Argonaute and Argonaute-Bound Small RNAs in Stem Cells
Abstract
:1. Introduction
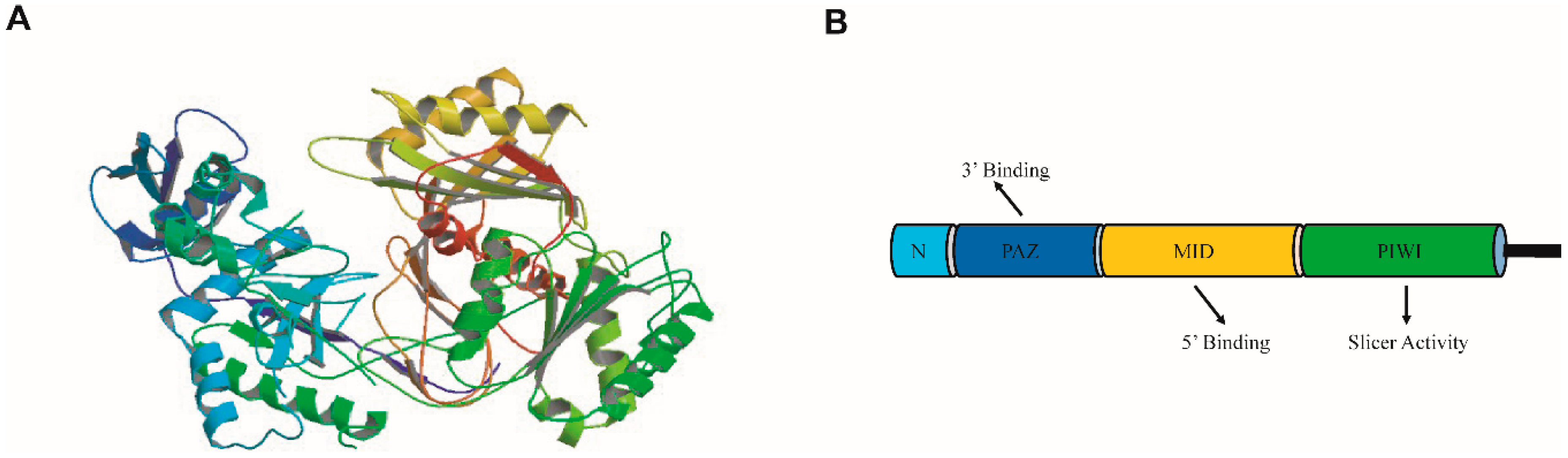
2. The Argonaute (AGO) Family
| Gene Name(Subfamily) | Molecular Function | sRNA Bound | Reference |
|---|---|---|---|
| Homo sapiens | |||
| EIF2C1/hAGO1 (AGO) | miRNA-directed target gene regulation, constitutive and alternative splicing, heterochromatin silencing | miRNAs | [29,30,31] |
| EIF2C2/hAGO2 (AGO) | miRNA-directed target gene regulation, heterochromatin silencing, RNAi | miRNAs, siRNA | [30,31,32] |
| EIF2C3/hAGO3 (AGO) | miRNA-directed target gene regulation | miRNAs | [30] |
| EIF2C4/hAGO4 (AGO) | miRNA-directed target gene regulation | miRNAs | [22] |
| Piwil1/Hiwi (Piwi) | stem cell self-renewal, division, gametogenesis, germ cell proliferation, and RNAi | piRNAs | [3,12] |
| Piwil2/Hili (Piwi) | signaling regulation | piRNAs | [33] |
| Piwil3 (Piwi) | n/d | piRNAs | [3,12] |
| Piwil4/Hiwi2 (Piwi) | transposon silencing | piRNAs | [34] |
| Drosophila melanogaster | |||
| dAGO1 (AGO) | miRNA-mediated gene silencing | 22–23-nt miRNAs | [35,36] |
| dAGO2 (AGO) | RNAi in embryos | miRNAs and 21-nt siRNAs | [35,36,37,38] |
| dAGO3 (Piwi) | transposon silencing | piRNAs | [39,40,41] |
| Aubergine/AUB (Piwi) | transposon silencing, stellate silencing, RNAi | piRNAs | [41,42] |
| Piwi (Piwi) | transposon silencing, germline stem-cell maintenance, RNAi | piRNAs, rasiRNAs | [41,42,43] |
| Arabidopsis thaliana | |||
| AtAGO1 | plant development regulation and stress responses | 21-nt miRNAs | [8,44,45] |
| AtAGO2 | antibacterial immunity, viral defense, DSB-induced sRNAs activity, and DNA repair | 21-nt miRNAs, vsiRNAs, diRNAs | [46,47,48] |
| AtAGO3 | n/d | n/d | |
| AtAGO4 | RdDM pathway | 24-nt siRNAs | [49,50] |
| AtAGO5 | initiation of megagametogenesis, antiviral RNA silencing | siRNAs | [46,51,52,53] |
| AtAGO6 | methylation of tasiRNA-generating loci and transcriptionally active TEs, shoot and root meristems | 24-nt siRNAs, 21–22-nt endo-siRNAs | [49,54,55,56] |
| AtAGO7/ZIPPY | TAS3-derived tasiRNA biogenesis, leaf development | miR390 | [57] |
| AtAGO8 | Proposed to be a pseudogene | n/d | [10] |
| AtAGO9 | germ cell fate repression in the somatic companion cells surrounding MMC, and DNA repair | 24-siRNAs | [46,58] |
| AtAGO10/PINHEAD/ZWILLE | regulation of shoot apical meristems | miR165/166, miR172 | [59,60,61,62,63] |
| Oryza sativa | |||
| OsAGO1a/b/c | miRNA-directed target gene regulation | 21-nt miRNAs | [64] |
| OsAGO5c/MEL1 | regulation of cell division of premeiotic germ cells | 21-nt siRNAs | [65,66] |
| OsAGO7 | TAS3-derived tasiRNA biogenesis | miR390 | [67] |
| OsAGO10/OsPNH1 | regulates leaf development and maintenance of the shoot apical meristem | n/d | [68] |
| OsAGO18 | broad-spectrum virus resistance | n/d | [69] |
| Zea mays | |||
| ZmAGO7/RGD2 | TAS3-derived tasiRNA biogenesis | miR390 | [70] |
| ZmAGO9/AGO104 | somatic cell fate repression in the germ cells | n/d | [71] |
| ZmAGO18b | tapetum and germ cell development | n/d | [72] |
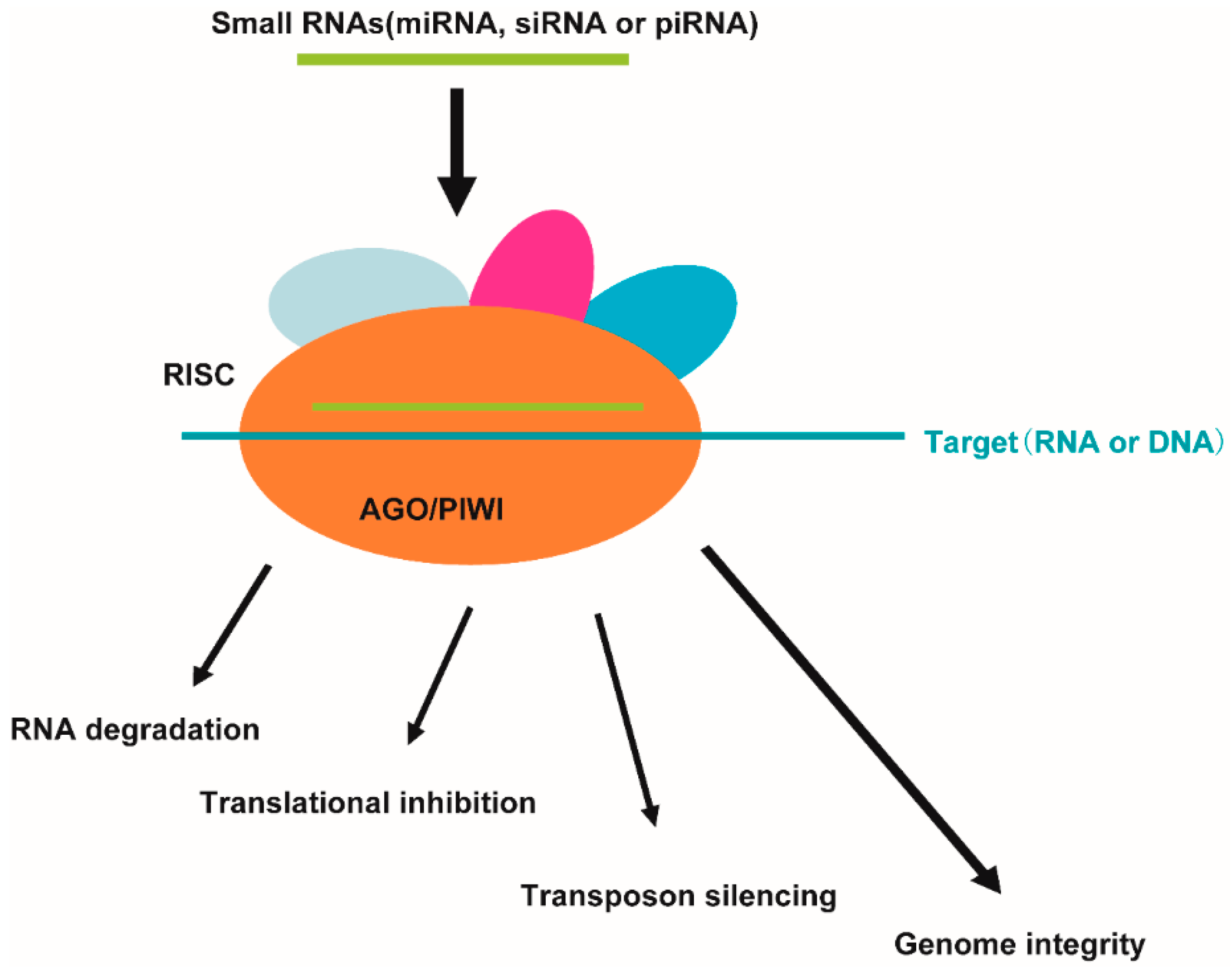
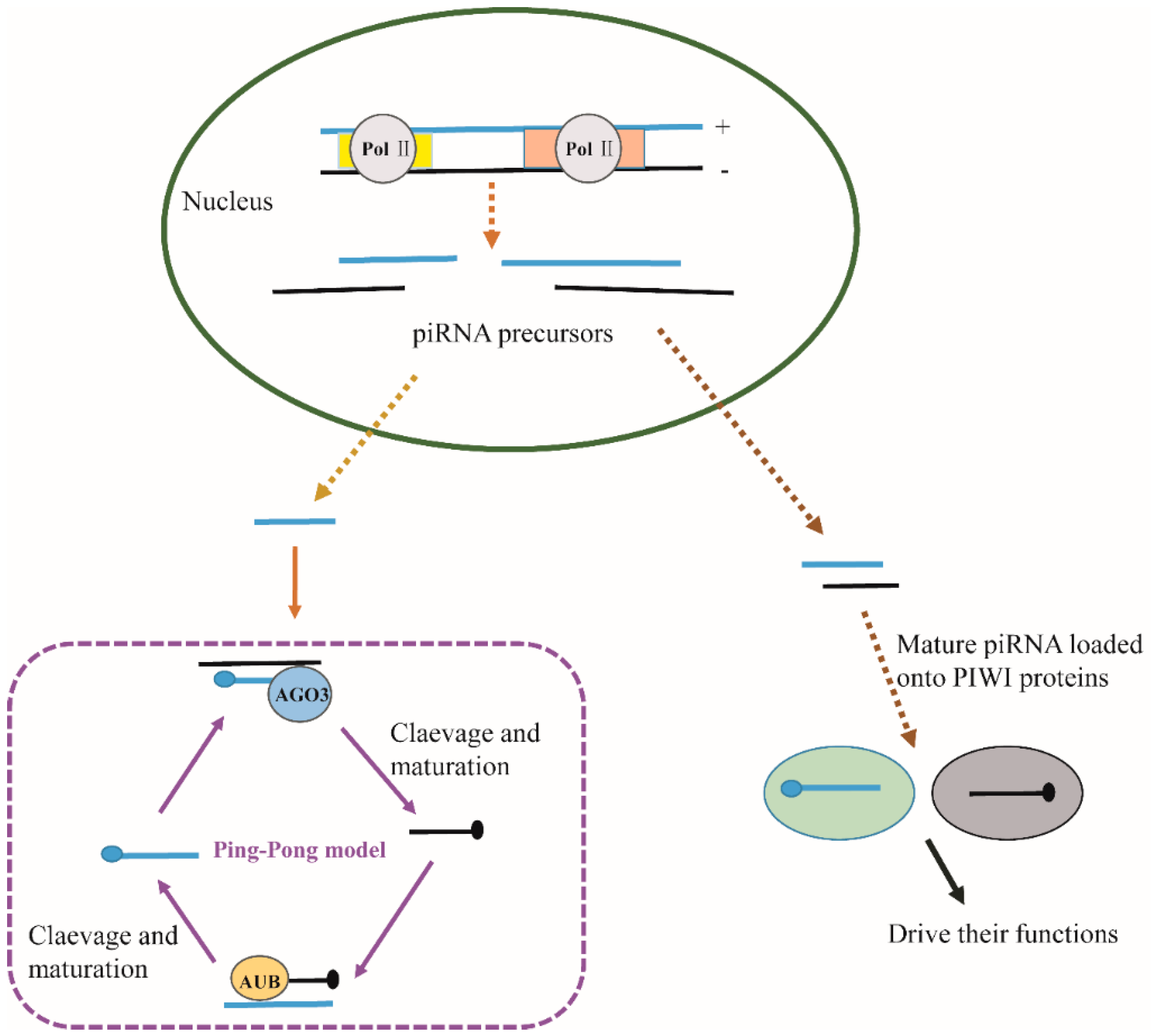
3. The Association of AGO Proteins and Small RNAs
4. Argonaute-Interacting Partners and Regulation of Argonaute Function
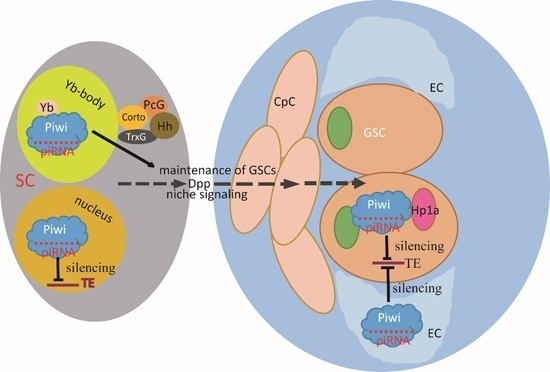
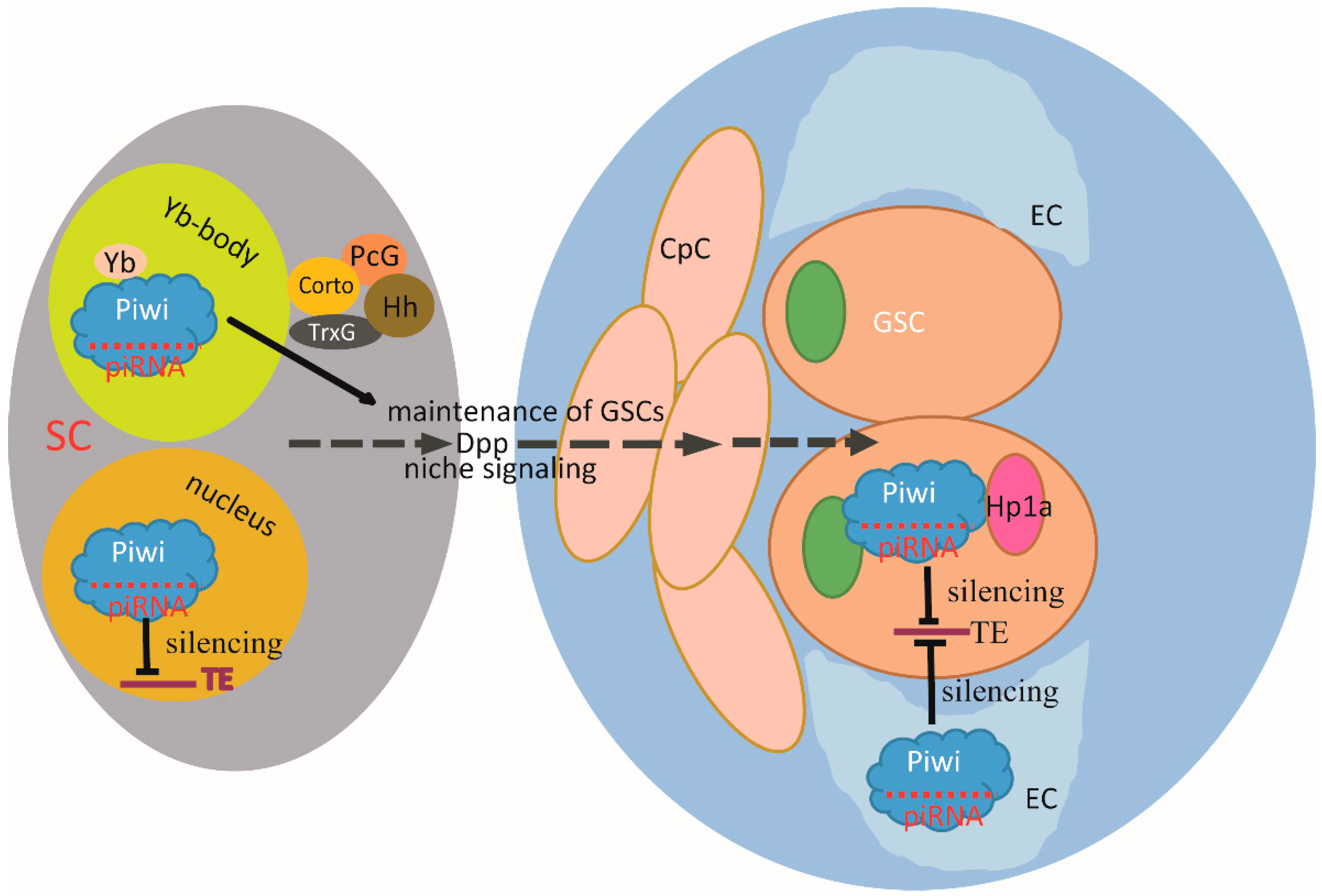
5. AGO Proteins and AGO-Bound Small RNAs in Stem Cells

6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Carthew, R.W.; Sontheimer, E.J. Origins and mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Farazi, T.A.; Juranek, S.A.; Tuschl, T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development 2008, 135, 1201–1214. [Google Scholar] [CrossRef] [PubMed]
- Höck, J.; Meister, G. The Argonaute protein family. Genome Biol. 2008, 9. [Google Scholar] [CrossRef] [PubMed]
- Carmell, M.A.; Xuan, Z.; Zhang, M.Q.; Hannon, G.J. The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002, 16, 2733–2742. [Google Scholar] [CrossRef] [PubMed]
- Unhavaithaya, Y.; Hao, Y.; Beyret, E.; Yin, H.; Kuramochi-Miyagawa, S.; Nakano, T.; Lin, H. MILI, a Piwi-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J. Biol. Chem. 2009, 284, 6507–6519. [Google Scholar] [CrossRef] [PubMed]
