Ras-Related Nuclear Protein Ran3B Gene Is Involved in Hormone Responses in the Embryogenic Callus of Dimocarpus longan Lour.
Abstract
:1. Introduction
2. Results
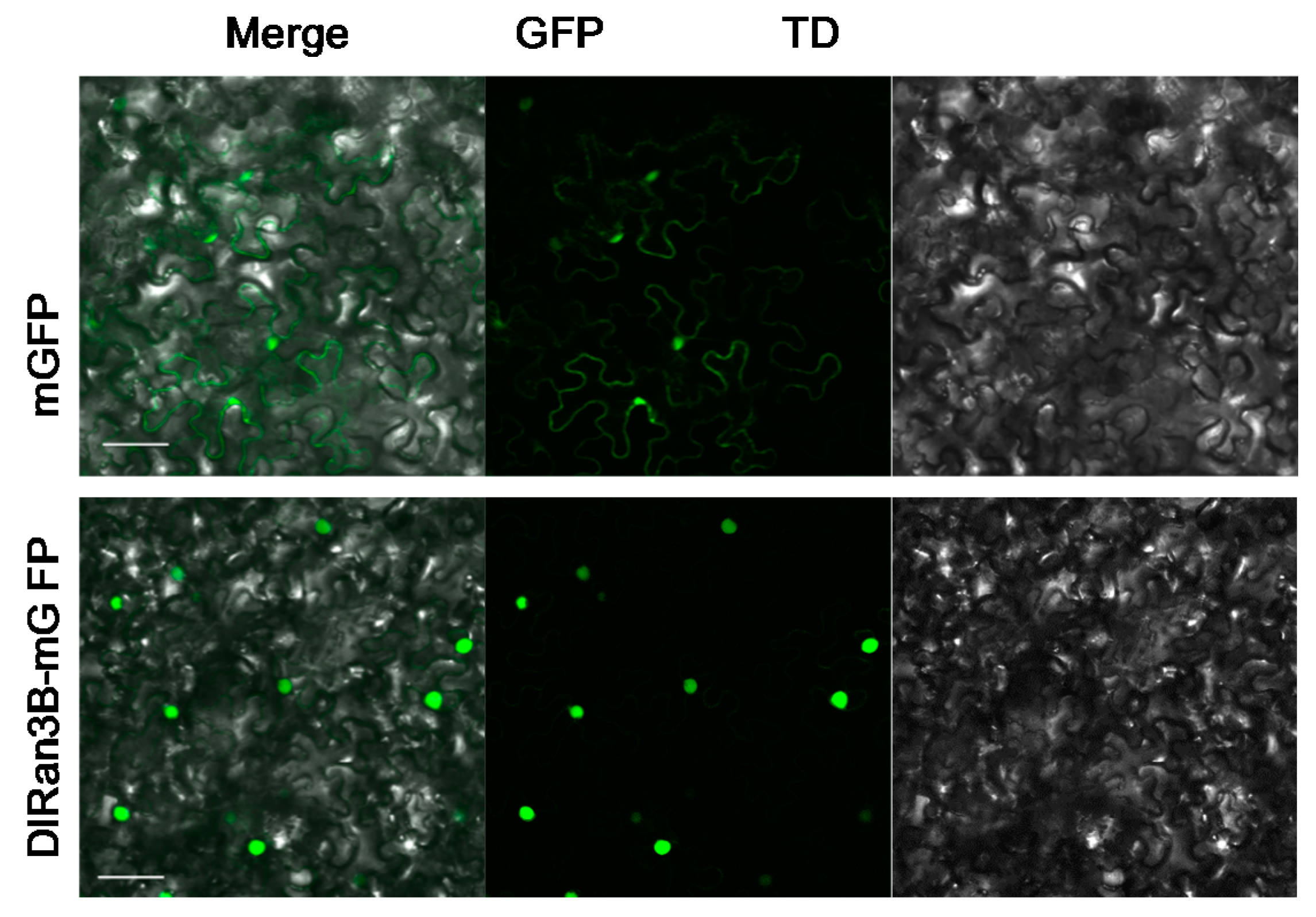
2.1. Subcellular Localization of DlRan3B
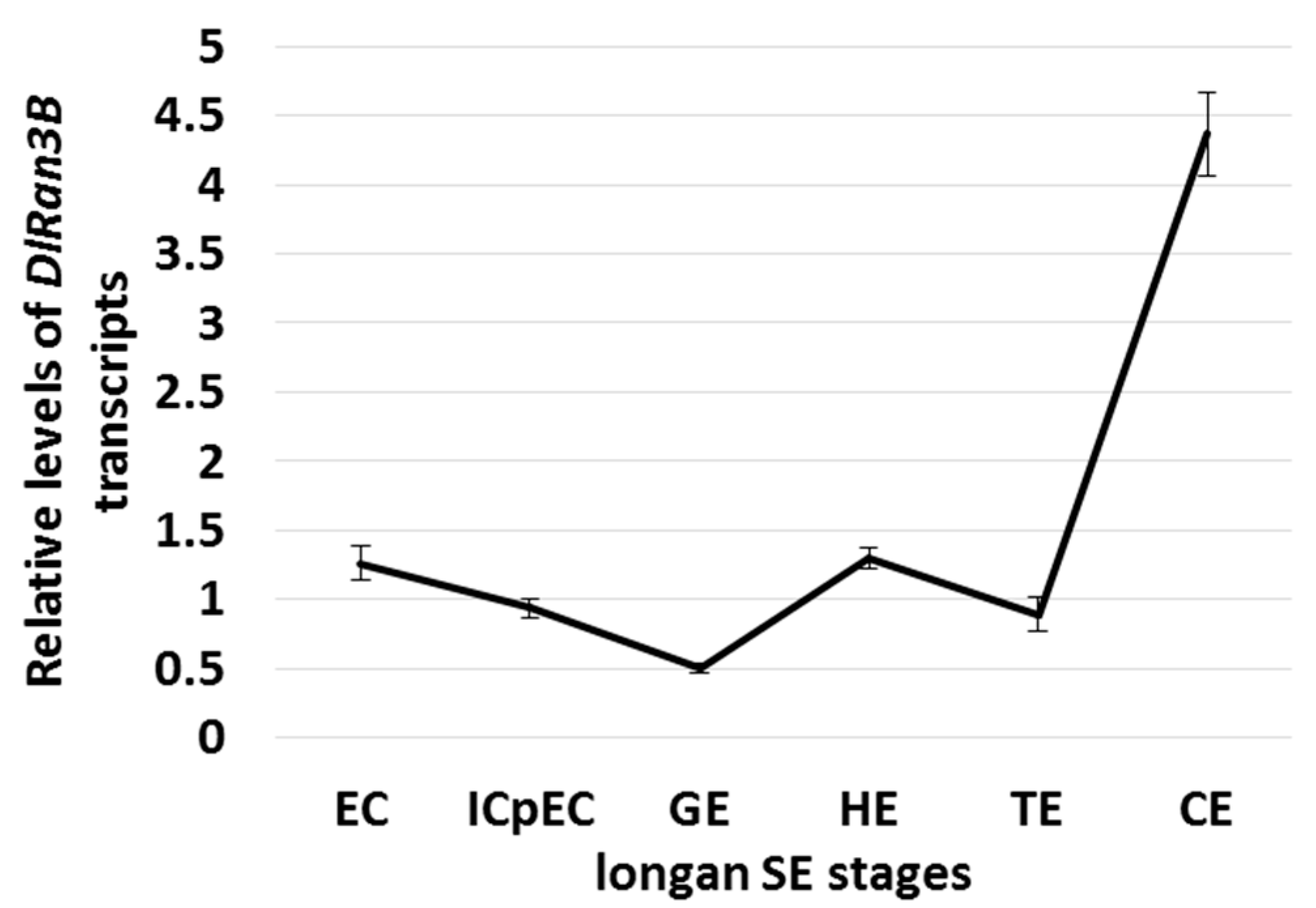
2.2. The Expression Profiling of DlRan3B During Longan Somatic Embryogenesis (SE)
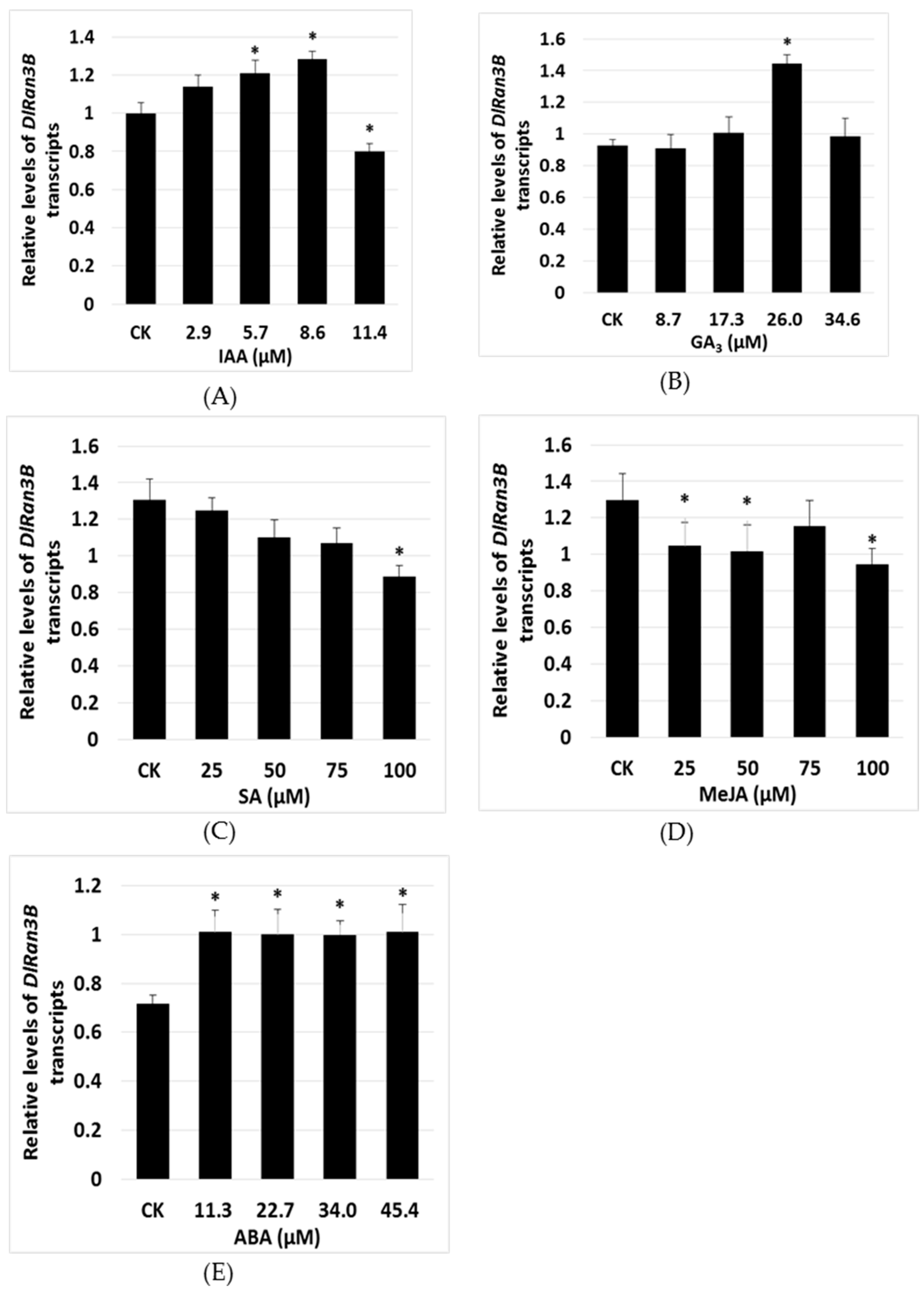
2.3. The Effect of Exogenous Plant Hormones on DlRan3B Expression
2.4. The Isolation and Bioinformatic Analysis of the Putative Promoter Region of the DlRan3B Gene
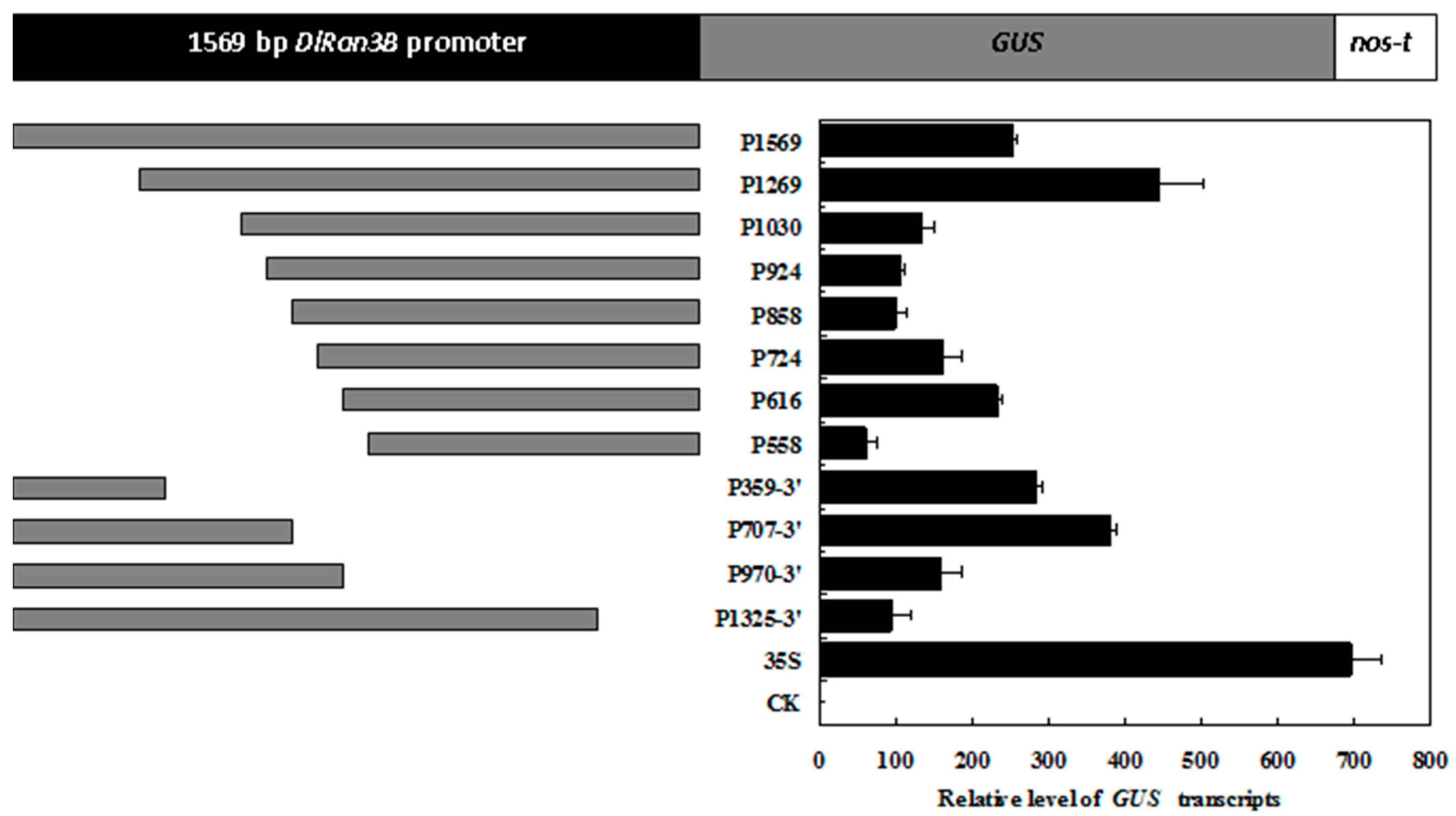
2.5. Deletion Analysis of the DlRan3B Promoter
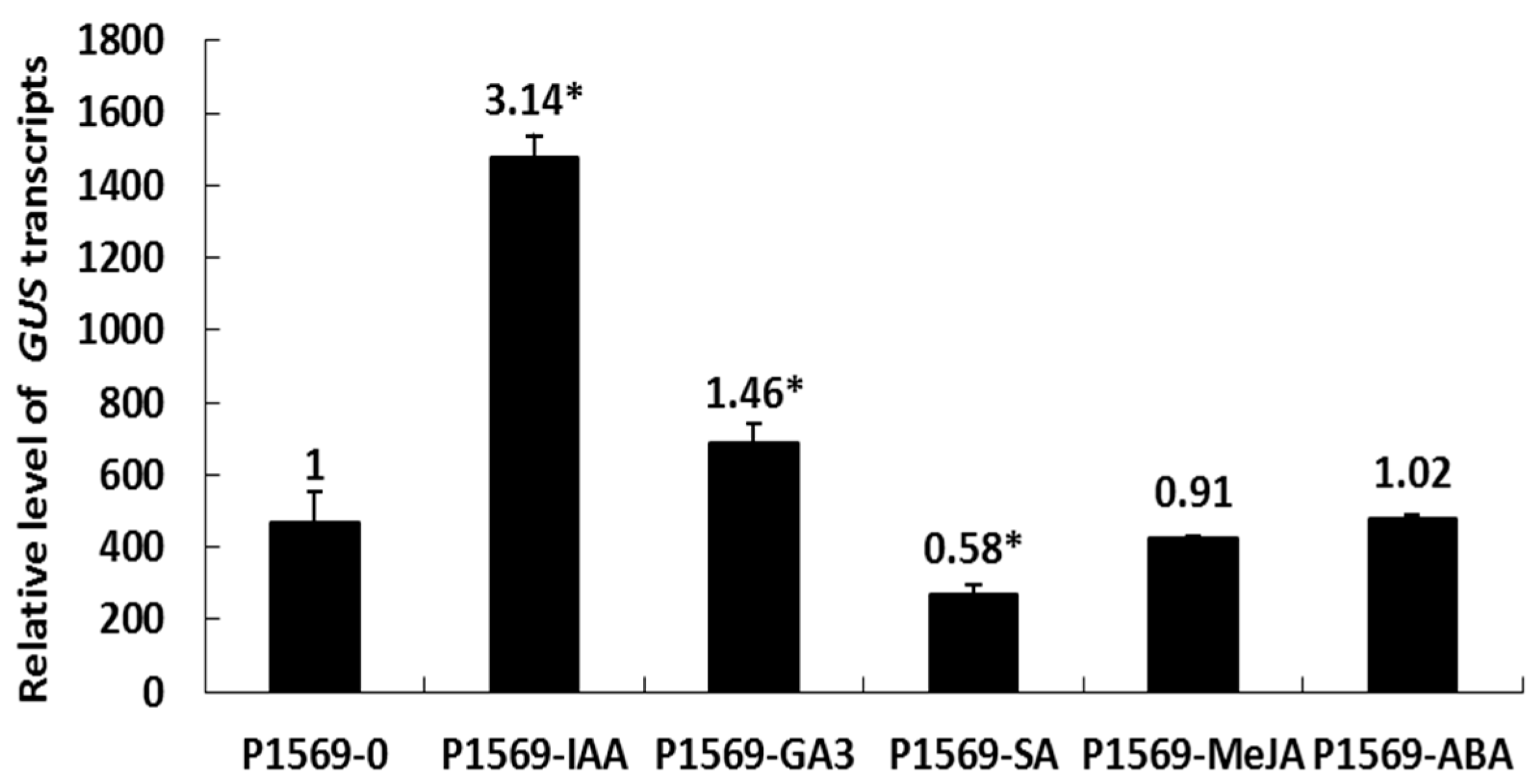
2.6. Responsiveness of the DlRan3B Promoter to Hormone Treatments
3. Discussion
3.1. DlRan3B Shares Similar Subcellular Localization with Its Longan Homolog and Animal Counterpart
3.2. DlRan3B Participates in the Auxin, Gibberellin, and ABA Signaling Pathways During Longan Late SE
3.3. The Potential Role of DlRan3B
4. Materials and Methods
4.1. Plant Materials and Nucleic Acid Extraction
4.2. Subcellular Localization
4.3. Phytohormone Treatments on Longan EC Samples
4.4. Isolation of DlRan3B Promoter and Bioinformatic Analysis
4.5. Construction of the PdlRan3B::GUS Fusion Vector and Agrobacterium-Mediated Transient Assay
4.6. Phytohormone Treatments of Tobacco Leaves
4.7. qPCR Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| Ran | Ras-related nuclear protein |
| GTP | guanosine triphosphate |
| NE | nuclear envelope |
| SE | somatic embryogenesis |
| IAA | indoleacetic acid |
| GA3 | gibberellin A3 |
| SA | salicylic acid |
| MeJA | methyl jasmonate |
| ABA | abscisic acid |
| qPCR | quantitative real-time PCR |
