Importance of Heat and Pressure for Solubilization of Recombinant Spider Silk Proteins in Aqueous Solution
Abstract
:1. Introduction
2. Diversity of Proteins and Fibers
3. Salt Influences Solubility
4. Solubilization without Microwaves
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Vollrath, F.; Knight, D.P. Liquid Crystalline Spinning of Spider Silk. Nature 2001, 410, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Dicko, C.; Kenney, J.; Knight, D.; Vollrath, F. Transition to a β-Sheet-Rich Structure in Spidroin In Vitro: The Effects of pH and Cations. Biochemistry 2004, 43, 14080–14087. [Google Scholar] [CrossRef] [PubMed]
- Eisoldt, L.; Thamm, C.; Scheibel, T. The Role of Terminal Domains during Storage and Assembly of Spider Silk Proteins. Biopolymers 2012, 97, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, C.Y.; Blackledge, T.A.; Lewis, R.V. Molecular and Mechanical Characterization of Aciniform Silk: Uniformity of Iterated Sequence Modules in a Novel Member of the Spider Silk Fibroin Gene Family. Mol. Biol. Evol. 2004, 21, 1950–1959. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Lewis, R.V. Structure of a Protein Superfiber: Spider Dragline Silk. Proc. Natl. Acad. Sci. USA 1990, 87, 7120–7124. [Google Scholar] [CrossRef] [PubMed]
- Hinman, M.B.; Lewis, R.V. Isolation of a Clone Encoding a Second Dragline Silk Fibroin. Nephila clavipes Dragline Silk Is a Two-Protein Fiber. J. Biol. Chem. 1992, 267, 19320–19324. [Google Scholar] [PubMed]
- Hayashi, C.Y.; Lewis, R.V. Spider Flagelliform Silk: Lessons in Protein Design, Gene Structure, and Molecular Evolution. BioEssays 2001, 23, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Perry, D.J.; Bittencourt, D.; Siltberg-Liberles, J.; Rech, E.L.; Lewis, R.V. Piriform Spider Silk Sequences Reveal Unique Repetitive Elements. Biomacromolecules 2010, 11, 3000–3006. