SARS Coronavirus Papain-Like Protease Inhibits the TLR7 Signaling Pathway through Removing Lys63-Linked Polyubiquitination of TRAF3 and TRAF6
Abstract
:1. Introduction
2. Results
2.1. PLPro Suppressed TLR7 Agonist-Induced Production of Type I IFNs in Human Promonocytes
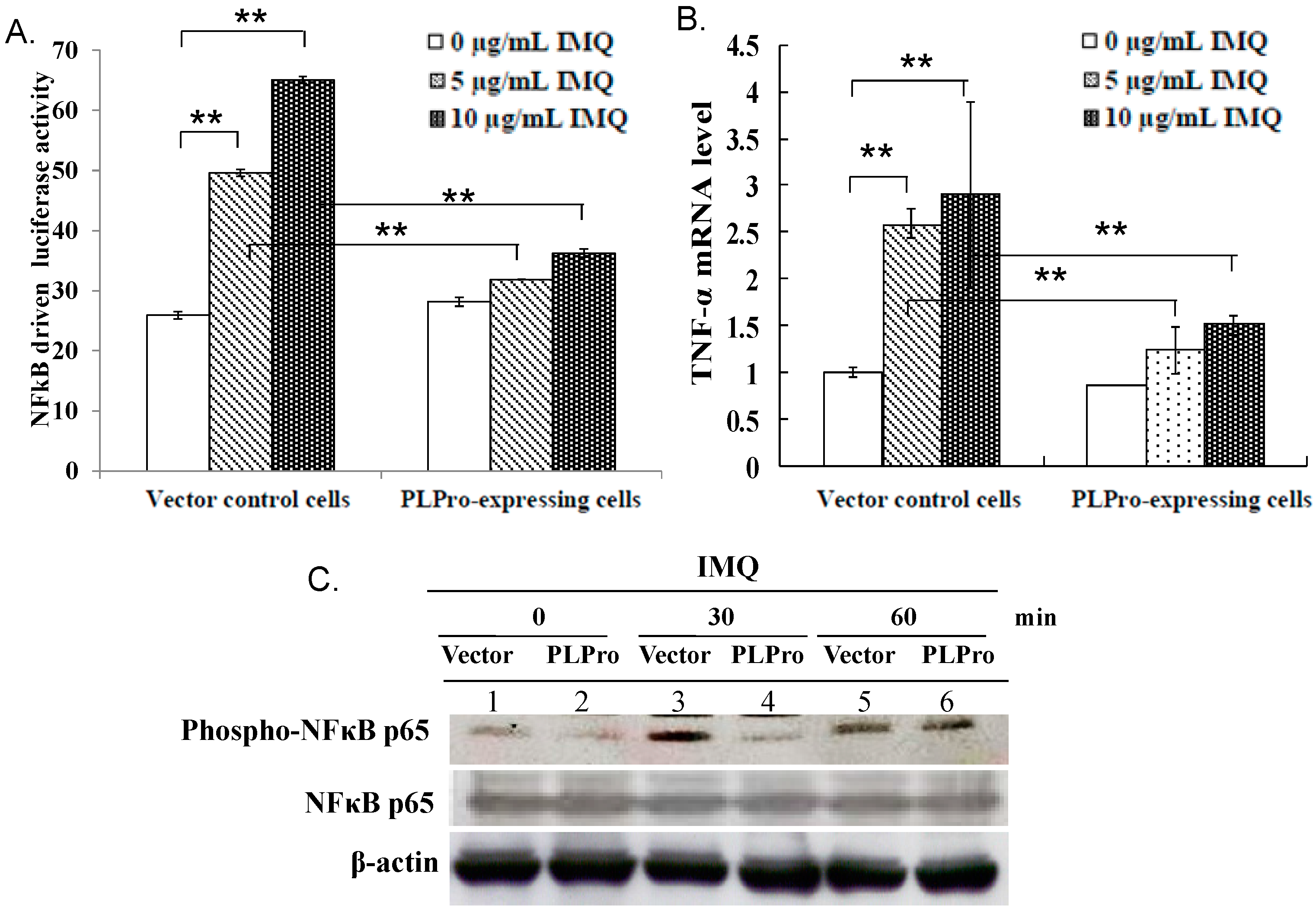
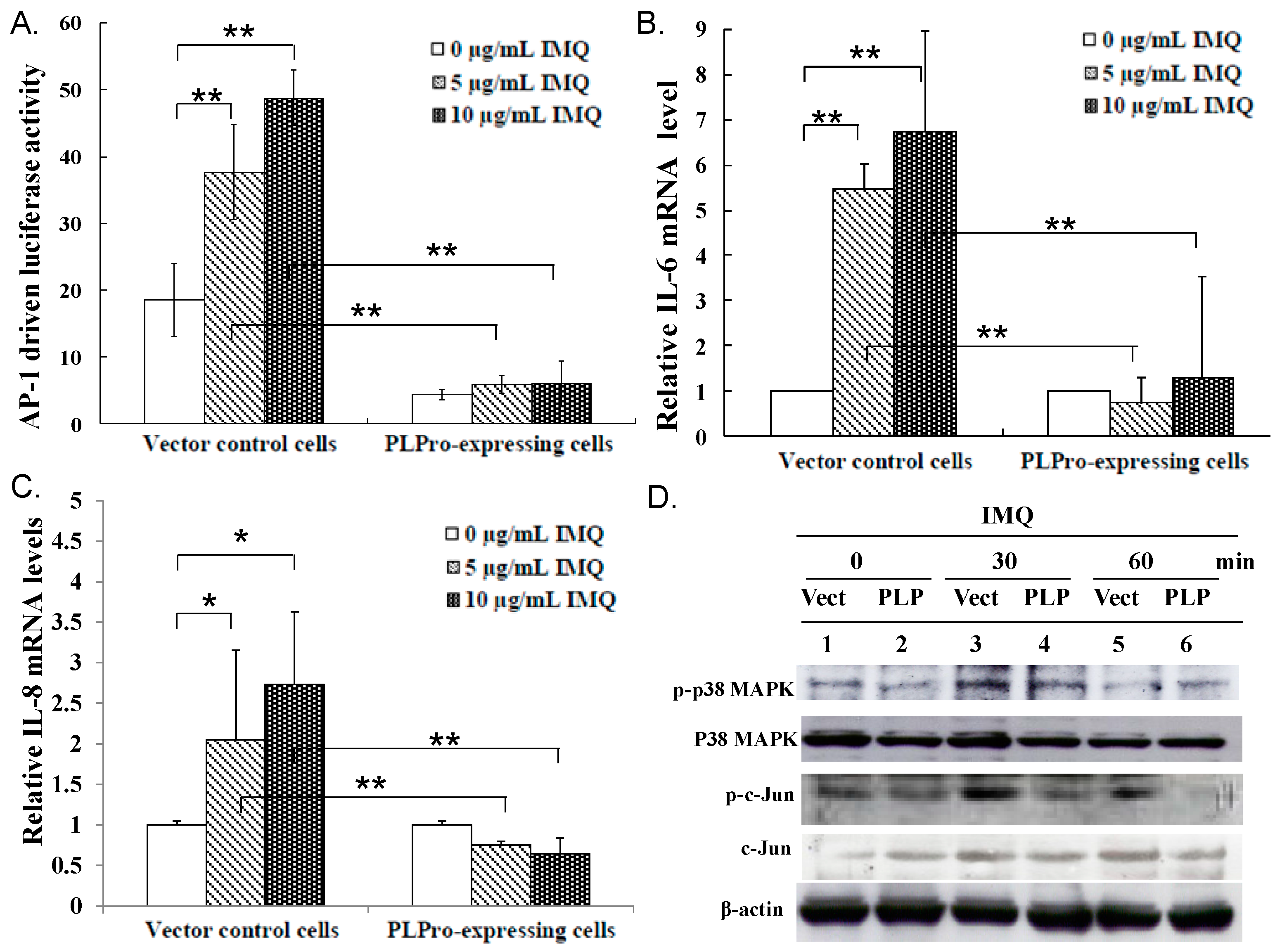
2.2. Inhibition of TLR7 Agonist-Induced Pro-Inflammatory Cytokines by SARS-CoV PLPro
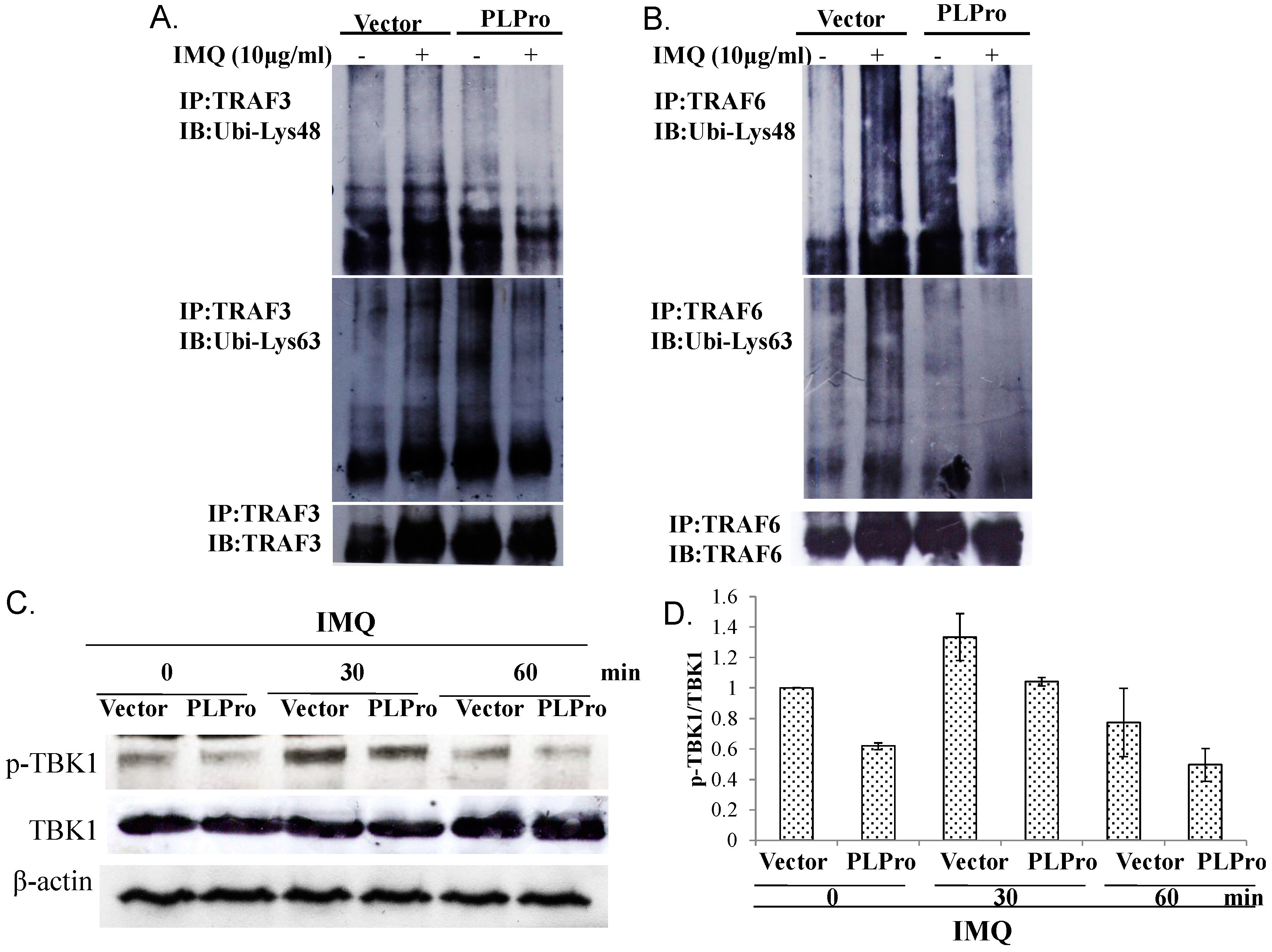
2.3. Ubiquitin Removal of TRAF3 and TRAF6 Responsible for the TLR7 Antagonism of SARS-CoV PLPro
3. Discussion
4. Materials and Methods
4.1. Cell Line and Western Blot Assay
4.2. Signaling Pathway Assays with the Dual Luciferase Reporter System
4.3. Quantitative Expression Analysis of Type I IFNs and Pro-Inflammatory Cytokines Using Real-Time PCR
4.4. Immunoprecipitation Assays
4.5. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Brown, J.; Wang, H.; Hajishengallis, G.N.; Martin, M. LR-signaling networks: An integration of adaptor molecules, kinases, and cross-talk. J. Dent. Res. 2011, 90, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Kawai, T.; Akira, S. Toll-like receptors and innate immunity. Biochem. Biophys. Res. Commun. 2009, 388, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Seki, E. Toll-like receptors in liver fibrosis: Cellular crosstalk and mechanisms. Front. Physiol. 2012, 3, 138. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Takeda, K. Current views of toll-like receptor signaling pathways. Gastroenterol. Res. Pract. 2010, 2010, 240365. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.J.; Su, I.J.; Theron, M.; Wu, Y.C.; Lai, S.K.; Liu, C.C.; Lei, H.Y. An interferon-γ-related cytokine storm in SARS patients. J. Med. Virol. 