- Grochola, L.F.; Greither, T.; Taubert, H.; Moller, P.; Knippschild, U.; Udelnow, A.; Henne-Bruns, D.; Wurl, P. The stem cell-associated Hiwi gene in human adenocarcinoma of the pancreas: Expression and risk of tumour-related death. Br. J. Cancer 2008, 99, 1083–1088. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Engel, W.; Nayernia, K. Stem cell protein Piwil2 modulates expression of murine spermatogonial stem cell expressed genes. Mol. Reprod. Dev. 2006, 73, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Bohmert, K.; Camus, I.; Bellini, C.; Bouchez, D.; Caboche, M.; Benning, C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998, 17, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Mallory, A.; Vaucheret, H. Form, function, and regulation of ARGONAUTE proteins. Plant Cell 2010, 22, 3879–3889. [Google Scholar] [CrossRef] [PubMed]
- Vaucheret, H. Plant Argonautes. Trends Plant Sci. 2008, 13, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Meister, G. Argonaute proteins: Functional insights and emerging roles. Nat. Rev. Genet. 2013, 14, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Hutvagner, G.; Simard, M.J. Argonaute proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008, 9, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Peters, L.; Meister, G. Argonaute proteins: Mediators of RNA silencing. Mol. Cell 2007, 26, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Shiohama, A.; Minoshima, S.; Shimizu, N. Identification of eight members of the Argonaute family in the human genome. Genomics 2003, 82, 323–330. [Google Scholar] [CrossRef]
- Gonzalez, J.; Qi, H.; Liu, N.; Lin, H. Piwi is a key regulator of both somatic and germline stem cells in the Drosophila testis. Cell Rep. 2015, 12, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.S.; Roe, S.M.; Barford, D. Structural insights into mRNA recognition from a Piwi domain-siRNA guide complex. Nature 2005, 434, 663–666. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.B.; Yuan, Y.R.; Meister, G.; Pei, Y.; Tuschl, T.; Patel, D.J. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 2005, 434, 666–670. [Google Scholar] [CrossRef] [PubMed]
- Bamezai, S.; Rawat, V.P.; Buske, C. Concise review: The Piwi-piRNA axis: Pivotal beyond transposon silencing. Stem Cells 2012, 30, 2603–2611. [Google Scholar] [CrossRef] [PubMed]
- Song, J.J.; Smith, S.K.; Hannon, G.J.; Joshua-Tor, L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 2004, 305, 1434–1437. [Google Scholar] [CrossRef] [PubMed]
- Rivas, F.V.; Tolia, N.H.; Song, J.-J.; Aragon, J.P.; Liu, J.; Hannon, G.J.; Joshua-Tor, L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005, 12, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, K.; Weinberg, D.E.; Bartel, D.P.; Patel, D.J. Structure of yeast Argonaute with guide RNA. Nature 2012, 486, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 2004, 15, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Schirle, N.T.; MacRae, I.J. The crystal structure of human Argonaute2. Science 2012, 336, 1037–1040. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Niu, D.; Carbonell, A.; Wang, A.; Lee, A.; Tun, V.; Wang, Z.; Carrington, J.C.; Chang, C.E.; Jin, H. ARGONAUTE Piwi domain and microRNA duplex structure regulate small RNA sorting in Arabidopsis. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tang, W.; Zhang, L.R.; Zhang, C.Y. FMRP regulates miR196a-mediated repression of HOXB8 via interaction with the AGO2 MID domain. Mol. Biosyst. 2014, 10, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Schirle, N.T.; Sheu-Gruttadauria, J.; MacRae, I.J. Structural basis for microRNA targeting. Science 2014, 346, 608–613. [Google Scholar] [CrossRef] [PubMed]
- Schirle, N.T.; Sheu-Gruttadauria, J.; Chandradoss, S.D.; Joo, C.; MacRae, I.J. Water-mediated recognition of t1-adenosine anchors Argonaute2 to microRNA targets. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Rashid, U.J.; Paterok, D.; Koglin, A.; Gohlke, H.; Piehler, J.; Chen, J.C. Structure of Aquifex aeolicus argonaute highlights conformational flexibility of the PAZ domain as a potential regulator of RNA-induced silencing complex function. J. Biol. Chem. 2007, 282, 13824–13832. [Google Scholar] [CrossRef] [PubMed]