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Lewis, R.V. Tubuliform Silk Protein: A Protein with Unique Molecular Characteristics and Mechanical Properties in the Spider Silk Fibroin Family. Appl. Phys. A 2006, 82, 265–273. [Google Scholar] [CrossRef]
- Stauffer, S.L.; Coguill, S.L.; Lewis, R.V. Comparison of Physical Properties of Three Silks from Nephila clavipes and Araneus gemmoides. J. Arachnol. 1994, 22, 5–11. [Google Scholar]
- Hayashi, C.Y.; Shipley, N.H.; Lewis, R.V. Hypotheses That Correlate the Sequence, Structure, and Mechanical Properties of Spider Silk Proteins. Int. J. Biol. Macromol. 1999, 24, 271–275. [Google Scholar] [CrossRef]
- Adrianos, S.L.; Teulé, F.; Hinman, M.B.; Jones, J.A.; Weber, W.S.; Yarger, J.L.; Lewis, R.V. Nephila clavipes Flagelliform Silk-Like GGX Motifs Contribute to Extensibility and Spacer Motifs Contribute to Strength in Synthetic Spider Silk Fibers. Biomacromolecules 2013, 14, 1751–1760. [Google Scholar] [CrossRef] [PubMed]
- Guinea, G.V.; Elices, M.; Plaza, G.R.; Perea, G.B.; Daza, R.; Riekel, C.; Agulló-Rueda, F.; Hayashi, C.; Zhao, Y.; Pérez-Rigueiro, J. Minor Ampullate Silks from Nephila and Argiope Spiders: Tensile Properties and Microstructural Characterization. Biomacromolecules 2012, 13, 2087–2098. [Google Scholar] [CrossRef] [PubMed]
- Colgin, M.A.; Lewis, R.V. Spider Minor Ampullate Silk Proteins Contain New Repetitive Sequences and Highly Conserved Non-Silk-Like “Spacer Regions”. Protein Sci. 1998, 7, 667–672. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Liu, X.; Zhang, Y.; Lin, S.; Yang, Z.; Johansson, J.; Rising, A.; Meng, Q. Full-Length Minor Ampullate Spidroin Gene Sequence. PLoS ONE 2012, 7, e52293. [Google Scholar] [CrossRef] [PubMed]
