Antibacterial Activity of Juglone against Staphylococcus aureus: From Apparent to Proteomic
Abstract
:1. Introduction
2. Results and Discussion
2.1. iTRAQ Analysis of the Proteome after Treatment with Juglone
2.2. Functional Annotation Analysis of Proteomic Differences
2.3. Upregulation of Glyoxalase, Potassium Uptake Protein, and Nitroreductase
2.4. Significance of the Downregulation of Thioredoxin, Threonine Dehydratase, and Ribulose-5-phosphate 3-Epimerase-epimerase
2.5. Downregulation of Proteins Related to DNA Replication and Transcription
2.6. Role of Juglone in Stimulating Stress Response in S. aureus
2.7. Role of Juglone on Protein Synthesis, Cell Wall Formation, and Permeability
3. Materials and Methods
3.1. Strain and Juglone
3.2. Culture Preparation
3.3. Protein Preparation
3.4. Protein Digestion and iTRAQ Labeling
3.5. Strong Cation-Exchange Chromatography (SCX) Fractionation
3.6. LC-ESI-MS/MS
3.7. Database Searching and Data Analysis
3.8. Statistical and Bioinformatics Analysis
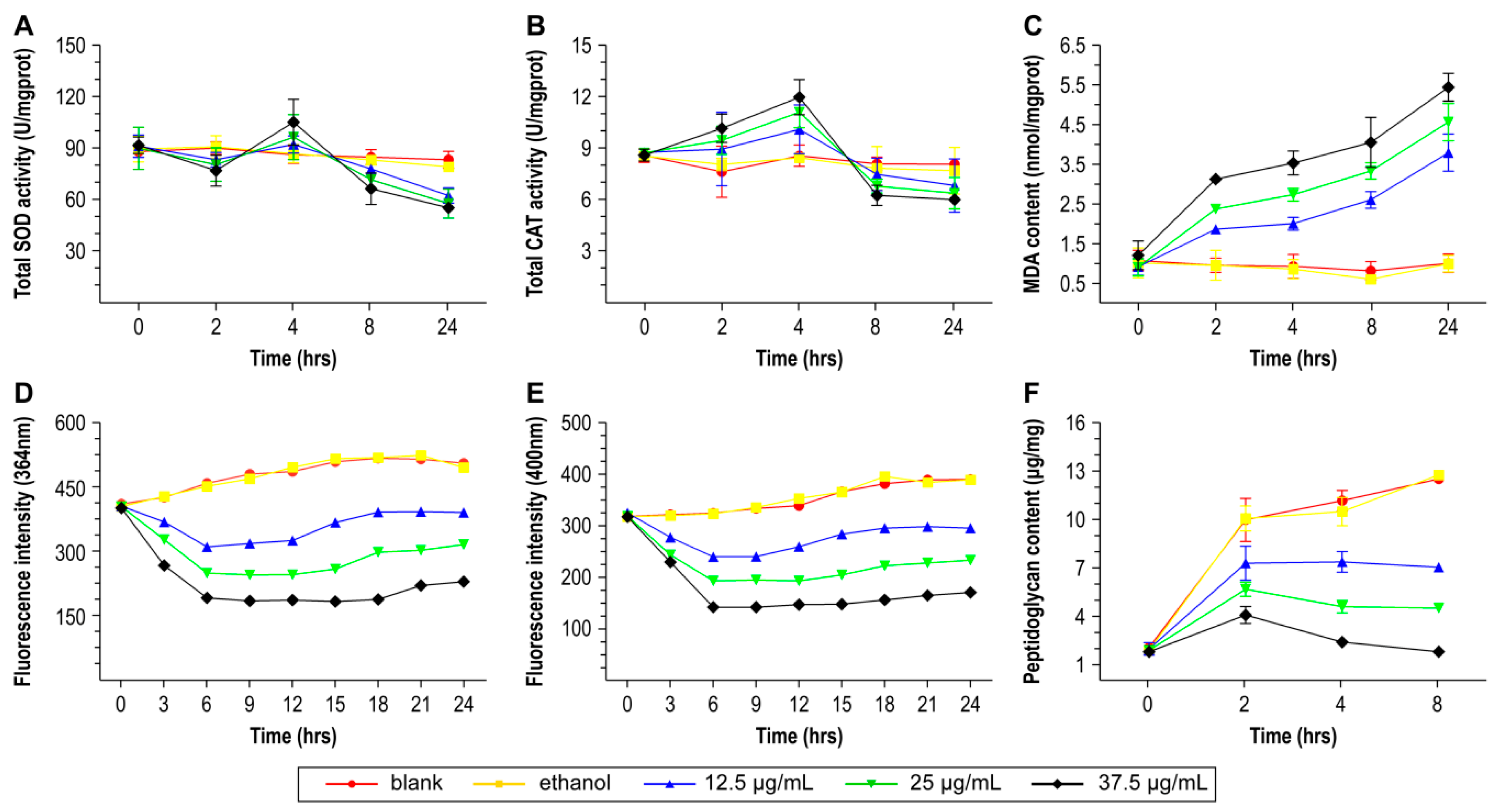
3.9. Detection of Changes in DNA and RNA Content
3.10. Peptidoglycan Content Determination
3.11. Determination of the Extent of Oxidative Damage
3.12. Phospholipid Extraction
3.13. Gas Chromatography Analysis of Fatty Acids
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| BCA | bicinchoninic acid |
| FASP | filter-aided proteome preparation |
| FBD | food-borne diseases |
| FDR | false discovery rate |
| GO | gene ontology |
| HCD | higher-energy collisional dissociation |
| iTRAQ | isobaric tags for relative and absolute quantitation |
| LC-ESI-MS/MS | liquid chromatography electrospray ionization tandem mass spectrometry |
| MDA | malondialdehyde |
| MIC | minimal inhibition concentration |
| MS2 | second-level mass spectrometry |
| ROS | reactive oxygen species |
| SEs | staphylococcal enterotoxins |
| SFA | saturated fatty acids |
| SFP | staphylococcal food poisoning |
| SOD | superoxide dismutase |
| TAG | triacylglycerols |
| UFA | unsaturated fatty acids |
| WHO | World Health Organization |
| GTP | guanosine triphosphate |
| CTP | cytidine triphosphate |
| CAT | catalase |
References
- Kadariya, J.; Smith, T.C.; Thapaliya, D. Staphylococcus aureus and staphylococcal food-borne disease: An ongoing challenge in public health. Biomed. Res. Int. 2014, 2014, 827965. [Google Scholar] [CrossRef] [PubMed]