- Allo, M.; Agirre, E.; Bessonov, S.; Bertucci, P.; Gomez Acuna, L.; Buggiano, V.; Bellora, N.; Singh, B.; Petrillo, E.; Blaustein, M.; et al. Argonaute-1 binds transcriptional enhancers and controls constitutive and alternative splicing in human cells. Proc. Natl. Acad. Sci. USA 2014, 111, 15622–15629. [Google Scholar] [CrossRef] [PubMed]
- Burroughs, A.M.; Ando, Y.; de Hoon, M.J.; Tomaru, Y.; Suzuki, H.; Hayashizaki, Y.; Daub, C.O. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 2011, 8, 158–177. [Google Scholar] [CrossRef] [PubMed]
- Janowski, B.A.; Huffman, K.E.; Schwartz, J.C.; Ram, R.; Nordsell, R.; Shames, D.S.; Minna, J.D.; Corey, D.R. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat. Struct. Mol. Biol. 2006, 13, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Goff, L.A.; Davila, J.; Swerdel, M.R.; Moore, J.C.; Cohen, R.I.; Wu, H.; Sun, Y.E.; Hart, R.P. Ago2 immunoprecipitation identifies predicted microRNAs in human embryonic stem cells and neural precursors. PLoS ONE 2009, 4, e7192. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Lu, Y.; Yang, P.; Li, C.; Sun, H.; Tao, D.; Liu, Y.; Zhang, S.; Ma, Y. Hili inhibits TGF-beta signaling by interacting with Hsp90 and promoting TbetaR degradation. PLoS ONE 2012, 7, e41973. [Google Scholar]
- Keam, S.P.; Young, P.E.; McCorkindale, A.L.; Dang, T.H.; Clancy, J.L.; Humphreys, D.T.; Preiss, T.; Hutvagner, G.; Martin, D.I.; Cropley, J.E.; et al. The human Piwi protein Hiwi2 associates with tRNA-derived piRNAs in somatic cells. Nucleic Acids Res. 2014, 42, 8984–8995. [Google Scholar] [CrossRef] [PubMed]
- Ghildiyal, M.; Xu, J.; Seitz, H.; Weng, Z.; Zamore, P.D. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA 2010, 16, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Okamura, K.; Ishizuka, A.; Siomi, H.; Siomi, M.C. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004, 18, 1655–1666. [Google Scholar] [CrossRef] [PubMed]
- Ameres, S.L.; Hung, J.H.; Xu, J.; Weng, Z.; Zamore, P.D. Target RNA-directed tailing and trimming purifies the sorting of endo-siRNAs between the two Drosophila Argonaute proteins. RNA 2011, 17, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M.; Boettcher, S.; Caudy, A.A.; Kobayashi, R.; Hannon, G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 2001, 293, 1146–1150. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.R.; Megosh, H.; Lin, H. Piwi proteins are essential for early Drosophila embryogenesis. Dev. Biol. 2014, 385, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Kalmykova, A.I.; Klenov, M.S.; Gvozdev, V.A. Argonaute protein Piwi controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res. 2005, 33, 2052–2059. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef] [PubMed]
- Vagin, V.V.; Sigova, A.; Li, C.; Seitz, H.; Gvozdev, V.; Zamore, P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 2006, 313, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Nishida, K.M.; Mori, T.; Kawamura, Y.; Miyoshi, K.; Nagami, T.; Siomi, H.; Siomi, M.C. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006, 20, 2214–2222. [Google Scholar] [CrossRef] [PubMed]
- Vaucheret, H.; Vazquez, F.; Crété, P.; Bartel, D.P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004, 18, 1187–1197. [Google Scholar] [CrossRef] [PubMed]
- Dzianott, A.; Sztuba-Solinska, J.; Bujarski, J.J. Mutations in the antiviral RNAi defense pathway modify Brome mosaic virus RNA recombinant profiles. MPMI 2012, 25, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Oliver, C.; Santos, J.L.; Pradillo, M. On the role of some ARGONAUTE proteins in meiosis and DNA repair in Arabidopsis thaliana. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Takeda, A.; Iwasaki, S.; Watanabe, T.; Utsumi, M.; Watanabe, Y. The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 2008, 49, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Jaubert, M.; Bhattacharjee, S.; Mello, A.F.; Perry, K.L.; Moffett, P. ARGONAUTE2 mediates RNA-silencing antiviral defenses against Potato virus X in Arabidopsis. Plant Physiol. 2011, 156, 1556–1564. [Google Scholar] [CrossRef] [PubMed]