| EC | EC: embryogenic callus |
| ICpEC | incomplete compact pro-embryogenic cultures |
| GE | globular embryos |
| HE | heart-shaped embryos |
| TE | torpedo-shaped embryos |
| CE | cotyledon embryos |
| GFP | green fluorescent protein |
| TSS | transcription start site |
| MYB | v-myb avian myeloblastosis viral oncogene homolog |
| 35S | CaMV35S |
| 2,4-D | 2,4-Dichlorophenoxyacetic acid |
| DAPI | 4',6-diamidino-2-phenylindole |
References
- Vernoud, V.; Horton, A.C.; Yang, Z.; Nielsen, E. Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 2003, 131, 1191–1208. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.; Gandhi, A.; Lim, P.-W.; Relles, D.; Sarosiek, K.; Kang, C.; Chipitsyna, G.; Sendecki, J.; Yeo, C.J.; Arafat, H.A. RAN GTPase and osteopontin in pancreatic cancer. Pancreat. Disord. Ther. 2013, 3, 113. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.; Collier, S.M.; Moffett, P.; Chai, J. Structural basis for the interaction between the potato virus X resistance protein (Rx) and its cofactor Ran GTPase-activating protein 2 (RanGAP2). J. Biol. Chem. 2013, 288, 35868–35876. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Lee, H.-S.; Lee, J.-S.; Kim, S.-K.; Kim, S.-H. Hormone-and light-regulated nucleocytoplasmic transport in plants: Current status. J. Exp. Bot. 2008, 59, 3229–3245. [Google Scholar] [CrossRef] [PubMed]
- García, A.V.; Parker, J.E. Heaven’s gate: Nuclear accessibility and activities of plant immune regulators. Trends Plant Sci. 2009, 14, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Lin, Y.; Yang, M.; Zhang, D.; Lai, R.; Lai, Z. DlRan3A is involved in hormone, light, and abiotic stress responses in embryogenic callus of Dimocarpus longan Lour. Gene 2015, 569, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, Y.; Han, Y.; Bao, S.; Du, J.; Yuan, M.; Xu, Z.; Chong, K. Overexpression of RAN1 in rice and Arabidopsis alters primordial meristem, mitotic progress, and sensitivity to auxin. Plant Physiol. 