2005, 75, 185–194. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Ding, Y.; Zhang, Q.; Che, X.; He, Y.; Shen, H.; Wang, H.; Li, Z.; Zhao, L.; Geng, J.; et al. Expression of elevated levels of pro-inflammatory cytokines in SARS-CoV-infected ACE2+ cells in SARS patients: Relation to the acute lung injury and pathogenesis of SARS. J. Pathol. 2006, 210, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Rota, P.A.; Oberste, M.S.; Monroe, S.S.; Nix, W.A.; Campagnoli, R.; Icenogle, J.P.; Peñaranda, S.; Bankamp, B.; Maher, K.; Chen, M.H.; et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 2003, 300, 1394–1399. [Google Scholar] [CrossRef] [PubMed]
- Ziebuhr, J. Molecular biology of severe acute respiratory syndrome coronavirus. Curr. Opin. Microbiol. 2004, 7, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Barretto, N.; Jukneliene, D.; Ratia, K.; Chen, Z.; Mesecar, A.D.; Baker, S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005, 79, 15189–15198. [Google Scholar] [CrossRef] [PubMed]
- Lindner, H.A.; Fotouhi-Ardakani, N.; Lytvyn, V.; Lachance, P.; Sulea, T.; Ménard, R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 2005, 79, 15199–15208. [Google Scholar] [CrossRef] [PubMed]
- Sulea, T.; Lindner, H.A.; Purisima, E.O.; Menard, R. Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain-like protease? J. Virol. 2005, 79, 4550–4551. [Google Scholar] [CrossRef] [PubMed]