- Havecker, E.R.; Wallbridge, L.M.; Hardcastle, T.J.; Bush, M.S.; Kelly, K.A.; Dunn, R.M.; Schwach, F.; Doonan, J.H.; Baulcombe, D.C. The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 2010, 22, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Matzke, M.A.; Mosher, R.A. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014, 15, 394–408. [Google Scholar] [CrossRef] [PubMed]
- Brosseau, C.; Moffett, P. Functional and genetic analysis identify a role for arabidopsis ARGONAUTE5 in antiviral RNA silencing. Plant Cell 2015, 27, 1742–1754. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Cai, T.; Hu, Y.; Chen, Y.; Hodges, E.; Ni, F.; Wu, L.; Li, S.; Zhou, H.; Long, C.; et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 2008, 133, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Tucker, M.R.; Okada, T.; Hu, Y.; Scholefield, A.; Taylor, J.M.; Koltunow, A.M. Somatic small RNA pathways promote the mitotic events of megagametogenesis during female reproductive development in Arabidopsis. Development 2012, 139, 1399–1404. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Zhu, J.; Kapoor, A.; Zhu, J.-K. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 2007, 26, 1691–1701. [Google Scholar] [CrossRef] [PubMed]
- Eun, C.; Lorkovic, Z.J.; Naumann, U.; Long, Q.; Havecker, E.R.; Simon, S.A.; Meyers, B.C.; Matzke, A.J.; Matzke, M. AGO6 functions in RNA-mediated transcriptional gene silencing in shoot and root meristems in Arabidopsis thaliana. PLoS ONE 2011, 6, e25730. [Google Scholar] [CrossRef] [PubMed]
- McCue, A.D.; Panda, K.; Nuthikattu, S.; Choudury, S.G.; Thomas, E.N.; Slotkin, R.K. ARGONAUTE 6 bridges transposable element mRNA-derived siRNAs to the establishment of DNA methylation. EMBO J. 2015, 34, 20–35. [Google Scholar] [CrossRef] [PubMed]
- Hunter, C.; Sun, H.; Poethig, R. The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr. Biol. 2003, 13, 1734–1739. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Hu, F.; Wang, R.; Zhou, X.; Sze, S.-H.H.; Liou, L.W.W.; Barefoot, A.; Dickman, M.; Zhang, X. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 2011, 145, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Moussian, B.; Schoof, H.; Haecker, A.; Jürgens, G.; Laux, T. Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 1998, 17, 1799–1809. [Google Scholar] [CrossRef] [PubMed]
- Lynn, K.; Fernandez, A.; Aida, M.; Sedbrook, J.; Tasaka, M.; Masson, P.; Barton, M.K. The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 1999, 126, 469–481. [Google Scholar] [PubMed]
- Liu, Q.; Yao, X.; Pi, L.; Wang, H.; Cui, X.; Huang, H. The ARGONAUTE10 gene modulates shoot apical meristem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis. Plant J. Cell Mol. Biol. 2009, 58, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Manavella, P.A.; Weigel, D.; Wu, L. Argonaute10 as a miRNA Locker. Cell 2011, 145, 173–174. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Liu, X.; Yan, J.; Wang, W.; Yumul, R.E.; Kim, Y.J.; Dinh, T.T.; Liu, J.; Cui, X.; Zheng, B. ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet. 2011, 7, e1001358. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhang, Q.; Zhou, H.; Ni, F.; Wu, X.; Qi, Y. Rice microRNA effector complexes and targets. Plant Cell Online 2009, 21, 3421–3435. [Google Scholar] [CrossRef] [PubMed]
- Nonomura, K.-I.; Morohoshi, A.; Nakano, M.; Eiguchi, M.; Miyao, A.; Hirochika, H.; Kurata, N. A germ cell-specific gene of the Argonaute family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell Online 2007, 19, 2583–2594. [Google Scholar] [CrossRef] [PubMed]
- Komiya, R.; Ohyanagi, H.; Niihama, M.; Watanabe, T.; Nakano, M.; Kurata, N.; Nonomura, K. Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J. Cell Mol. Biol. 2014, 78, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Wang, J.; Wan, X.; Shen, G.; Wang, X.; Zhang, J. Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit. Planta 2007, 226, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, A.; Ito, M.; Kamiya, N.; Sato, Y.; Matsuoka, M. OsPNH1 regulates leaf development and maintenance of the shoot apical meristem in rice. Plant J. Cell Mol. Biol. 2002, 30, 189–201. [Google Scholar] [CrossRef]
