Characterization of Stripe Rust Resistance Genes in the Wheat Cultivar Chuanmai45
Abstract
:1. Introduction
2. Results
2.1. Assessment of Stripe Rust Resistance
2.2. Genetic Analysis of Resistance in Chuanmai45 (CH45)
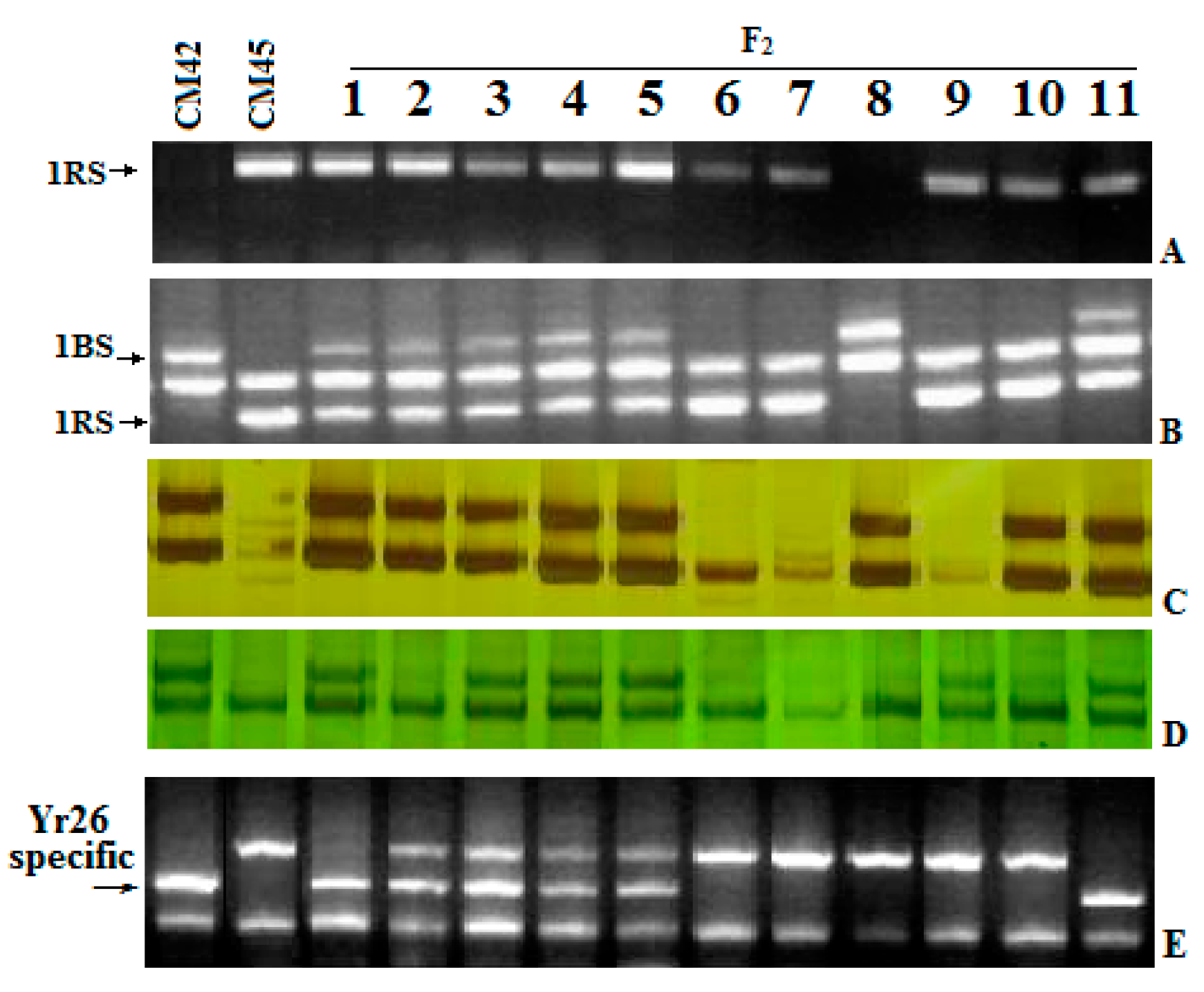
2.3. Bulk Segregate Analysis (BSA) of Resistance
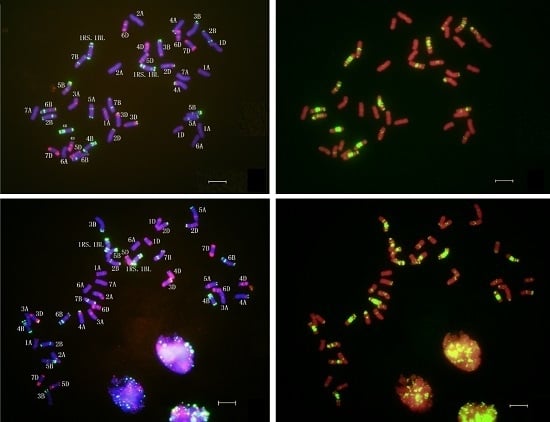
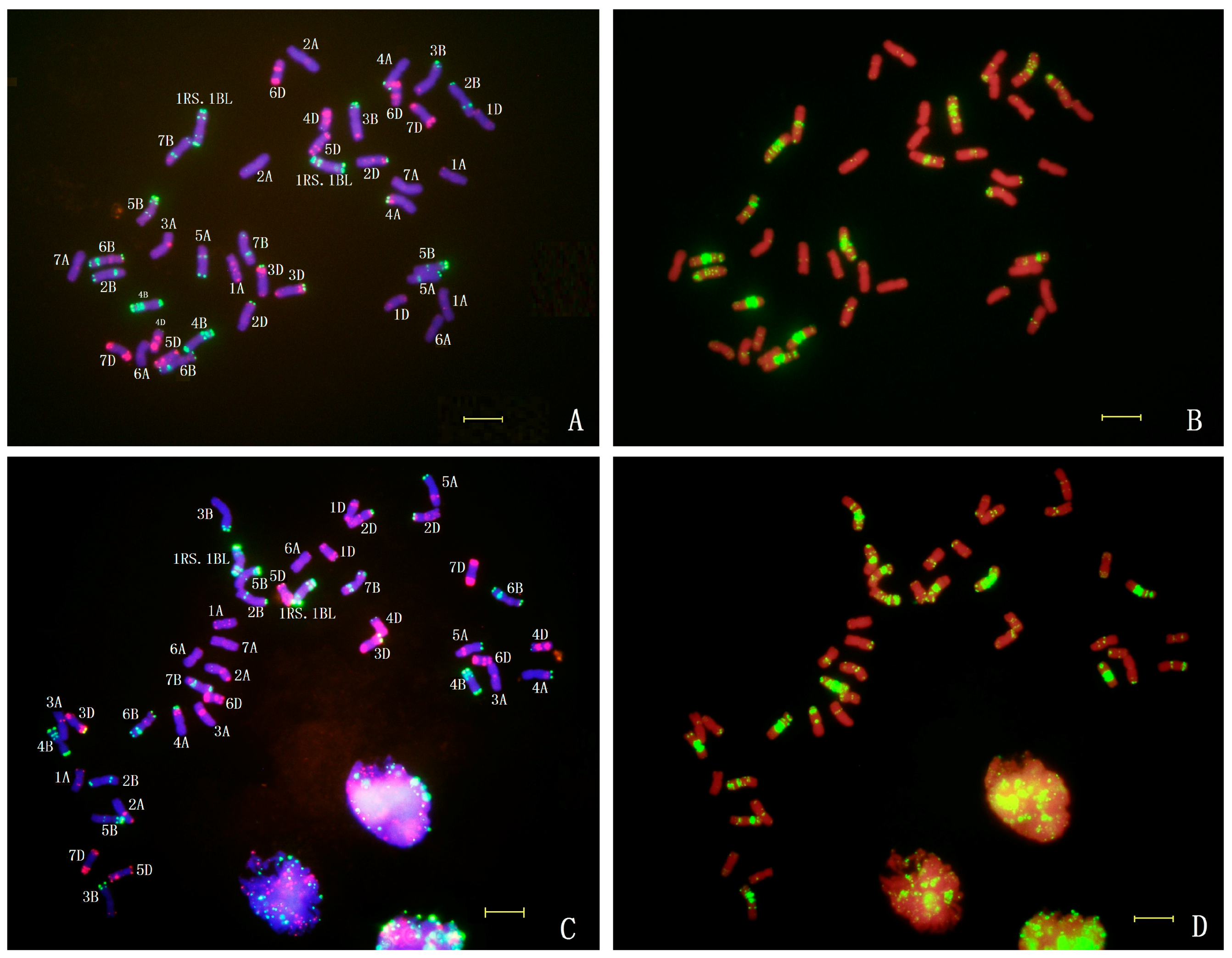
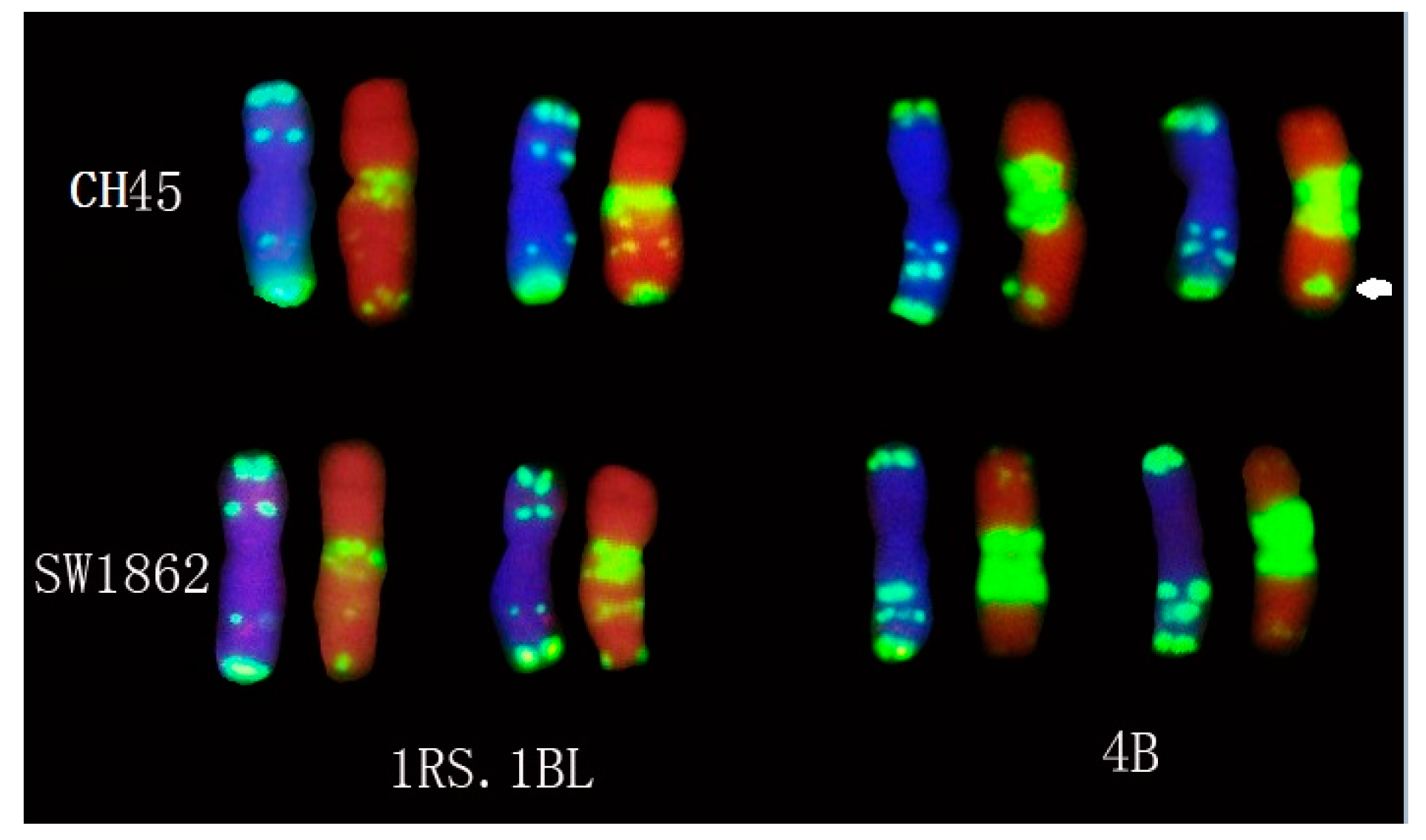
2.4. In Situ Hybridization of CH45
2.5. Recombination of Yr26 in CH45
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Disease Resistance Screening
4.3. Cytological Analysis
4.4. Molecular Marker Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Roelfs, A.P.; Singh, R.P.; Saari, E.E. Rust Diseases of Wheat: Concepts and Methods of Disease Management; CIMMYT: Mexico City, Mexico, 1992. [Google Scholar]
- Wellings, C.R. Global status of stripe rust: A review of historical and current threats. Euphytica 2011, 179, 129–141. [Google Scholar] [CrossRef]
- McIntosh, R.A.; Dubcovsky, J.; Rogers, J.; Morris, C.; Appels, R.; Xia, X. 2010: Catalogue of Gene Symbols for Wheat: 2010 Supplement. Available online: http://www.shigen.nig.ac.jp/wheat/komugi/genes/ (accessed on 30 July 2010).
- Ellis, J.G.; Lagudah, E.S.; Spielmeyer, W.; Dodds, P.N. The past, present and future of breeding rust resistant wheat. Front Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.Q.; Wu, L.R.; Liu, T.G.; Xu, S.C.; Jin, S.L.; Peng, Y.L.; Wang, B.T. Race dynamics, diversity, and virulence evolution in Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust in China from 2003 to 2007. Plant Dis. 2009, 93, 1093–1101. [Google Scholar] [CrossRef]