- Lefèvre, T.; Paquet-Mercier, F.; Rioux-Dubé, J.; Pézolet, M. Structure of Silk by Raman Spectromicroscopy: From the Spinning Glands to the Fibers. Biopolymers 2012, 97, 322–336. [Google Scholar] [CrossRef] [PubMed]
- Motriuk-Smith, D.; Smith, A.; Hayashi, C.Y.; Lewis, R.V. Analysis of the Conserved N-terminal Domains in Major Ampullate Spider Silk Proteins. Biomacromolecules 2005, 6, 3152–3159. [Google Scholar] [CrossRef] [PubMed]
- Hagn, F.; Eisoldt, L.; Hardy, J.G.; Vendrely, C.; Coles, M.; Scheibel, T.; Kessler, H. A Conserved Spider Silk Domain Acts as a Molecular Switch That Controls Fibre Assembly. Nature 2010, 465, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Fahnestock, S.R.; Bedzyk, L.A. Production of Synthetic Spider Dragline Silk Protein in Pichia pastoris. Appl. Microbiol. Biotechnol. 1997, 47, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Barr, L.A.; Fahnestock, S.R.; Liu, Z. High Yield Recombinant Silk-like Protein Production in Transgenic Plants through Protein Targeting. Transgenic Res. 2005, 14, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Scheller, J.; Gührs, K.; Grosse, F.; Conrad, U. Production of Spider Silk Proteins in Tobacco and Potato. Nat. Biotechnol. 2001, 19, 573–577. [Google Scholar] [CrossRef] [PubMed]
- Barr, L.A.; Fahnestock, S.R.; Yang, J. Production and Purification of Recombinant DP1B Silk-like Protein in Plants. Mol. Breed. 2004, 13, 345–356. [Google Scholar] [CrossRef]