- Wu, J.; Yang, Z.; Wang, Y.; Zheng, L.; Ye, R.; Ji, Y.; Zhao, S.; Ji, S.; Liu, R.; Xu, L.; et al. Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Douglas, R.N.; Wiley, D.; Sarkar, A.; Springer, N.; Timmermans, M.C.; Scanlon, M.J. ragged seedling2 encodes an ARGONAUTE7-like protein required for mediolateral expansion, but not dorsiventrality, of maize leaves. Plant Cell Online 2010, 22, 1441–1451. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Goel, S.; Meeley, R.B.; Dantec, C.; Parrinello, H.; Michaud, C.; Leblanc, O.; Grimanelli, D. Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell Online 2011, 23, 443–458. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.; Sun, W.; Zhang, K.; Jia, H.; Liu, L.; Liu, Z.; Teng, F.; Zhang, Z. Identification and characterization of Argonaute gene family and meiosis-enriched Argonaute during sporogenesis in maize. J. Integr. Plant Biol. 2014, 56, 1042–1052. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xia, R.; Meyers, B.C.; Walbot, V. Evolution, functions, and mysteries of plant ARGONAUTE proteins. Curr. Opin. Plant Biol. 2015, 27, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Ender, C.; Meister, G. Argonaute proteins at a glance. J. Cell Sci. 2010, 123, 1819–1823. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Tomari, Y. RISC assembly: Coordination between small RNAs and Argonaute proteins. Biochim. Biophys. Acta 2016, 1859, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Hannon, G.J. RNA interference. Nature 2002, 418, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Tomari, Y.; Zamore, P.D. Perspective: Machines for RNAi. Genes Dev. 2005, 19, 517–529. [Google Scholar] [CrossRef] [PubMed]
- Lim, R.S.; Kai, T. A piece of the pi(e): The diverse roles of animal piRNAs and their Piwi partners. Semin. Cell Dev. Biol. 2015, 47–48, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Nishida, K.M.; Saito, K.; Mori, T.; Kawamura, Y.; Nagami-Okada, T.; Inagaki, S.; Siomi, H.; Siomi, M.C. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA 2007, 13, 1911–1922. [Google Scholar] [CrossRef] [PubMed]
- Barckmann, B.; Pierson, S.; Dufourt, J.; Papin, C.; Armenise, C.; Port, F.; Grentzinger, T.; Chambeyron, S.; Baronian, G.; Desvignes, J.P.; et al. Aubergine iCLIP reveals piRNA-dependent decay of mRNAs involved in germ cell development in the early embryo. Cell Rep. 2015, 12, 1205–1216. [Google Scholar] [CrossRef] [PubMed]
- Klenov, M.S.; Lavrov, S.A.; Stolyarenko, A.D.; Ryazansky, S.S.; Aravin, A.A.; Tuschl, T.; Gvozdev, V.A. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007, 35, 5430–5438. [Google Scholar] [CrossRef] [PubMed]
- Toth, K.F.; Pezic, D.; Stuwe, E.; Webster, A. The piRNA pathway guards the germline genome against transposable elements. Adv. Exp. Med. Biol. 2016, 886, 51–77. [Google Scholar] [PubMed]
- Senti, K.A.; Jurczak, D.; Sachidanandam, R.; Brennecke, J. piRNA-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear piRNA repertoire. Genes Dev. 2015, 29, 1747–1762. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Song, L.; Liu, C.; Lv, X.; Li, X.; Jie, J.; Zhao, D.; Li, D. piR-55490 inhibits the growth of lung carcinoma by suppressing mTOR signaling. Tumour Biol. 2015, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Bao, H.; Wu, Q.; Wang, Y. Transcriptome-wide identification of miRNA targets under nitrogen deficiency in populus tomentosa using degradome sequencing. Int. J. Mol. Sci. 2015, 16, 13937–13958. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Wang, Y.; Liu, X.; Lv, S.; Feng, C.; Qi, M.; Li, T. Small RNA and degradome sequencing reveals microRNAs and their targets involved in tomato pedicel abscission. Planta 2015, 242, 963–984. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.; Liu, Z.; Zou, X.; Jiang, Y.; Qiu, F.; Zheng, Y.; Zhang, Z. Genome-wide identification and analysis of microRNA responding to long-term waterlogging in crown roots of maize seedlings. Physiol. Plant. 2013, 147, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Chi, S.W.; Zang, J.B.; Mele, A.; Darnell, R.B. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 2009, 460, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.D.; Huang, H.Y.; Chou, C.H.; Sun, Y.M.; Hsu, M.T.; Tsou, A.P. Integrated analyses to reconstruct microRNA-mediated regulatory networks in mouse liver using high-throughput profiling. BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Haecker, I.; Renne, R. HITS-CLIP and PAR-CLIP advance viral miRNA targetome analysis. Crit. Rev. Eukaryot. Gene Expr. 2014, 24, 101–116. [Google Scholar] [CrossRef] [PubMed]