2006, 140, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Lü, S.; Fan, Y.; Jin, C. Overexpression of a Ran GTPase homologous gene, FaRan from tall fescue, in transgenic Arabidopsis. Biol. Plant. 2011, 55, 331–334. [Google Scholar] [CrossRef]
- Zang, A.; Xu, X.; Neill, S.; Cai, W. Overexpression of OsRAN2 in rice and Arabidopsis renders transgenic plants hypersensitive to salinity and osmotic stress. J. Exp. Bot. 2010, 61, 777–789. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Xu, Y.; Wang, X.; Du, C.; Du, J.; Yuan, M.; Xu, Z.; Chong, K. OsRAN2, essential for mitosis, enhances cold tolerance in rice by promoting export of intranuclear tubulin and maintaining cell division under cold stress. Plant Cell Environ. 2011, 34, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Prasad, K.N.; Hao, J.; Shi, J.; Liu, T.; Li, J.; Wei, X.; Qiu, S.; Xue, S.; Jiang, Y. Antioxidant and anticancer activities of high pressure-assisted extract of longan (Dimocarpus longan Lour.) fruit pericarp. Innov. Food Sci. Emerg. Technol. 2009, 10, 413–419. [Google Scholar] [CrossRef]
- Sudjaroen, Y.; Hull, W.E.; Erben, G.; Würtele, G.; Changbumrung, S.; Ulrich, C.M.; Owen, R.W. Isolation and characterization of ellagitannins as the major polyphenolic components of longan (Dimocarpus longan Lour.) seeds. Phytochemistry 2012, 77, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Jiang, Y.; Shi, J.; Chen, F.; Ashraf, M. Extraction and pharmacological properties of bioactive compounds from longan (Dimocarpus longan Lour.) fruit—A review. Food Res. Int. 2011, 44, 1837–1842. [Google Scholar] [CrossRef]