- Lazaris, A.; Arcidiacono, S.; Huang, Y.; Zhou, J.; Duguay, F.; Chretien, N.; Welsh, E.A.; Soares, J.W.; Karatzas, C.N. Spider Silk Fibers Spun from Soluble Recombinant Silk Produced in Mammalian Cells. Science 2002, 295, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.V.; Hinman, M.; Kothakota, S.; Fournier, M.J. Expression and Purification of a Spider Silk Protein: A New Strategy for Producing Repetitive Proteins. Protein Expr. Purif. 1996, 7, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Teulé, F.; Cooper, A.R.; Furin, W.A.; Bittencourt, D.; Rech, E.L.; Brooks, A.; Lewis, R.V. A Protocol for the Production of Recombinant Spider Silk-like Proteins for Artificial Fiber Spinning. Nat. Protoc. 2009, 4, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Karatzas, C.N.; Turner, J.D.; Lazaris-Karatzas, A. Production of Biofilaments in Transgenic Animals. U.S. Patent 7157615, 27 June 2001. [Google Scholar]
- Huang, Y.; Karatzas, C.N.; Turcotte, C. Recovery of Biofilament Proteins from Biological Fluids. Patent WO2003057720 A2, 17 July 2003. [Google Scholar]
- Xia, X.-X.; Qian, Z.; Ki, C.S.; Park, Y.H.; Kaplan, D.L.; Lee, S.Y. Native-Sized Recombinant Spider Silk Protein Produced in Metabolically Engineered Escherichia coli Results in a Strong Fiber. Proc. Natl. Acad. Sci. USA 2010, 107, 14059–14063. [Google Scholar] [CrossRef] [PubMed]
- Tucker, C.L.; Jones, J.A.; Bringhurst, H.N.; Copeland, C.G.; Addison, J.B.; Weber, W.S.; Mou, Q.; Yarger, J.L.; Lewis, R.V. Mechanical and Physical Properties of Recombinant Spider Silk Films Using Organic and Aqueous Solvents. Biomacromolecules 2014, 15, 3158–3170. [Google Scholar] [CrossRef] [PubMed]
- Scheibel, T. Spider Silks: Recombinant Synthesis, Assembly, Spinning, and Engineering of Synthetic Proteins. Microb. Cell Fact. 2004, 3, 14. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.A.; Harris, T.I.; Tucker, C.L.; Berg, K.R.; Christy, S.Y.; Day, B.A.; Gaztambide, D.A.; Needham, N.J.; Ruben, A.L.; Oliveira, P.F.; et al. More than Just Fibers: An Aqueous Method for the Production of Innovative Recombinant Spider Silk Protein Materials. Biomacromolecules 2015, 16, 1418–1425. [Google Scholar] [CrossRef] [PubMed]
