 | 48,325 | 108 | 3555 | 19,744 | ||
| 8 | 28,966 | 204 | 1467 | 7904 | ||
| C−S− | 5 | 39,276 | 100 | 1 | 2 | |
| 6 | 22,648 | 110 | 1 | 1 | ||
| 7 | 30,608 | 66 | - | 3 | ||
| 8 | 18,702 | 63 | 6 | - |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Staudacher, H.; Schimmel, B.C.J.; Lamers, M.M.; Wybouw, N.; Groot, A.T.; Kant, M.R. Independent Effects of a Herbivore’s Bacterial Symbionts on Its Performance and Induced Plant Defences. Int. J. Mol. Sci. 2017, 18, 182. https://doi.org/10.3390/ijms18010182
Staudacher H, Schimmel BCJ, Lamers MM, Wybouw N, Groot AT, Kant MR. Independent Effects of a Herbivore’s Bacterial Symbionts on Its Performance and Induced Plant Defences. International Journal of Molecular Sciences. 2017; 18(1):182. https://doi.org/10.3390/ijms18010182
Chicago/Turabian StyleStaudacher, Heike, Bernardus C. J. Schimmel, Mart M. Lamers, Nicky Wybouw, Astrid T. Groot, and Merijn R. Kant. 2017. "Independent Effects of a Herbivore’s Bacterial Symbionts on Its Performance and Induced Plant Defences" International Journal of Molecular Sciences 18, no. 1: 182. https://doi.org/10.3390/ijms18010182
APA StyleStaudacher, H., Schimmel, B. C. J., Lamers, M. M., Wybouw, N., Groot, A. T., & Kant, M. R. (2017). Independent Effects of a Herbivore’s Bacterial Symbionts on Its Performance and Induced Plant Defences. International Journal of Molecular Sciences, 18(1), 182. https://doi.org/10.3390/ijms18010182