- Wan, A.M.; Zhao, Z.H.; Chen, X.M.; He, Z.H.; Jin, S.L.; Jia, Q.Z.; Yao, G.; Yang, J.X.; Wang, B.T.; Li, G.B.; et al. Wheat stripe rust epidemic and virulence of Puccinia striiformis f. sp. tritici in China in 2002. Plant Dis. 2004, 88, 896–904. [Google Scholar]
- Kang, Z.; Zhao, J.; Han, D.; Zhang, H.; Wang, X.; Wang, C.; Han, Q.; Guo, J.; Huang, L. Status of Wheat Rust Research and Control in China; BGRI 2010 Technical Workshop: St. Petersburg, Russia, 2010; pp. 50–69. [Google Scholar]
- He, Z.H.; Xia, X.C.; Chen, X.M.; Zhuang, Q.S. Progress of wheat breeding in China and the future perspective. Acta Agron. Sin. 2011, 37, 202–215. [Google Scholar] [CrossRef]
- Liu, G.T.; Peng, Y.L.; Chen, W.Q.; Zhang, Z.Y. First detection of virulence in Puccinia striiformis f. sp. tritici in China to resistance genes Yr24 (=Yr26) present in wheat cultivar Chuanmai 42. Plant Dis. 2010, 94. [Google Scholar] [CrossRef]
- Li, G.Q.; Li, Z.F.; Yang, W.Y.; Zhang, Y.; He, Z.H.; Xu, S.C.; Singh, R.P.; Qu, Y.Y.; Xia, X.C. Molecular mapping of stripe rust resistance gene YrCH42 in Chinese wheat cultivar Chuanmai 42 and its allelism with Yr24 and Yr26. Theor. Appl. Genet. 2006, 112, 1434–1440. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.E.; Li, G.Q.; He, Z.H.; Yang, W.Y.; Xu, M.L.; Xia, X.C. Development of an STS marker tightly linked to Yr26 against wheat stripe rust using the resistance gene-analog polymorphism (RGAP) technique. Mol. Breed. 2008, 22, 507–515. [Google Scholar] [CrossRef]