- Friedman, M.; Rasooly, R. Review of the inhibition of biological activities of food-related selected toxins by natural compounds. Toxins 2013, 5, 743–775. [Google Scholar] [CrossRef] [PubMed]
- Hennekinne, J.A.; de Buyser, M.L.; Dragacci, S. Staphylococcus aureus and its food poisoning toxins: Characterization and outbreak investigation. FEMS Microbiol. Rev. 2012, 36, 815–836. [Google Scholar] [CrossRef] [PubMed]
- Gallina, S.; Bianchi, D.M.; Bellio, A.; Nogarol, C.; Macori, G.; Zaccaria, T.; Biorci, F.; Carraro, E.; Decastelli, L. Staphylococcal poisoning foodborne outbreak: Epidemiological investigation and strain genotyping. J. Food Prot. 2013, 76, 2093–2098. [Google Scholar] [CrossRef] [PubMed]
- Pinchuk, I.V.; Beswick, E.J.; Reyes, V.E. Staphylococcal enterotoxins. Toxins 2010, 2, 2177–2197. [Google Scholar] [CrossRef] [PubMed]
- Babula, P.; Adam, V.; Havel, L.; Kizek, R. Noteworthy secondary metabolites naphthoquinones—Their occurrence, pharmacological properties and analysis. Curr. Pharm. Anal. 2009, 5, 47–68. [Google Scholar] [CrossRef]
- Wu, Z.; Chen, G.; Wang, Y. Inhibition effect of juglong on several food deterioration microorganisms. China Brew. 2009, 8, 76–78. [Google Scholar]
- Padayatty, S.J.; Katz, A.; Wang, Y.; Eck, P.; Kwon, O.; Lee, J.-H.; Chen, S.; Corpe, C.; Dutta, A.; Dutta, S.K.; et al. Vitamin c as an antioxidant: Evaluation of its role in disease prevention. J. Am. Coll. Nutr. 2003, 22, 18–35. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.L. Antioxidants and oxidative stress in exercise. Exp. Biol. Med. 1999, 222, 283–292. [Google Scholar] [CrossRef]
- Paiva, S.A.R.; Russell, R.M. Β-carotene and other carotenoids as antioxidants. J. Am. Coll. Nutr. 1999, 18, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Ferland, G. The discovery of vitamin k and its clinical applications. Ann. Nutr. Metab. 2012, 61, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Hofmeister, A.E.; Grabowski, R.; Linder, D.; Buckler, W. L-serine and l-threonine dehydratase from clostridium propionicum two enzymes with different prosthetic groups. Eur. J. Biochem. 1993, 215, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Sobota, J.M.; Imlay, J.A. Iron enzyme ribulose-5-phosphate 3-epimerase in escherichia coli is rapidly damaged by hydrogen peroxide but can be protected by manganese. Proc. Natl. Acad. Sci. USA 2011, 108, 5402–5407. [Google Scholar] [CrossRef] [PubMed]