- Teule, F.; Furin, W.A.; Cooper, A.R.; Duncan, J.R.; Lewis, R.V. Modifications of Spider Silk Sequences in an Attempt to Control the Mechanical Properties of the Synthetic Fibers. J. Mater. Sci. 2007, 42, 8974–8985. [Google Scholar] [CrossRef]
- Seidel, A.; Liivak, O.; Jelinski, L.W. Artificial Spinning of Spider Silk. Macromolecules 1998, 31, 6733–6736. [Google Scholar] [CrossRef]
- Doblhofer, E.; Heidebrecht, A.; Scheibel, T. To Spin or Not to Spin: Spider Silk Fibers and More. Appl. Microbiol. Biotechnol. 2015, 99, 9361–9380. [Google Scholar] [CrossRef] [PubMed]
- Heim, M.; Keerl, D.; Scheibel, T. Spider Silk: From Soluble Protein to Extraordinary Fiber. Angew. Chem. Int. Ed. 2009, 48, 3584–3596. [Google Scholar] [CrossRef] [PubMed]
- Bogush, V.G.; Sokolova, O.S.; Davydova, L.I.; Klinov, D.V.; Sidoruk, K.V.; Esipova, N.G.; Neretina, T.V.; Orchanskyi, I.A.; Makeev, V.Y.; Tumanyan, V.G.; et al. A Novel Model System for Design of Biomaterials Based on Recombinant Analogs of Spider Silk Proteins. J. Neuroimmune Pharmacol. 2009, 4, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Copeland, C.G.; Bell, B.E.; Christensen, C.D.; Lewis, R.V. Development of a Process for the Spinning of Synthetic Spider Silk. ACS Biomater. Sci. Eng. 2015, 1, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Heidebrecht, A.; Eisoldt, L.; Diehl, J.; Schmidt, A.; Geffers, M.; Lang, G.; Scheibel, T. Biomimetic Fibers Made of Recombinant Spidroins with the Same Toughness as Natural Spider Silk. Adv. Mater. 2015, 27, 2189–2194. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Guo, C.; Holland, G.P. Probing the Impact of Acidification on Spider Silk Assembly Kinetics. Biomacromolecules 2015, 16, 2072–2079. [Google Scholar] [CrossRef] [PubMed]