- Xue, S.; Zhang, Z.; Lin, F.; Kong, Z.; Cao, Y.; Li, C.; Yi, H.; Mei, M.; Zhu, H.; Wu, J.; et al. A high-density intervarietal map of the wheat genome enriched with markers derived from expressed sequence tags. Theor. Appl. Genet. 2008, 117, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, G.; Nakamura, T.; Ashida, T.; Saito, M.; Nasuda, S.; Endo, T.; Wu, J.; Matsumoto, T. Localization of anchor loci representing five hundred annotated rice genes to wheat chromosomes using PLUG markers. Theor. Appl. Genet. 2009, 118, 499–514. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Chen, L.; Wang, Y.; Li, M.; Yang, Z.; Qiu, L.; Yan, B.; Ren, Z.; Tang, Z. Oligonucleotide probes for ND-FISH analysis to identify rye and wheat chromosomes. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.M.; Zhang, Y.P.; Han, D.J.; Kang, Z.S.; Li, G.P.; Cao, A.Z.; Chen, P.D. SSR and STS markers for wheat stripe rust resistance gene Yr26. Euphytica 2008, 159, 359–366. [Google Scholar] [CrossRef]
- Singh, R.P.; Huerta-Espino, J.; Rajaram, S.; Crossa, J. Agronomic effects from chromosome translocations 7DL.7Ag and 1RS.1BL in spring wheat. Crop Sci. 1998, 38, 27–33. [Google Scholar] [CrossRef]
- Villareal, R.L.; Rajaram, S.; Mujeeb-Kazi, A.; Del Toro, E. The effect of chromosome 1RS.1BL translocation on the yield potential of certain spring wheats (Triticum aestivum L.). Plant Breed. 1991, 106, 77–81. [Google Scholar] [CrossRef]
- Lelley, T.; Eder, C.; Grausgruber, H. Influence of 1RS.1BL wheat-rye chromosome translocation on genotype by environment interaction. J. Cereal. Sci. 2004, 39, 313–320. [Google Scholar] [CrossRef]