- Vourekas, A.; Mourelatos, Z. HITS-CLIP (CLIP-Seq) for mouse Piwi proteins. Methods Mol. Biol. 2014, 1093, 73–95. [Google Scholar] [PubMed]
- Montgomery, T.A.; Howell, M.D.; Cuperus, J.T.; Li, D.; Hansen, J.E.; Alexander, A.L.; Chapman, E.J.; Fahlgren, N.; Allen, E.; Carrington, J.C. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 2008, 133, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Durán-Figueroa, N.; Vielle-Calzada, J.-P. ARGONAUTE9-dependent silencing of transposable elements in pericentromeric regions of Arabidopsis. Plant Signal. Behav. 2010, 5, 1476–1479. [Google Scholar] [CrossRef] [PubMed]
- Girard, A.; Sachidanandam, R.; Hannon, G.J.; Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006, 442, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Aravin, A.; Gaidatzis, D.; Pfeffer, S.; Lagos-Quintana, M.; Landgraf, P.; Iovino, N.; Morris, P.; Brownstein, M.J.; Kuramochi-Miyagawa, S.; Nakano, T.; et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 2006, 442, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Beitzinger, M.; Peters, L.; Zhu, J.Y.; Kremmer, E.; Meister, G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007, 4, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Hafner, M.; Landthaler, M.; Burger, L.; Khorshid, M.; Hausser, J.; Berninger, P.; Rothballer, A.; Ascano, M., Jr.; Jungkamp, A.-C.; Munschauer, M. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010, 141, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Darnell, R.B. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat. Biotechnol. 2011, 29, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.K.; Young, A.G.; Bhutkar, A.; Zheng, G.X.; Bosson, A.D.; Nielsen, C.B.; Sharp, P.A. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat. Struct. Mol. Biol. 2011, 18, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Clark, P.M.; Loher, P.; Quann, K.; Brody, J.; Londin, E.R.; Rigoutsos, I. Argonaute CLIP-Seq reveals miRNA targetome diversity across tissue types. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Pfaff, J.; Hennig, J.; Herzog, F.; Aebersold, R.; Sattler, M.; Niessing, D.; Meister, G. Structural features of Argonaute-GW182 protein interactions. Proc. Natl. Acad. Sci. USA 2013, 110, E3770–E3779. [Google Scholar] [CrossRef] [PubMed]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Yao, B.; Li, S.; Chan, E.K. Function of GW182 and GW bodies in siRNA and miRNA pathways. Adv. Exp. Med. Biol. 2013, 768, 71–96. [Google Scholar] [PubMed]
- Braun, J.E.; Huntzinger, E.; Izaurralde, E. The role of GW182 proteins in miRNA-mediated gene silencing. Adv. Exp. Med. Biol. 2013, 768, 147–163. [Google Scholar] [PubMed]
- Pfaff, J.; Meister, G. Argonaute and GW182 proteins: An effective alliance in gene silencing. Biochem. Soc. Trans. 2013, 41, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Miles, W.O.; Tschop, K.; Herr, A.; Ji, J.Y.; Dyson, N.J. Pumilio facilitates miRNA regulation of the E2F3 oncogene. Genes Dev. 2012, 26, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Weinmann, L.; Hock, J.; Ivacevic, T.; Ohrt, T.; Mutze, J.; Schwille, P.; Kremmer, E.; Benes, V.; Urlaub, H.; Meister, G. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell 2009, 136, 496–507. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Suh, M.R.; Han, J.; Yeom, K.H.; Lee, Y.; Heo, I.; Ha, M.; Hyun, S.; Kim, V.N. Human UPF1 participates in small RNA-induced mRNA downregulation. Mol. Cell. Biol. 2009, 29, 5789–5799. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Meng, S.; Lu, Y.; Trombly, M.I.; Chen, J.; Lin, C.; Turk, A.; Wang, X. Mammalian hyperplastic discs homolog EDD regulates miRNA-mediated gene silencing. Mol. Cell 2011, 43, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Sienski, G.; Batki, J.; Senti, K.A.; Donertas, D.; Tirian, L.; Meixner, K.; Brennecke, J. Silencio/CG9754 connects the Piwi-piRNA complex to the cellular heterochromatin machinery. Genes Dev. 2015, 29, 2258–2271. [Google Scholar] [CrossRef] [PubMed]