- Askjaer, P.; Galy, V.; Hannak, E.; Mattaj, I.W. Ran GTPase cycle and importins α and β are essential for spindle formation and nuclear envelope assembly in living Caenorhabditis elegans embryos. Mol. Biol. Cell 2002, 13, 4355–4370. [Google Scholar] [CrossRef] [PubMed]
- Bamba, C.; Bobinnec, Y.; Fukuda, M.; Nishida, E. The GTPase Ran regulates chromosome positioning and nuclear envelope assembly in vivo. Curr. Biol. 2002, 12, 503–507. [Google Scholar] [CrossRef]
- Onuma, Y.; Nishihara, R.; Takahashi, S.; Tanegashima, K.; Fukui, A.; Asashima, M. Expression of the Xenopus GTP-binding protein gene Ran during embryogenesis. Dev. Genes Evol. 2000, 210, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Lin, Y.; Lai, Z.; Wang, T. Cloning and bioinformatic analysis of the promoters of DlRan3A and DlRan3B from embryogenic callus in Dimocarpus longan. Chin. J. Trop. Crops 2014, 35, 82–89. [Google Scholar]
- Fang, Z.; Lai, C.; Zhang, Y.; Lai, Z. Molecular cloning, structural and expression profiling of DlRan genes during somatic embryogenesis in Dimocarpus longan Lour. SpringerPlus 2016, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ren, M.; Drivas, G.; D’Eustachio, P.; Rush, M.G. Ran/tc4: A small nuclear GTP-binding protein that regulates DNA synthesis. J. Cell Biol. 1993, 120, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Melchior, F.; Gerace, L. Two-way trafficking with Ran. Trends Cell Biol. 1998, 8, 175–179. [Google Scholar] [CrossRef]
- Lai, Z.; Chen, C. Changes of endogenous phytohormones in the process of somatic embryogenesis in longan (Dimocarpus longan Lour.). Chin. J. Trop. Crops 2001, 23, 41–47. [Google Scholar]