- Ren, T.H.; Chen, F.; Yan, B.J.; Zhang, H.Q.; Ren, Z.L. Genetic diversity of wheat-rye 1RS.1BL translocation lines derived from different wheat and rye sources. Euphytica 2012, 183, 133–146. [Google Scholar] [CrossRef]
- Luo, P.G.; Zhan, H.Y.; Shu, K.; Zhang, H.Q.; Luo, H.Y.; Ren, Z.L. Stripe rust (Puccinia striiformis f. sp. tritici) resistance in wheat with wheat-rye 1BL/1RS chromosomal translocation. Can. J. Plant Pathol. 2008, 30, 254–259. [Google Scholar] [CrossRef]
- Rabinovich, S.V. Importance of wheat-rye translocations for breeding modern cultivars of Triticum aestivum L. Euphytica 1998, 100, 323–340. [Google Scholar] [CrossRef]
- Fluch, S.; Kopecky, D.; Burg, K.; Šimková, H.; Taudien, S.; Petzold, A.; Kubaláková, M.; Platzer, M.; Berenyi, M.; Krainer, S.; et al. Sequence composition and gene content of the short arm of rye (Secale cereale) chromosome 1. PLoS ONE 2012, 7, e30784. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chang, Z.; Zhang, X.; Yang, Z.; Li, X.; Jia, J.; Zhan, H.; Guo, H.; Wang, J. Putative Thinopyrum intermedium-derived stripe rust resistance gene Yr50 maps on wheat chromosome arm 4BL. Theor. Appl. Genet. 2013, 126, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.L.; Rosewarne, G.M.; Li, C.S.; Wu, X.L.; Yang, W.Y.; Wu, C. Physiological factors underpinning grain yield improvements of synthetic derived wheat in south western China. Crop Sci. 2014, 55, 98–122. [Google Scholar] [CrossRef]
- Ma, J.X.; Zhou, R.H.; Dong, Y.S.; Wang, L.F.; Wang, X.M.; Jia, J.Z. Molecular mapping and detection of the yellow rust resistance gene Yr26 in wheat transferred from Triticum turgidum L. using microsatellite markers. Euphytica 2001, 120, 219–226. [Google Scholar] [CrossRef]