- Iwata-Reuyl, D. Biosynthesis of the 7-deazaguanosine hypermodified nucleosides of transfer rna. Bioorg. Chem. 2003, 31, 24–43. [Google Scholar] [CrossRef]
- Zhang, R.L.; Hirsch, O.; Mohsen, M.; Samuni, A. Effects of nitroxide stable radicals on juglone cytotoxicity. Arch. Biochem. Biophys. 1994, 312, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Klaus, V.; Hartmann, T.; Gambini, J.; Graf, P.; Stahl, W.; Hartwig, A.; Klotz, L.O. 1,4-naphthoquinones as inducers of oxidative damage and stress signaling in hacat human keratinocytes. Arch. Biochem. Biophys. 2010, 496, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Saify, Z.S.; Mushtaq, N.; Noor, F.; Takween, S.; Arif, M. Role of quinone moiety as antitumour agents: A review. Pak. J. Pharm. Sci. 1999, 12, 21–31. [Google Scholar] [PubMed]
- Alvarez, H.M.; Steinbuchel, A. Triacylglycerols in prokaryotic microorganisms. Appl. Microbiol. Biotechnol. 2002, 60, 367–376. [Google Scholar] [PubMed]
- Hecker, M.; Volker, U. General stress response of bacillus subtilis and other bacteria. Adv. Microb. Physiol. 2001, 44, 35–91. [Google Scholar] [PubMed]
- Wozniak, D.J.; Tiwari, K.B.; Soufan, R.; Jayaswal, R.K. The mcsb gene of the clpc operon is required for stress tolerance and virulence in staphylococcus aureus. Microbiology 2012, 158, 2568–2576. [Google Scholar] [CrossRef] [PubMed]
- Overton, T.W.; Justino, M.C.; Li, Y.; Baptista, J.M.; Melo, A.M.; Cole, J.A.; Saraiva, L.M. Widespread distribution in pathogenic bacteria of di-iron proteins that repair oxidative and nitrosative damage to iron-sulfur centers. J. Bacteriol. 2008, 190, 2004–2013. [Google Scholar] [CrossRef] [PubMed]
- Brunskill, E.W.; de Jonge, B.L.; Bayles, K.W. The staphylococcus aureus scda gene: A novel locus that affects cell division and morphogenesis. Microbiology 1997, 143, 2877–2882. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.; Small, D.A.; Toghrol, F.; Bentley, W.E. Global transcriptome analysis of staphylococcus aureus response to hydrogen peroxide. J. Bacteriol. 2006, 188, 1648–1659. [Google Scholar] [CrossRef] [PubMed]
- Freiberg, C.; Brunner, N.A.; Schiffer, G.; Lampe, T.; Pohlmann, J.; Brands, M.; Raabe, M.; Habich, D.; Ziegelbauer, K. Identification and characterization of the first class of potent bacterial acetyl-coa carboxylase inhibitors with antibacterial activity. J. Biol. Chem. 2004, 279, 26066–26073. [Google Scholar] [CrossRef] [PubMed]