- Leonov, G.; Shah, K.; Yee, D.; Timmis, J.; Sharp, T.V.; Lagos, D. Suppression of AGO2 by miR-132 as a determinant of miRNA-mediated silencing in human primary endothelial cells. Int. J. Biochem. Cell Biol. 2015, 69, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Lv, J.; Liu, M.; Tang, H. miR-346 Up-regulates Argonaute 2 (AGO2) protein expression to augment the activity of other microRNAs (miRNAs) and contributes to cervical cancer cell malignancy. J. Biol. Chem. 2015, 290, 30342–30350. [Google Scholar] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.N.; Chao, A.; Baker, J.; Chang, L.; Qiao, D.; Lin, H. A novel class of evolutionarily conserved genes defined by Piwi are essential for stem cell self-renewal. Genes Dev. 1998, 12, 3715–3727. [Google Scholar] [CrossRef] [PubMed]
- Baumberger, N.; Baulcombe, D. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 2005, 102, 11928–11933. [Google Scholar] [CrossRef] [PubMed]
- Mallory, A.C.; Hinze, A.; Tucker, M.R.; Bouche, N.; Gasciolli, V.; Elmayan, T.; Lauressergues, D.; Jauvion, V.; Vaucheret, H.; Laux, T. Redundant and specific roles of the ARGONAUTE proteins AGO1 and ZLL in development and small RNA-directed gene silencing. PLoS Genet. 2009, 5, e1000646. [Google Scholar] [CrossRef] [PubMed]
- Szakmary, A.; Cox, D.N.; Wang, Z.; Lin, H. Regulatory relationship among Piwi, pumilio, and bag-of-marbles in Drosophila germline stem cell self-renewal and differentiation. Curr. Biol. 2005, 15, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Flynt, A.S.; Lai, E.C. Drosophila Piwi mutants exhibit germline stem cell tumors that are sustained by elevated Dpp signaling. Curr. Biol. 2013, 23, 1442–1448. [Google Scholar] [CrossRef] [PubMed]
- Ishizu, H.; Siomi, H.; Siomi, M.C. Biology of Piwi-interacting RNAs: New insights into biogenesis and function inside and outside of germlines. Genes Dev. 2012, 26, 2361–2373. [Google Scholar] [CrossRef] [PubMed]
- Klenov, M.S.; Sokolova, O.A.; Yakushev, E.Y.; Stolyarenko, A.D.; Mikhaleva, E.A.; Lavrov, S.A.; Gvozdev, V.A. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc. Natl. Acad. Sci. USA 2011, 108, 18760–18765. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Wang, S.; Do, T.; Song, X.; Inaba, M.; Nishimoto, Y.; Liu, L.; Gao, Y.; Mao, Y.; Li, H.; et al. Piwi is required in multiple cell types to control germline stem cell lineage development in the Drosophila ovary. PLoS ONE 2014, 9, e90267. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Chen, D.; Duan, R.; Xia, L.; Wang, J.; Qurashi, A.; Jin, P. Argonaute 1 regulates the fate of germline stem cells in Drosophila. Development 2007, 134, 4265–4272. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Vagin, V.V.; Lee, S.; Xu, J.; Ma, S.; Xi, H.; Seitz, H.; Horwich, M.D.; Syrzycka, M.; Honda, B.M.; et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 2009, 137, 509–521. [Google Scholar] [CrossRef] [PubMed]
- Visvader, J.E.; Lindeman, G.J. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat. Rev. Cancer 2008, 8, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.J.; Shen, R.; Chen, L.; Ye, Y.; He, G.; Hua, K.; Jarjoura, D.; Nakano, T.; Ramesh, G.K.; Shapiro, C.L.; et al. Piwil2 is expressed in various stages of breast cancers and has the potential to be used as a novel biomarker. Int. J. Clin. Exp. Pathol. 2010, 3, 328–337. [Google Scholar] [PubMed]
- Shahali, M.; Kabir-Salmani, M.; Nayernia, K.; Soleimanpour-Lichaei, H.R.; Vasei, M.; Mowla, S.J.; Ranaie, E.; Shakibaie, M.; Modaresi, M.H. A novel in vitro model for cancer stem cell culture using ectopically expressed piwil2 stable cell line. Cell J. 2013, 15, 250–257. [Google Scholar] [PubMed]
- Zhang, H.; Ren, Y.; Xu, H.; Pang, D.; Duan, C.; Liu, C. The expression of stem cell protein Piwil2 and piR-932 in breast cancer. Surg. Oncol. 2013, 22, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Litwin, M.; Dubis, J.; Arczynska, K.; Piotrowska, A.; Frydlewicz, A.; Karczewski, M.; Dziegiel, P.; Witkiewicz, W. Correlation of Hiwi and Hili expression with cancer stem cell markers in colorectal cancer. Anticancer Res. 2015, 35, 3317–3324. [Google Scholar] [PubMed]