- Qi, L.L.; Chen, P.D.; Liu, D.J.; Zhou, B.; Zhang, S.Z. The gene Pm21: A new source for resistance to wheat powdery mildew. Acta Agron. Sin. 1995, 21, 257–262. [Google Scholar]
- Zhang, X.; Han, D.; Zeng, Q.; Duan, Y.; Yuan, F.; Shi, J.; Wang, Q.; Wu, J.; Huang, L.; Kang, Z. Fine mapping of wheat stripe rust resistance gene Yr26 based on collinearity of wheat with Brachypodium distachyon and rice. PLoS ONE 2013, 8, e57885. [Google Scholar] [CrossRef] [PubMed]
- Bariana, H.S.; McIntosh, R.A. Cytogenetic studies in wheat. XV. Location of rust resistance genes in VPM1 and their genetic linkage with other disease resistance genes in chromosome 2A. Genome 1993, 36, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Han, F.P.; Lamb, J.C.; Birchler, J.A. High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc. Natl. Acad. Sci. USA 2006, 103, 3238–3243. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.X.; Yang, Z.J.; Fu, S.L. Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 2014, 55, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.J.; Li, G.R.; Jia, J.Q.; Zeng, X.; Lei, M.P.; Zeng, Z.X.; Zhang, T.; Ren, Z.L. Molecular cytogenetic characterization of wheat—Secale africanum amphiploids and derived introgression lines with stripe rust resistance. Euphytica 2009, 167, 197–202. [Google Scholar] [CrossRef]
- Bartos, J.; Paux, E.; Kofler, R.; Havrankova, M.; Kopecky, D.; Suchankova, P.; Safar, J.; Simkova, H.; Town, C.D.; Lelley, T.; et al. A first survey of the rye (Secale cereale) genome composition through BAC end sequencing of the short arm of chromosome 1R. BMC Plant Biol. 2008, 8. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, M.; Fukushima, T.; Nasuda, S.; Masoudi-Nejad, A.; Ishikawa, G.; Nakamura, T.; Endo, T.R. Dissection of rye chromosome 1R in common wheat. Genes Genet. Syst. 2008, 83, 43–53. [Google Scholar] [CrossRef] [PubMed]