- De Smet, M.J.; Kingma, J.; Witholt, B. The effect of toluene on the structure and permeability of the outer and cytoplasmic membranes of escherichia coli. BBA Biomembr. 1978, 506, 64–80. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Meth. 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, A.; Lindell, G.; Källström, B.N.; Branca, R.M.; Danielsson, K.G.; Dahlberg, M.; Larson, B.; Forshed, J.; Lehtiö, J. Tumor proteomics by multivariate analysis on individual pathway data for characterization of vulvar cancer phenotypes. Mol. Cell. Proteom. MCP 2012, 11. [Google Scholar] [CrossRef] [PubMed]
- Gan, C.S.; Chong, P.K.; Pham, T.K.; Wright, P.C. Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (itraq). J. Proteome Res. 2007, 6, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talon, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the blast2go suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
| Accession | Protein Name | Gene id | Unique Peptides | Total Peptides | Mw (kDa) | Fold Change | p-Value |
|---|---|---|---|---|---|---|---|
| Oxidative damage | |||||||
| Q2FVA5 | glyoxalase | SAOUHSC_02828 | 5 | 5 | 30.00 | 1.824 | 0.012 |
| Q2FZH1 | potassium uptake protein | SAOUHSC_01034 | 2 | 2 | 24.30 | 1.401 | 0.037 |
| Q2FUW1 | serine rich repeat containing protein | sasA | 1 | 1 | 227.90 | 1.406 | 0.027 |
| Q2FWN0 | nitroreductase | SAOUHSC_02258 | 4 | 4 | 24.00 | 1.203 | 0.005 |
| Q2FZD2 | thioredoxin | trxA | 9 | 9 | 11.40 | 0.654 | 0.001 |
| Q2G000 | thioredoxin | SAOUHSC_00834 | 6 | 6 | 12.10 | 0.758 | 0.046 |
| Q2FXI6 | thioredoxin | SAOUHSC_01860 | 8 | 8 | 11.80 | 0.781 | 0.003 |
| Q2FZ62 | ribulose-phosphate 3-epimerase | SAOUHSC_01189 | 2 | 2 | 23.60 | 0.750 | 0.038 |
| Q2FWJ9 | threonine dehydratase biosynthetic | ilvA | 3 | 3 | 46.90 | 0.751 | 0.016 |
| Q2FV58 | 4,4′-diaponeurosporenoate glycosyltransferase | crtQ | 2 | 2 | 42.50 | 0.782 | 0.016 |
| DNA replication and transcription | |||||||
| Q2FWH6 | DNA-binding response regulator | SAOUHSC_02315 | 2 | 2 | 26.50 | 0.787 | 0.034 |
| Q2FXW6 | uridine kinase | Udk | 3 | 3 | 23.50 | 0.798 | 0.022 |
| Q2FV02 | anaerobic ribonucleoside-triphosphate | SAOUHSC_02942 | 3 | 3 | 70.40 | 0.711 | 0.030 |
| Q2G112 | single-stranded DNA-binding protein | SAOUHSC_00349 | 7 | 7 | 18.50 | 0.816 | 0.014 |
| Q2FZ97 | transcriptional regulator MraZ | mraZ | 3 | 3 | 17.20 | 0.819 | 0.006 |