- Kim, S.-H.; Roux, S.J. An Arabidopsis Ran-binding protein, AtRanBP1c, is a co-activator of Ran GTPase-activating protein and requires the C-terminus for its cytoplasmic localization. Planta 2003, 216, 1047–1052. [Google Scholar] [PubMed]
- Chen, M.; Du, X.; Zhu, Y.; Wang, Z.; Hua, S.; Li, Z.; Guo, W.; Zhang, G.; Peng, J.; Jiang, L. Seed fatty acid reducer acts downstream of gibberellin signalling pathway to lower seed fatty acid storage in Arabidopsis. Plant Cell Environ. 2012, 35, 2155–2169. [Google Scholar] [CrossRef] [PubMed]
- Kępczyńska, E.; Zielińska, S. Regulation of Medicago sativa L. Somatic embryos regeneration by gibberellin A3 and abscisic acid in relation to starch content and α-amylase activity. Plant Growth Regul. 2006, 49, 209–217. [Google Scholar] [CrossRef]
- Kosová, K.; Vítámvás, P.; Prášil, I.T.; Renaut, J. Plant proteome changes under abiotic stress—Contribution of proteomics studies to understanding plant stress response. J. Proteom. 2011, 74, 1301–1322. [Google Scholar] [CrossRef] [PubMed]
- Ralevski, A. The Interaction of Early Salt Stress-Induced 2 (ESI2) and the Ran G Protein in Arabidopsis; Concordia University: Irvine, CA, USA, 2013. [Google Scholar]
- Xu, P.; Cai, W. RAN1 is involved in plant cold resistance and development in rice (Oryza sativa). J. Exp. Bot. 2014, 65, 3277–3287. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Chen, C.; Zeng, L.; Chen, Z. Somatic embryogenesis in longan (Dimocarpus longan Lour.). In Somatic Embryogenesis in Woody Plants; Jain, S.M., Gupta, P., Newton, R., Eds.; Springer: Dordrecht, The Netherlands, 2000; Volume 67, pp. 415–431. [Google Scholar]
- Lai, Z.; Chen, Z. Somatic embryogenesis of high frequency from longan embryogenic calli. J. Fujian Agric. Univ. 1997, 26, 271–276. [Google Scholar]