| Genotypes/Lines | Yr Gene Location | Infection Types |
|---|---|---|
| Avocet | - | 4 |
| Avocet+Yr1 | 2A | 0 |
| Kalyansona (Yr2, Yr29) | 7B, 1B | 3 |
| Avocet+Yr5 | 2B | 0 |
| Avocet+Yr6 | 7B | 0 |
| Avocet+Yr7 | 2B | 4 |
| Avocet+Yr8 | 2M/2B | 4 |
| Avocet+Yr10 | 1B | 4 |
| Avocet+Yr15 | 1B | 0 |
| Avocet+Yr17, Yr18 | 2A, 7D | 3 |
| Avocet+Yr18 | 7D | 4 |
| Avocet+Yr24 | 1B | 4 |
| Avocet+Yr26 | 1B | 4 |
| Avocet+Yr27 | 2B | 3 |
| Avocet+Yr28 | 4D | 0 |
| Avocet+Yr29 | 1B | 4 |
| Avocet+Yr31 | 2B | 3 |
| Avocet+YrA | unkmown | 4 |
| Avocet+YrCV | 2AL | 0 |
| Avocet+YrSp | 2B | 0 |
| Pavon 76 (Yr6, Yr7, Yr29, Yr30, +) | 1B, 2B, 7B | 1 |
| PBW 343 (Yr9, Yr27, Yr29, +) | 1B, 2B | 4 |
| Seri 82 (Yr2, Yr9, Yr29, Yr30, +) | 1R/1B, 1B, 3B, 7B | 3 |
| Super Kauz (Yr9, Yr27, Yr18, Yr30, +) | 1R/1B, 2B, 3B, 7D | 3 |
| Chuanmai45 | - | 0 |
| SW1862 | - | 0 |
| CN19 (Yr41) | 2B | 4 |
| Chuanmai42 (YrCH42=Yr26) | 1B | 4 |
| Materials | No. of Plants | Infection Type (R) | Infection Type (S) | Expected Ratio | Chi-Square Test (χ2) | p Value * | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 1 | 2 | 3 | 4 | |||||
| CH45 | 10 | 10 | - | - | - | - | - | - | - | - |
| CH42 | 10 | - | - | - | - | - | 10 | - | - | - |
| CN19 | 10 | - | - | - | - | - | 10 | - | - | - |
| CH45/CH42F1 | 10 | - | - | - | - | - | 10 | - | - | - |
| CH45/CH42F2 | 192 | 13 | 48 | 23 | 29 | 18 | 61 | 7:9 | 0 | 1 |