| Q2FV69 | family transcriptional regulator | SAOUHSC_02867 | 1 | 2 | 21.90 | 0.623 | 0.034 |
| Q2FZK9 | family transcriptional regulator | SAOUHSC_00992 | 2 | 2 | 16.40 | 0.650 | 0.003 |
| Q2G273 | urease accessory protein ureg | ureG | 3 | 3 | 22.30 | 0.796 | 0.021 |
| Q2FXX1 | acetyl- biotin carboxylase subunit | SAOUHSC_01709 | 4 | 4 | 50.20 | 0.761 | 0.022 |
| Q2G1X5 | queuosine biosynthesis protein | SAOUHSC_00720 | 3 | 3 | 16.00 | 0.831 | 0.000 |
| Q2FXT6 | queuine tRNA-ribosyltransferase | tgt | 2 | 2 | 43.30 | 0.603 | 0.008 |
| Protein synthesis | |||||||
| Q2FW17 | 50s ribosomal protein l24 | rplX | 2 | 2 | 11.50 | 1.225 | 0.001 |
| Q2FW29 | 50s ribosomal protein l36 | rpmJ | 1 | 1 | 4.30 | 0.704 | 0.002 |
| Q2FW16 | 50s ribosomal protein l14 | rplN | 4 | 4 | 13.10 | 0.734 | 0.010 |
| Q2FY22 | 50S ribosomal protein L33 2 | rpmG2 | 1 | 1 | 5.90 | 0.459 | 0.003 |
| Stress response | |||||||
| Q2FUU5 | lipase 1 | lipA | 2 | 3 | 76.60 | 1.255 | 0.004 |
| Q2FZS8 | chaperone protein clpB | clpB | 23 | 24 | 98.30 | 1.211 | 0.000 |
| Q2G222 | N-acetylmuramoyl-l-alanine amidase | SAOUHSC_02979 | 11 | 11 | 69.20 | 1.259 | 0.000 |
| Q2G0P6 | ATP:guanido phosphotransferase | mcsB | 2 | 2 | 38.60 | 1.234 | 0.010 |
| P72360 | iron-sulfur cluster repair di-iron protein | scdA | 1 | 2 | 25.50 | 0.796 | 0.028 |
| Q2G112 | single-stranded DNA-binding protein | SAOUHSC_00349 | 7 | 7 | 18.50 | 0.816 | 0.014 |
| Cell wall synthesis and cell division | |||||||
| Q2FUW7 | accessory sec system glycosylation | gtf1 | 1 | 1 | 58.20 | 1.591 | 0.031 |
| Q2G222 | N-acetylmuramoyl-l-alanine amidase | SAOUHSC_02979 | 11 | 11 | 69.20 | 1.259 | 0.000 |
| P72360 | iron-sulfur cluster repair di-iron protein | scdA | 1 | 2 | 25.50 | 0.796 | 0.028 |
| Q2FZ97 | transcriptional regulator MraZ | mraZ | 3 | 3 | 17.20 | 0.819 | 0.006 |
| Q2FVQ1 | gnat family acetyltransferase | SAOUHSC_02651 | 1 | 1 | 20.10 | 0.669 | 0.019 |
| Membrane permeability and formation | |||||||
| Q2FZH1 | potassium uptake protein | SAOUHSC_01034 | 2 | 2 | 24.30 | 1.401 | 0.037 |
| Q2FZS8 | chaperone protein clpB | clpB | 23 | 24 | 98.30 | 1.211 | 0.000 |
| Q2FZD2 | Thioredoxin | trxA | 9 | 9 | 11.40 | 0.654 | 0.001 |
| Q2FXI6 | Thioredoxin | SAOUHSC_01860 | 8 | 8 | 11.80 | 0.781 | 0.003 |
| Q2FXX1 | acetyl- biotin carboxylase subunit | SAOUHSC_01709 | 4 | 4 | 50.20 | 0.761 | 0.022 |
| Others | |||||||