- Harris, T.I.; Gaztambide, D.A.; Day, B.A.; Brock, C.L.; Ruben, A.L.; Jones, J.A.; Lewis, R.V. A Sticky Situation: An Investigation of Robust Aqueous-Based Recombinant Spider Silk Protein Coatings and Adhesives. Biomacromolecules 2016. [Google Scholar] [CrossRef] [PubMed]
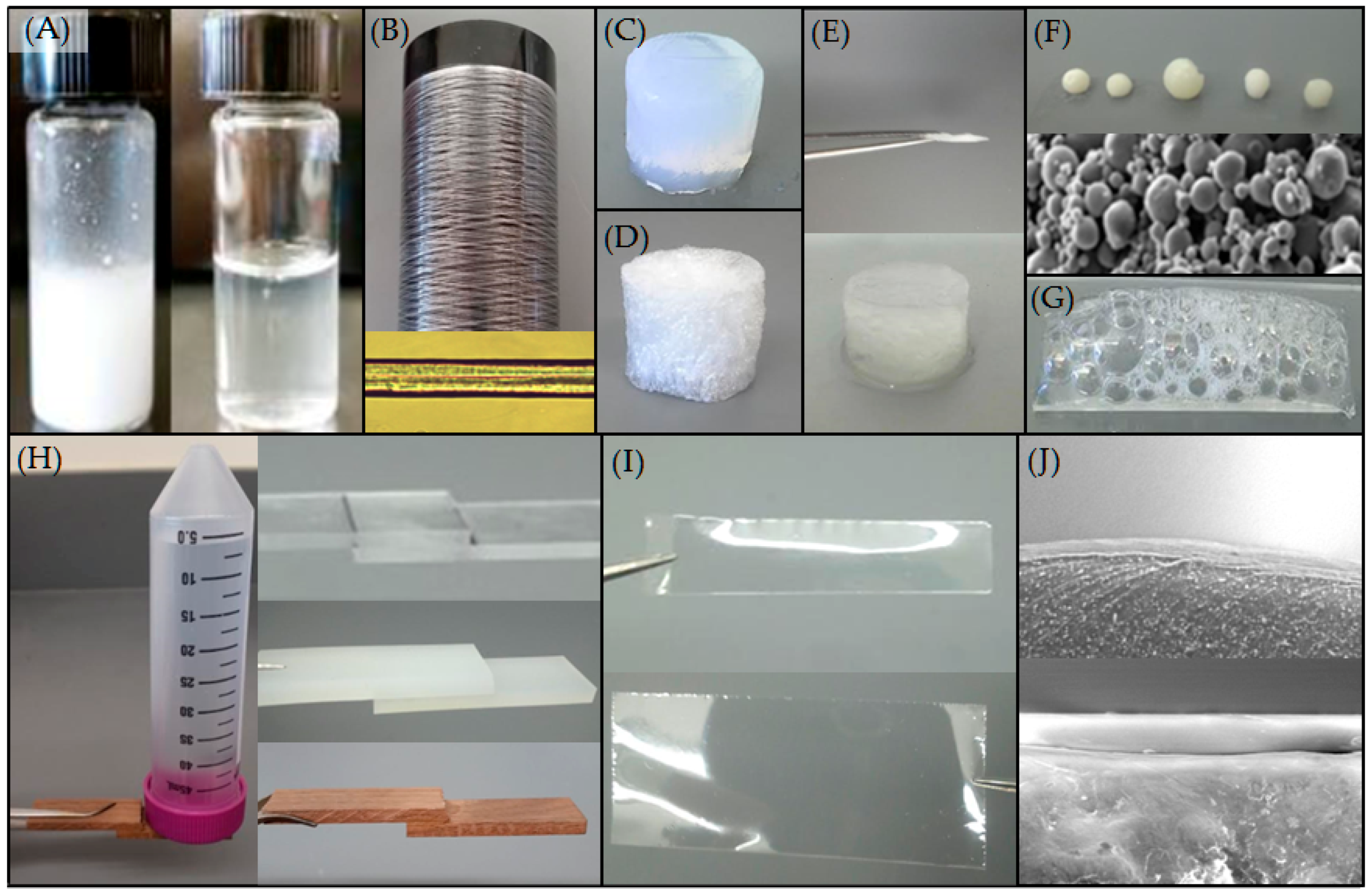
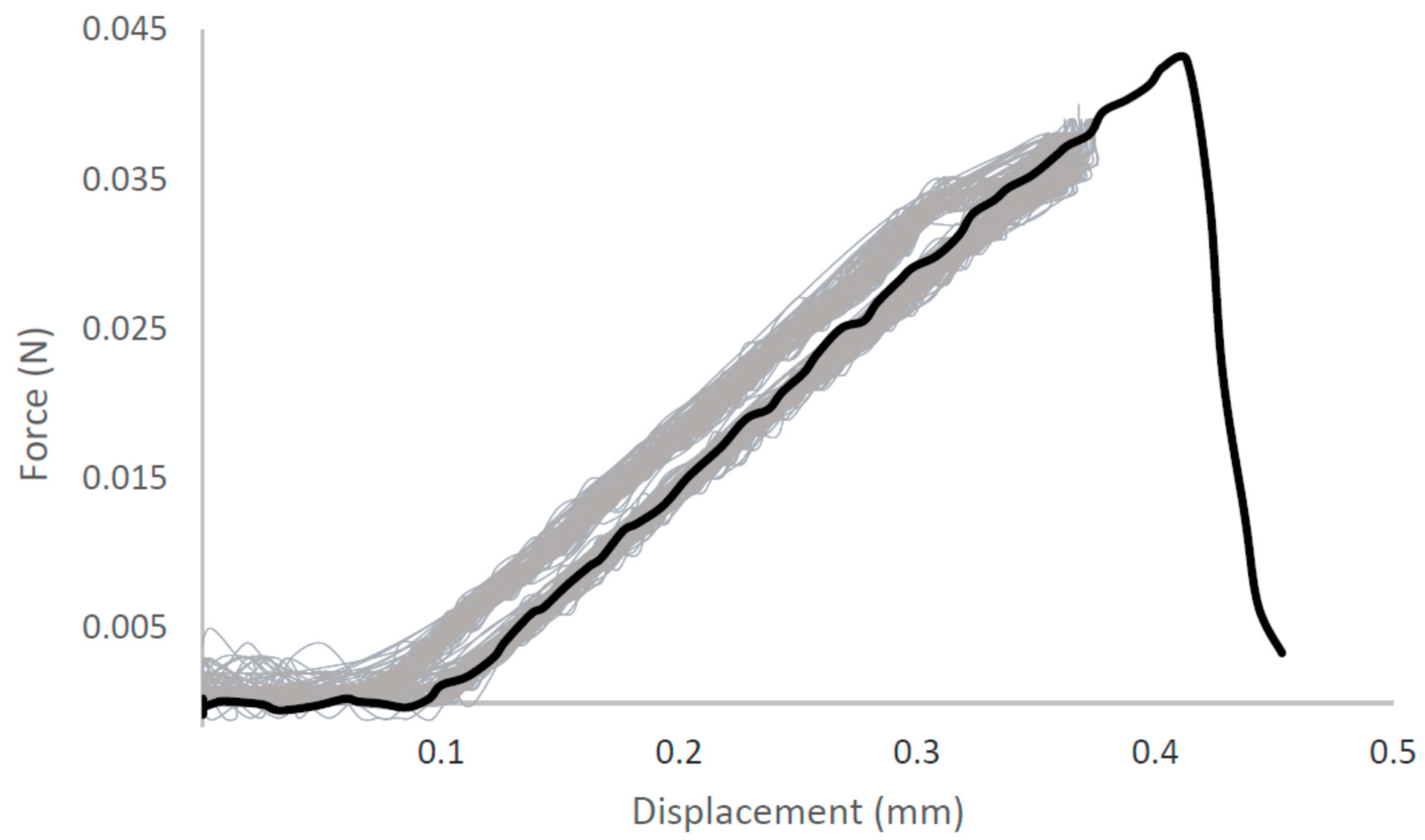
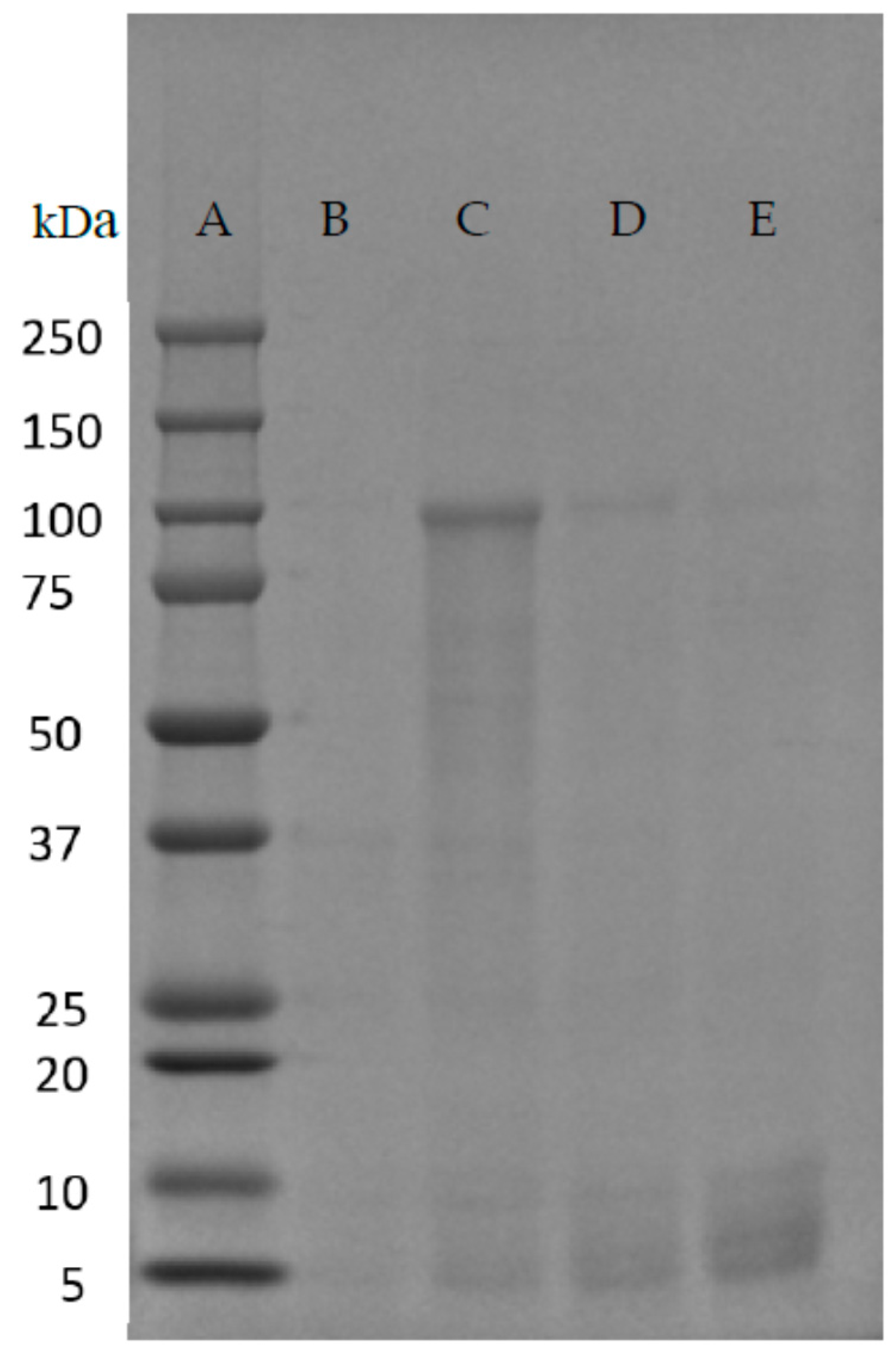
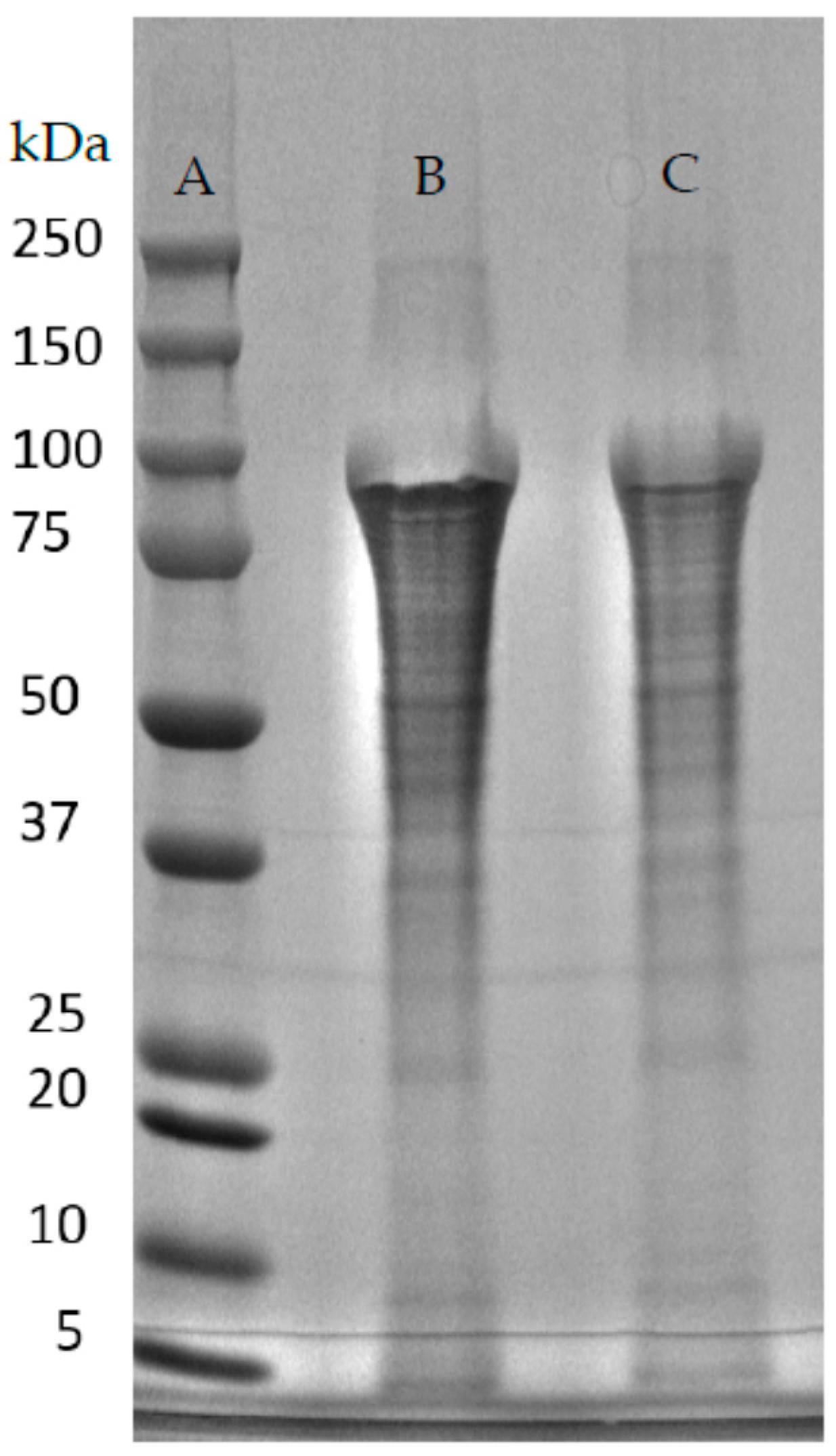
| Native-Like Sequences (Host Organism and Construct) |
|---|
| C-Term: |
| SRLSSPQASSRLSSAVSNLVATGPTNSAALSSTISNVVSQIGASNPGLSGCDVLIQALLEVVSALIQILGSSSIGQVNYGSAGQATQIVGQSVYQALG |
| Goat MaSp1 with C-Term: |
| QGAGAAAAAAGGAGQGGYGGLGGQGAGQGGYGGLGGQGAGQGAGAAAAAAAGGAGQGGYGGLGSQGAGRGGQGAGAAAAAAGGAGQGGYGGLGSQGAGRGGLGGQGAGAAAAAAAGGAGQGGYGGLGNQGAGRGGQGAAAAAAGGAGQGGYGGLGSQGAGRGGLGGQGAGAAAAAAGGAGQGGYGGLGGQGAGQGGYGGLGSQGAGRGGLGGQGAGAAAAAAAGGAGQGGLGGQGAGQGAGASAAAAGGAGQGGYGGLGSQGAGRGGEGAGAAAAAAGGAGQGGYGGLGGQGAGQGGYGGLGSQGAGRGGLGGQGAGAAAAGGAGQGGLGGQGAGQGAGAAAAAAGGAGQGGYGGLGSQGAGRGGLGGQGAGAVAAAAAGGAGQGGYGGLGSQGAGRGGQGAGAAAAAAGGAGQRGYGGLGNQGAGRGGLGGQGAGAAAAAAAGGAGQGGYGGLGNQGAGRGGQGAAAAAGGAGQGGYGGLGSQGAGRGGQGAGAAAAAAVGAGQEGIRGQGAGQGGYGGLGSQGSGRGGLGGQGAGAAAAAAGGAGQGGLGGQGAGQGAGAAAAAAGGVRQGGYGGLGSQGAGRGGQGAGAAAAAAGGAGQGGYGGLGGQGVGRGGLGGQGAGAAAAGGAGQGGYGGVGSGASAASAAASRLSSPQASSRLSSAVSNLVATGPTNSAALSSTISNVVSQIGASNPGLSGCDVLIQALLEVVSALIQILGSSSIGQVNYGSAGQATQIVGQSVYQALG |
| Goat MaSp2 with C-term: |