- Ratia, K.; Saikatendu, K.S.; Santarsiero, B.D.; Barretto, N.; Baker, S.C.; Stevens, R.C.; Mesecar, A.D. Severe acute respiratory syndrome coronavirus papain-like protease: Structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. USA 2006, 103, 5717–5722. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, M.; Pichlmair, A.; Martínez-Sobrido, L.; Cros, J.; García-Sastre, A.; Haller, O.; Weber, F. Inhibition of β interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 2005, 79, 2079–2086. [Google Scholar] [CrossRef] [PubMed]
- Frieman, M.; Ratia, K.; Johnston, R.E.; Mesecar, A.D.; Baric, R.S. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-κB signaling. J. Virol. 2009, 83, 6689–6705. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Xing, Y.; Chen, X.; Zheng, Y.; Yang, Y.; Nichols, D.B.; Clementz, M.A.; Banach, B.S.; Li, K.; Baker, S.C.; et al. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS ONE 2012, 7, e30802. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yang, X.; Zheng, Y.; Yang, Y.; Xing, Y.; Chen, Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell 2014, 5, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Kovalenko, A.; Chable-Bessia, C.; Cantarella, G.; Israël, A.; Wallach, D.; Courtois, G. The tumour suppressor CYLD negatively regulates NF-κB signalling by deubiquitination. Nature 2003, 424, 801–805. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Lv, J.; Han, L.; Ichikawa, T.; Wang, W.; Li, S.; Wang, X.L.; Tang, D.; Cui, T. A pro-inflammatory role of deubiquitinating enzyme cylindromatosis (CYLD) in vascular smooth muscle cells. Biochem. Biophys. Res. Commun. 2012, 420, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Parvatiyar, K.; Harhaj, E.W. Regulation of inflammatory and antiviral signaling by A20. Microbes Infect. 