Abstract
All living organisms synchronize biological functions with environmental changes; melatonin plays a vital role in regulating daily and seasonal variations. Due to rhythmic activity of the timezyme aralkylamine N-acetyltransferase (AANAT), the blood level of melatonin increases at night and decreases during daytime. Whereas other vertebrates have a single form of AANAT, bony fishes possess various isoforms of aanat genes, though the reasons are still unclear. Here, we have taken advantage of multiple unpublished teleost aanat sequences to explore and expand our understanding of the molecular evolution of aanat in fish. Our results confirm that two rounds of whole-genome duplication (WGD) led to the existence of three fish isoforms of aanat, i.e., aanat1a, aanat1b, and aanat2; in addition, gene loss led to the absence of some forms from certain special fish species. Furthermore, we suggest the different roles of two aanat1s in amphibious mudskippers, and speculate that the loss of aanat1a, may be related to terrestrial vision change. Several important sites of AANAT proteins and regulatory elements of aanat genes were analyzed for structural comparison and functional forecasting, respectively, which provides insights into the molecular evolution of the differences between AANAT1 and AANAT2.
1. Introduction
Light stimulus is a key factor for maintaining the physiological balance of fish. The day/night variation may exert an influence over their entire lifetime. Several organs participate in receiving external light signals. In all vertebrates, the retina is the main photoreceptive organ for reception of photic input [1]. In contrast to mammals, the pineal gland of lower vertebrates, such as fish, can also detect changes in environmental light levels. The primary function of the pineal gland is to produce and release melatonin in response to circadian and lighting stimulation. Circulating melatonin levels are much higher at night than during daytime in essentially all vertebrates. The difference in blood melatonin levels between daytime and night provides information to the rest of the organism regarding the time of day and duration of the night period. Melatonin can regulate the endocrine activities through binding to its receptors throughout the fish body [2], hence it is considered a key component of the circadian system. For instance, we demonstrated that melatonin regulates reproduction in blue-spotted mudskipper (Boleophthalmus pectinirostris, a semilunar spawning fish) by combining with receptors of the hypothalamic-pituitary-gonad (HPG) axis or directly acting on gonadal tissues [3]. Melatonin can also influence the appetite of fish because of melatonin binding sites in gastrointestinal tract [4,5,6]. Melatonin is thought to influence wide-ranging physiologies including reproduction, sleep, the immune system, and so on [7,8,9,10].
Melatonin is synthesized from tryptophan (Trp) through four enzyme-catalyzed reactions [11]. The first step converts Trp to 5-hydroxytryptophan (5-HTrp) by tryptophan hydroxylase (TPH, EC 1.14.16.4). The subsequent reaction converts 5-HTrp to 5-hydroxytryptamine (5-HT, serotonin) through the catalysis by of aromatic-l-amino-acid decarboxylase (AAAD, EC 4.1.1.28). The third product is N-acetylserotonin (NAS), formed by acetylation of 5-HT by aralkylamine N-acetyltransferase (AANAT, EC 2.3.1.87). The last reaction is catalyzed by acetylserotonin-O-methyltransferase (ASMT, EC 2.1.1.4) to generate melatonin.
AANAT, also known as serotonin N-acetyltransferase (SNAT), is the penultimate enzyme in the synthesis of melatonin [12,13]. As a result of catalysis by AANAT, 5-HT receives an acetyl group from acetyl CoA (AcCoA) to become NAS. The large daily rhythms in melatonin production in the pineal gland are driven by changes in AANAT activity; these changes may be due to both regulation at the transcriptional level and through post-translational modifications. The last enzyme in melatonin synthesis, ASMT, usually shows little day/night variation, but may only provide an upper limit, or cap, on the maximum amount of melatonin produced. Thus, to better understand control of melatonin synthesis in fish, we sought to explore the evolution of aanat and to explain the reason for the existence of multiple aanat isoforms in fish.
The vertebrate-type of AANAT evolved from a more-primitive non-vertebrate-type of AANAT [14]. The primitive AANAT is widely-known from organisms evolutionarily more ancient than Agnathans, including amphioxus [15]. During early vertebrate evolution, this gene was duplicated and one of the paralogs apparently diverged to become the vertebrate-type AANAT. The non-vertebrate-type was lost from almost all vertebrate lineages, but the two forms coexist in certain chondrichthyes (specifically the elephant shark, Callorhinchus milii [14]). In teleosts and tetrapods, only the vertebrate-type exists, and the multiple forms of this gene are the subject of this report. Prior to the divergence of Gnathostomes, the vertebrate-type AANAT was duplicated, but only one of these duplicates was retained in the tetrapods. In teleosts, the two isoforms were retained and are called aanat1 and aanat2. Subsequently aanat1 was again duplicated to yield aanat1a and aanat1b. Throughout teleost evolution, the timing of these duplication, and selective losses of isoforms, have influenced regulation of melatonin production. Additionally, numerous teleost species are tetraploid, adding further complexity.