| PGGYGPGQQGPGGYGPGQQGPSGPGSAAAAAAAAAAGPGGYGPGQQGPGGYGPGQQGPGRYGPGQQGPSGPGSAAAAAAGSGQQGPGGYGPRQQGPGGYGQGQQGPSGPGSAAAASAAASAESGQQGPGGYGPGQQGPGGYGPGQQGPGGYGPGQQGPSGPGSAAAAAAAASGPGQQGPGGYGPGQQGPGGYGPGQQGPSGPGSAAAAAAAASGPGQQGPGGYGPGQQGPGGYGPGQQGLSGPGSAAAAAAAGPGQQGPGGYGPGQQGPSGPGSAAAAAAAAAGPGGYGPGQQGPGGYGPGQQGPSGAGSAAAAAAAGPGQQGLGGYGPGQQGPGGYGPGQQGPGGYGPGSASAAAAAAGPGQQGPGGYGPGQQGPSGPGSASAAAAAAAAGPGGYGPGQQGPGGYAPGQQGPSGPGSASAAAAAAAAGPGGYGPGQQGPGGYAPGQQGPSGPGSAAAAAAAAAGPGGYGPAQQGPSGPGIAASAASAGPGGYGPAQQGPAGYGPGSAVAASAGAGSAGYGPGSQASAAASRLASPDSGARVASAVSNLVSSGPTSSAALSSVISNAVSQIGASNPGLSGCDVLIQALLEIVSACVTILSSSSIGQVNYGAASQFAQVVGQSVLSAF |
| E. coli MaSp1 with and without C-term: |
| (GAGQGGYGGLGSQGAGRGGLGGQGAGAAAAAAAAGGAGQGGYGGLGSQGAGRGGLGGQGAGAAAAAAAAAGQGGYGGLGSQGGGAGQGGYGGLGSQGAGRGGLGGQGAGAAAAAAAAGGAGQGGYGGLGSQGAGRGGLGGQGAGAAAAAAAAGGAGQGGYGGLGSQGAGRGGLGGQGAGAAAAAAAAGGAGQGGYGGLGSQGAGRGGLGGQGAGAAAAAAAAAGQGGYGGLGSQGGGAGQGGYGGLGSQGAGRGGLGGQGAGAAAAAAAAGGAGQGGYGGLGSQGAGRGGLGGQGAGAAAAAAAA)2 |
| E. coli MaSp2 with and without C-term: |
| (GGPGQQGPGGYGPGQQGPSGPGSAAAAAAAAGPGQQGPGGYGPGQQGPGGYGPGQQGPSGPGSAAAAAAAAGPGGYGPGQQGPGGYGPGQQGPGGYGPGQQGPSGPGSAAAAAAAAGPGQQGPGGYGPGQQGPGGYGPGQQGPSGPGSAAAAAAAAGPGQQGPGGYGPGQQGPSGPGSAAAAAAAAGPGQQGPGGYGPGQQGPGGYGPGQQGPSGPGSAAAAAAAAGPGGYGPGQQGPGGYGPGQQGPGGYGPGQQGPSGPGSAAAAAAAAGPGQQGPGGYGPGQQGPGGYGPGQQGPSGPGSAAAAAAAAGPGQQGPGGYGPGQQGPSGPGSAAAAAAAA)4 |
| E. coli Piriform with and without C-term: |
| (VSQVQQASIQQAQSSSAQSRQSSVAQQASISQSQQASVSQSQQASVSQSQQASVSQSQQSSNAYSAASNAASSVSQASSDSSYFNSQVVQSALSSSLQSSSALSSIAYGQTSANINDVAAAVARSVSQSLGVSQQAAQSVISQQLASAGSGASAQTLAQLISSAVSSLVQQSGTVSAGQEQSISQSLSSSILSSLSQVVAQRPLPVPAPRPLPAPLPAPRPIPAPLPRPVPI)4 |
| Chimeric Sequences (Host Organism and Construct) |
| E. coli FlAS3 with and without C-term: |