2011, 13, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, M.; Cao, H.; Zhu, Y.; Zheng, J.; Zhou, H. Extraordinary GU-rich single-strand RNA identified from SARS coronavirus contributes an excessive innate immune response. Microbes Infect. 2013, 15, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Cervantes-Barragan, L.; Züst, R.; Weber, F.; Spiegel, M.; Lang, K.S.; Akira, S.; Thiel, V.; Ludewig, B. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood 2007, 109, 1131–1137. [Google Scholar] [CrossRef] [PubMed]
- Devaraj, S.G.; Wang, N.; Chen, Z.; Chen, Z.; Tseng, M.; Barretto, N.; Lin, R.; Peters, C.J.; Tseng, C.T.; Baker, S.C.; et al. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2007, 282, 32208–32221. [Google Scholar] [CrossRef] [PubMed]
- Li, S.W.; Lai, C.C.; Ping, J.F.; Tsai, F.J.; Wan, L.; Lin, Y.J.; Kung, S.H.; Lin, C.W. Severe acute respiratory syndrome coronavirus papain-like protease suppressed α interferon-induced responses through downregulation of extracellular signal-regulated kinase 1-mediated signalling pathways. J. Gen. Virol. 2011, 92, 1127–1140. [Google Scholar] [CrossRef] [PubMed]
- Li, S.W.; Yang, T.C.; Wan, L.; Lin, Y.J.; Tsai, F.J.; Lai, C.C.; Lin, C.W. Correlation between TGF-β1 expression and proteomic profiling induced by severe acute respiratory syndrome coronavirus papain-like protease. Proteomics 2012, 12, 3193–3205. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.; Plater, L.; Peggie, M.; Cohen, P. Use of the pharmacological inhibitor BX795 to Study the Regulation and Physiological Roles of TBK1 and IκB Kinase ε: A distinct upstream kinase mediates Ser-172 phosphorylation and activation. J. Biol. Chem. 2009, 284, 14136–14146. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.C.; Li, S.W.; Lai, C.C.; Lu, K.Z.; Chiu, M.T.; Hsieh, T.H.; Wan, L.; Lin, C.W. Proteomic analysis for Type I interferon antagonism of Japanese encephalitis virus NS5 protein. Proteomics 2013, 13, 3442–3456. [Google Scholar] [CrossRef] [PubMed]


© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.-W.; Wang, C.-Y.; Jou, Y.-J.; Huang, S.-H.; Hsiao, L.-H.; Wan, L.; Lin, Y.-J.; Kung, S.-H.; Lin, C.-W. SARS Coronavirus Papain-Like Protease Inhibits the TLR7 Signaling Pathway through Removing Lys63-Linked Polyubiquitination of TRAF3 and TRAF6. Int. J. Mol. Sci. 2016, 17, 678. https://doi.org/10.3390/ijms17050678
Li S-W, Wang C-Y, Jou Y-J, Huang S-H, Hsiao L-H, Wan L, Lin Y-J, Kung S-H, Lin C-W. SARS Coronavirus Papain-Like Protease Inhibits the TLR7 Signaling Pathway through Removing Lys63-Linked Polyubiquitination of TRAF3 and TRAF6. International Journal of Molecular Sciences. 2016; 17(5):678. https://doi.org/10.3390/ijms17050678
Chicago/Turabian StyleLi, Shih-Wen, Ching-Ying Wang, Yu-Jen Jou, Su-Hua Huang, Li-Hsin Hsiao, Lei Wan, Ying-Ju Lin, Szu-Hao Kung, and Cheng-Wen Lin. 2016. "SARS Coronavirus Papain-Like Protease Inhibits the TLR7 Signaling Pathway through Removing Lys63-Linked Polyubiquitination of TRAF3 and TRAF6" International Journal of Molecular Sciences 17, no. 5: 678. https://doi.org/10.3390/ijms17050678
APA StyleLi, S.-W., Wang, C.-Y., Jou, Y.-J., Huang, S.-H., Hsiao, L.-H., Wan, L., Lin, Y.-J., Kung, S.-H., & Lin, C.-W. (2016). SARS Coronavirus Papain-Like Protease Inhibits the TLR7 Signaling Pathway through Removing Lys63-Linked Polyubiquitination of TRAF3 and TRAF6. International Journal of Molecular Sciences, 17(5), 678. https://doi.org/10.3390/ijms17050678