- Zhao, Y.M.; Zhou, J.M.; Wang, L.R.; He, H.W.; Wang, X.L.; Tao, Z.H.; Sun, H.C.; Wu, W.Z.; Fan, J.; Tang, Z.Y.; et al. Hiwi is associated with prognosis in patients with hepatocellular carcinoma after curative resection. Cancer 2012, 118, 2708–2717. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Gao, Q.; Chen, K.; Xue, X.; Li, M.; Chen, Q.; Zhu, G.; Gao, Y. Hiwi facilitates chemoresistance as a cancer stem cell marker in cervical cancer. Oncol. Rep. 2014, 32, 1853–1860. [Google Scholar] [CrossRef] [PubMed]
- Taubert, H.; Greither, T.; Kaushal, D.; Wurl, P.; Bache, M.; Bartel, F.; Kehlen, A.; Lautenschlager, C.; Harris, L.; Kraemer, K.; et al. Expression of the stem cell self-renewal gene Hiwi and risk of tumour-related death in patients with soft-tissue sarcoma. Oncogene 2007, 26, 1098–1100. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.K.; Nelson, M.C.; Brandt, J.E.; Wessman, M.; Mahmud, N.; Weller, K.P.; Hoffman, R. Human CD34+ stem cells express the Hiwi gene, a human homologue of the Drosophila gene Piwi. Blood 2001, 97, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Nolde, M.J.; Cheng, E.C.; Guo, S.; Lin, H. Piwi genes are dispensable for normal hematopoiesis in mice. PLoS ONE 2013, 8, e71950. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Zhang, Y.; Hao, L.; Wang, F.; Liu, D.; Su, Y.; Sun, H. More insight into mesenchymal stem cells and their effects inside the body. Expert Opin. Biol. Ther. 2010, 10, 215–230. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Ma, Q.; Shehadeh, L.A.; Wilson, A.; Xia, L.; Yu, H.; Webster, K.A. Expression of the Argonaute protein PiwiL2 and piRNAs in adult mouse mesenchymal stem cells. Biochem. Biophys. Res. Commun. 2010, 396, 915–920. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Lin, H. Miwi, a murine homolog of Piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell 2002, 2, 819–830. [Google Scholar] [CrossRef]
- Aravin, A.A.; Sachidanandam, R.; Bourc’his, D.; Schaefer, C.; Pezic, D.; Toth, K.F.; Bestor, T.; Hannon, G.J. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 2008, 31, 785–799. [Google Scholar] [CrossRef] [PubMed]
- Cheng, E.C.; Kang, D.; Wang, Z.; Lin, H. Piwi proteins are dispensable for mouse somatic development and reprogramming of fibroblasts into pluripotent stem cells. PLoS ONE 2014, 9, e97821. [Google Scholar] [CrossRef] [PubMed]
- Shekar, P.C.; Naim, A.; Sarathi, D.P.; Kumar, S. Argonaute-2-null embryonic stem cells are retarded in self-renewal and differentiation. J. Biosci. 2011, 36, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Kaneda, M.; Tang, F.; O’Carroll, D.; Lao, K.; Surani, M.A. Essential role for Argonaute2 protein in mouse oogenesis. Epigenet. Chromatin. 2009, 2. [Google Scholar] [CrossRef] [PubMed]
- Stein, P.; Rozhkov, N.V.; Li, F.; Cardenas, F.L.; Davydenko, O.; Vandivier, L.E.; Gregory, B.D.; Hannon, G.J.; Schultz, R.M. Essential Role for endogenous siRNAs during meiosis in mouse oocytes. PLoS Genet. 2015, 11, e1005013. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, G.; Calhoun, G.; Schedl, P. Drosophila argonaute-2 is required early in embryogenesis for the assembly of centric/centromeric heterochromatin, nuclear division, nuclear migration, and germ-cell formation. Genes Dev. 2005, 19, 1680–1685. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhai, L.; Wang, L.; Teng, F.; Zhou, L.; Zhang, W.; Xiao, J.; Liu, Y.; Deng, W. Argonaute and Argonaute-Bound Small RNAs in Stem Cells. Int. J. Mol. Sci. 2016, 17, 208. https://doi.org/10.3390/ijms17020208
Zhai L, Wang L, Teng F, Zhou L, Zhang W, Xiao J, Liu Y, Deng W. Argonaute and Argonaute-Bound Small RNAs in Stem Cells. International Journal of Molecular Sciences. 2016; 17(2):208. https://doi.org/10.3390/ijms17020208
Chicago/Turabian StyleZhai, Lihong, Lin Wang, Feng Teng, Lanting Zhou, Wenjing Zhang, Juan Xiao, Ying Liu, and Wenbin Deng. 2016. "Argonaute and Argonaute-Bound Small RNAs in Stem Cells" International Journal of Molecular Sciences 17, no. 2: 208. https://doi.org/10.3390/ijms17020208
APA StyleZhai, L., Wang, L., Teng, F., Zhou, L., Zhang, W., Xiao, J., Liu, Y., & Deng, W. (2016). Argonaute and Argonaute-Bound Small RNAs in Stem Cells. International Journal of Molecular Sciences, 17(2), 208. https://doi.org/10.3390/ijms17020208