| (GPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPSGPGSAAAAAAAAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPSGPGSAAAAAAAAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPSGPGSAAAAAAAAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPSGPGSAAAAAAAAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPGGAGPSGPGSAAAAAAAA)3 |
| E. coli chimeric A4S8 with and without C-term: |
| (GGAGPGGAGPGGAGPGGAGP4GGPSGPGSAAAAAAAAGP8)8 |
| Temperature | Max Pressure | Time to Target Temperature/Max Pressure |
|---|---|---|
| 120 °C | 127 psi | 16 min |
| 130 °C | 131 psi | 16 min |
| 135 °C | 140 psi | 17 min |
| 140 °C | 155 psi | 20 min |
| 150 °C | 161 psi | 25 min |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jones, J.A.; Harris, T.I.; Oliveira, P.F.; Bell, B.E.; Alhabib, A.; Lewis, R.V. Importance of Heat and Pressure for Solubilization of Recombinant Spider Silk Proteins in Aqueous Solution. Int. J. Mol. Sci. 2016, 17, 1955. https://doi.org/10.3390/ijms17111955
Jones JA, Harris TI, Oliveira PF, Bell BE, Alhabib A, Lewis RV. Importance of Heat and Pressure for Solubilization of Recombinant Spider Silk Proteins in Aqueous Solution. International Journal of Molecular Sciences. 2016; 17(11):1955. https://doi.org/10.3390/ijms17111955
Chicago/Turabian StyleJones, Justin A., Thomas I. Harris, Paula F. Oliveira, Brianne E. Bell, Abdulrahman Alhabib, and Randolph V. Lewis. 2016. "Importance of Heat and Pressure for Solubilization of Recombinant Spider Silk Proteins in Aqueous Solution" International Journal of Molecular Sciences 17, no. 11: 1955. https://doi.org/10.3390/ijms17111955
APA StyleJones, J. A., Harris, T. I., Oliveira, P. F., Bell, B. E., Alhabib, A., & Lewis, R. V. (2016). Importance of Heat and Pressure for Solubilization of Recombinant Spider Silk Proteins in Aqueous Solution. International Journal of Molecular Sciences, 17(11), 1955. https://doi.org/10.3390/ijms17111955