- Lin, Y.-L.; Lai, Z.-X. Superoxide dismutase multigene family in longan somatic embryos: A comparison of CuZn-SOD, Fe-SOD, and Mn-SOD gene structure, splicing, phylogeny, and expression. Mol. Breed. 2013, 32, 595–615. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; van de Peer, Y.; Rouzé, P.; Rombauts, S. Plantcare, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Lu, L.; Lai, Z.; Pan, D. Extraction and purification of genomic DNA in Nai (Prunus salicina Lindl. var. cordata). Acta Agric. Univ. Jiangxiensis 2004, 26, 329–333. [Google Scholar]
- Lin, Y.; Lai, Z. Reference gene selection for qPCR analysis during somatic embryogenesis in longan tree. Plant Sci. 2010, 178, 359–365. [Google Scholar] [CrossRef]

| Regulatory Element | Function | Sequence |
|---|---|---|
| 5UTR Py-rich stretch | cis-acting element conferring high transcription levels | TTTCTTCTCT/TTTCTCTCTCTCTC |
| A-box | sequence conserved in alpha-amylase promoters | AATAACAAACTCC |
| AACA-motif | involved in endosperm-specific negative expression | TAACAAACTCCA |
| ABRE | cis-acting element involved in the abscisic acid responsiveness | TACGTG |
| ARE | cis-acting regulatory element essential for the anaerobic induction | TGGTTT |
| ACE | cis-acting regulatory element involved in light responsiveness | CTAACGTATT |
| AuxRR-core | cis-acting regulatory element involved in auxin responsiveness | GGTCCAT |
| Box 4 | part of a conserved DNA module involved in light responsiveness | ATTAAT |
| Box I | light responsive element | TTTCAAA |
| CAAT-box | common cis-acting element in promoter and enhancer regions | CAAT/CAAAT |
| CGTCA-motif | cis-acting regulatory element involved in the MeJA-responsiveness | CGTCA |