| CH45/CN19F1 | 10 | - | - | - | - | 8 | 2 | - | - | - |
| CH45/CN19F2 | 206 | 25 | 43 | 28 | 9 | 5 | 96 | 7:9 | 0.7 | 0.4 |
| No Plant of Homozygous 1RS | No Plant of Non Homozygous 1RS | Expected Ratio | Chi-Square Test (χ2) | p Values * | ||
|---|---|---|---|---|---|---|
| 44 | 148 | 1:3 | 0.4 | 0.50 | ||
| Resistance | Susceptible | Resistance | Susceptible | - | - | - |
| 44 | 0 | - | - | - | - | - |
| - | - | 40 | 108 | 1:3 | 0.32 | 0.57 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, E.; Li, G.; Li, L.; Zhang, Z.; Yang, W.; Peng, Y.; Zhu, Y.; Yang, Z.; Rosewarne, G.M. Characterization of Stripe Rust Resistance Genes in the Wheat Cultivar Chuanmai45. Int. J. Mol. Sci. 2016, 17, 601. https://doi.org/10.3390/ijms17040601
Yang E, Li G, Li L, Zhang Z, Yang W, Peng Y, Zhu Y, Yang Z, Rosewarne GM. Characterization of Stripe Rust Resistance Genes in the Wheat Cultivar Chuanmai45. International Journal of Molecular Sciences. 2016; 17(4):601. https://doi.org/10.3390/ijms17040601
Chicago/Turabian StyleYang, Ennian, Guangrong Li, Liping Li, Zhenyu Zhang, Wuyun Yang, Yunliang Peng, Yongqing Zhu, Zujun Yang, and Garry M. Rosewarne. 2016. "Characterization of Stripe Rust Resistance Genes in the Wheat Cultivar Chuanmai45" International Journal of Molecular Sciences 17, no. 4: 601. https://doi.org/10.3390/ijms17040601
APA StyleYang, E., Li, G., Li, L., Zhang, Z., Yang, W., Peng, Y., Zhu, Y., Yang, Z., & Rosewarne, G. M. (2016). Characterization of Stripe Rust Resistance Genes in the Wheat Cultivar Chuanmai45. International Journal of Molecular Sciences, 17(4), 601. https://doi.org/10.3390/ijms17040601