| Q2FVB3 | antibiotic transport system permease | SAOUHSC_02821 | 1 | 1 | 28.90 | 1.299 | 0.007 |
| Q2FWL8 | transcriptional regulator | SAOUHSC_02271 | 2 | 2 | 8.20 | 1.448 | 0.001 |
| Q2G2L6 | pf09954 family protein | SAOUHSC_02812 | 1 | 1 | 16.00 | 1.383 | 0.019 |
| Q2FZ07 | uncharacterized protein | SAOUHSC_01264 | 2 | 2 | 8.20 | 0.809 | 0.000 |
| Q2FV28 | uncharacterized conserved protein | SAOUHSC_02911 | 2 | 2 | 27.80 | 0.827 | 0.023 |
| Q2G2Q9 | conserved hypothetical family protein | SAOUHSC_00274 | 1 | 2 | 20.10 | 0.586 | 0.001 |
| Q2G297 | metal-dependent phosphohydrolase | SAOUHSC_01696 | 5 | 5 | 22.40 | 0.807 | 0.002 |
| Fatty Acid Species | Control (%) | 12.5 μg/mL (%) | 25 μg/mL (%) | 37.5 μg/mL (%) |
|---|---|---|---|---|
| C8:0 | 1.16 | 1.45 | 1.31 | 1.49 |
| C10:0 | 2.27 | 2.06 | 1.97 | 1.93 |
| C12:0 | 1.61 | 1.53 | 1.45 | 1.55 |
| C14:0 | 2.96 | 2.98 | 3.06 | 3.16 |
| C16:1 | 20.34 | 21.03 | 21.56 | 21.93 |
| C16:0 | 17.21 | 17.03 | 16.52 | 16.4 |
| C18:2 | 6.29 | 6.14 | 5.62 | 5.21 |
| C18:1 | 7.64 | 7.85 | 7.92 | 7.96 |
| C18:0 | 21.03 | 20.36 | 19.14 | 18.64 |
| UFA | 27.98 | 28.88 | 29.48 | 29.89 |
| SFA | 38.24 | 37.39 | 35.66 | 35.01 |
| SFA/UFA | 1.37 | 1.3 | 1.21 | 1.17 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Cheng, Y.; Wu, R.; Jiang, D.; Bai, B.; Tan, D.; Yan, T.; Sun, X.; Zhang, Q.; Wu, Z. Antibacterial Activity of Juglone against Staphylococcus aureus: From Apparent to Proteomic. Int. J. Mol. Sci. 2016, 17, 965. https://doi.org/10.3390/ijms17060965
Wang J, Cheng Y, Wu R, Jiang D, Bai B, Tan D, Yan T, Sun X, Zhang Q, Wu Z. Antibacterial Activity of Juglone against Staphylococcus aureus: From Apparent to Proteomic. International Journal of Molecular Sciences. 2016; 17(6):965. https://doi.org/10.3390/ijms17060965
Chicago/Turabian StyleWang, Jiayi, Yuhuan Cheng, Rina Wu, Donghua Jiang, Bing Bai, Dehong Tan, Tingcai Yan, Xiyun Sun, Qi Zhang, and Zhaoxia Wu. 2016. "Antibacterial Activity of Juglone against Staphylococcus aureus: From Apparent to Proteomic" International Journal of Molecular Sciences 17, no. 6: 965. https://doi.org/10.3390/ijms17060965
APA StyleWang, J., Cheng, Y., Wu, R., Jiang, D., Bai, B., Tan, D., Yan, T., Sun, X., Zhang, Q., & Wu, Z. (2016). Antibacterial Activity of Juglone against Staphylococcus aureus: From Apparent to Proteomic. International Journal of Molecular Sciences, 17(6), 965. https://doi.org/10.3390/ijms17060965