| G-box | cis-acting regulatory element involved in light responsiveness | CACGTA |
| GAG-motif | part of a light responsive element | AGAGAGT |
| GARE-motif | gibberellin-responsive element | AAACAGA |
| GATA-motif | part of a light responsive element | GATAGGA |
| GCN4-motif | cis-acting element involved in endosperm expression | CAAGCCA/TGTGTCA |
| GT1-motif | light responsive element | GGTTAA |
| I-box | part of a light responsive element | GATATGG/GATAAGATA |
| LTR | cis-acting element involved in low-temperature responsiveness | CCGAAA |
| MBS | v-myb avian myeloblastosis viral oncogene homolog (MYB) binding site involved in drought-inducibility | CAACTG |
| Skn-1-motif | cis-acting regulatory element required for endosperm expression | GTCAT |
| Sp1 | light responsive element | CC(G/A)CCC |
| TATA-box | core promoter element around −30 of transcription start | TATA/TAATA/TTTTA |
| TC-rich repeats | cis-acting element involved in defense and stress responsiveness | ATTTTCTTCA |
| TCA-element | cis-acting element involved in salicylic acid responsiveness | CCATCTTTTT/CAGAAAAGGA |
| TCCC-motif | part of a light responsive element | TCTCCCT |
| TGACG-motif | cis-acting regulatory element involved in the MeJA-responsiveness | TGACG |
| Primer | Sequence (5′ to 3′) | Corresponding Construct | Transgenic Line |
|---|---|---|---|
| Del-F1 | TGATTACGCCAAGCTTAACAACTACCTACCAATGAGCG | Full-length (P1569) | P1569 |
| Del-R1 | GACCACCCGGGGATCCGAGAGCTGTCCTTGAGAAAGCG | Full-length (P1569) | P1569 |
| Del-F2 | TGATTACGCCAAGCTTCCAATAGTGACGAGTGTGAATC | 5′Δ (P1269) | P1269 |
| Del-F3 | TGATTACGCCAAGCTTATTAGTGTAATTTTCAATCAGGTGG | 5′Δ (P1030) | P1030 |
| Del-F4 | TGATTACGCCAAGCTTCAAATTGAGATTTCCCCTCCTG | 5′Δ (P924) | P924 |
| Del-F5 | TGATTACGCCAAGCTTCACCAACTGTTTGAGCTTCCAAC | 5′Δ (P858) | P858 |
| Del-F6 | TGATTACGCCAAGCTTTTTTTTATGACACAAATTTTGGTG | 5′Δ (P724) | P724 |
| Del-F7 | TGATTACGCCAAGCTTAGGGTGGGAGGTAAACTACG | 5′Δ (P616) | P616 |
| Del-F8 | TGATTACGCCAAGCTTCCCAACCAATTTGAAAAGACAACC | 5′Δ (P558) | P558 |
| Del-R2 | GACCACCCGGGGATCCATTTTTTTCAAGACTCTCTCGACC | 3′Δ (P1325-3′) | P1325-3′ |
| Del-R3 | GACCACCCGGGGATCCAGTTTACCTCCCACCCTTTCG | 3′Δ (P970-3′) | P970-3′ |
| Del-R4 | GACCACCCGGGGATCCGGCAAACCTTCGAACAAGTGTC | 3′Δ (P707-3′) | P707-3′ |
| Del-R5 | GACCACCCGGGGATCCACATGGCACCACTAGTCACAC | 3′Δ (P359-3′) | P359-3′ |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, Q.; Lin, Y.; Zhang, D.; Lai, R.; Lai, Z. Ras-Related Nuclear Protein Ran3B Gene Is Involved in Hormone Responses in the Embryogenic Callus of Dimocarpus longan Lour. Int. J. Mol. Sci. 2016, 17, 873. https://doi.org/10.3390/ijms17060873
Tian Q, Lin Y, Zhang D, Lai R, Lai Z. Ras-Related Nuclear Protein Ran3B Gene Is Involved in Hormone Responses in the Embryogenic Callus of Dimocarpus longan Lour. International Journal of Molecular Sciences. 2016; 17(6):873. https://doi.org/10.3390/ijms17060873
Chicago/Turabian StyleTian, Qilin, Yuling Lin, Dongmin Zhang, Ruilian Lai, and Zhongxiong Lai. 2016. "Ras-Related Nuclear Protein Ran3B Gene Is Involved in Hormone Responses in the Embryogenic Callus of Dimocarpus longan Lour." International Journal of Molecular Sciences 17, no. 6: 873. https://doi.org/10.3390/ijms17060873
APA StyleTian, Q., Lin, Y., Zhang, D., Lai, R., & Lai, Z. (2016). Ras-Related Nuclear Protein Ran3B Gene Is Involved in Hormone Responses in the Embryogenic Callus of Dimocarpus longan Lour. International Journal of Molecular Sciences, 17(6), 873. https://doi.org/10.3390/ijms17060873