In addition to the presence or absence of aanat isoforms, levels of expression can influence melatonin production. Several important DNA regulatory elements can influence the expression of aanat. Specifically, photoreceptor conserved element (PCE, TAATT/C) is the most common regulatory element of aanat. In zebrafish, PCE plays a role in pineal-specific expression of aanat2 [16]. Also, E-box (CACGTG) and E-box-like element (CATGTG or TACGTG) can strengthen pineal-specific expression of aanat2 and suppress non-pineal gland expression [17,18]. Meanwhile, some circadian genes can also direct regulation through the E-box [19]. The cis-acting regulatory element (CAATC), which is also called pineal expression-promoting element (PIPE), directs pineal-specific gene expression in zebrafish [20].
In general, the synthesis of melatonin in fish depends on output from the circadian clock and input from external signals (Figure 1), primarily through effects on AANAT activity [21,22]. The circadian clock directly drives transcription of aanat2 at night. The translated AANAT2 is phosphorylated by cyclic-AMP-dependent activation of protein kinase A (PKA). Phosphorylated AANAT2 (pAANAT2) binds to 14-3-3 proteins which protects it from proteasomal proteolysis. This results in elevated AANAT2 activity and rapid melatonin synthesis. External input, for instance light during the daytime, decreases the cyclic-AMP-dependent activation of protein kinase A (PKA) as well as cellular calcium levels; both effects lead to decreased phosphorylation of AANAT and subsequent destruction by the proteasome. This decreases melatonin production [23].
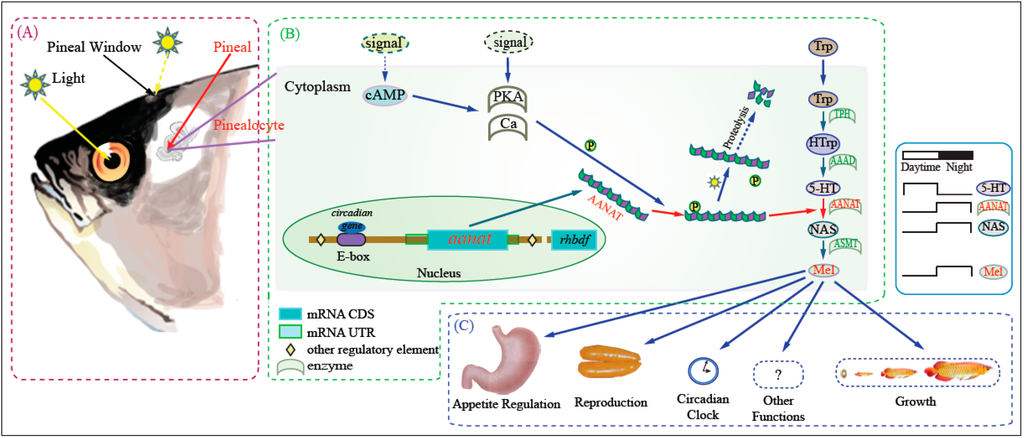
Figure 1.
Outline of melatonin biosynthesis and function. Amphibians, reptiles, and fishes can detect light by photoreceptor cells of the retinae and pinealocytes of the pineal glands. (A) Photic detecting and transforming system includes the retina photoreceptor cells and the pinealocytes; (B) Melatonin synthesis pathway is summarized in the frame; (C) Melatonin performs its various biological functions in different tissues and at different growth/development phases through binding to its receptors. In mammals the photoreceptive role of the pineal gland is lost.
Melatonin is transported through the circulatory system and binds to melatonin receptors in different organs (for example ovary) to exert its effects. Thus, melatonin, synthesized in the pineal gland, plays an important endocrine role in regulating physiological activities [24]. However, melatonin secreted by the retina may play a paracrine role [25], and aanat1 may acetylate retinal dopamine [25,26,27]. Furthermore, aanat gene expression is not restricted to the retina and the pineal gland. In some organisms, like goldfish, low levels of AANAT expression can also be detected in other organs, like skin, the gastrointestinal tract and brain [28,29].
With development of the next-generation sequencing technologies, the entire genomic sequences of many fish species have become publicly available. In addition, we have access to, and include here, many unpublished fish aanat gene and protein sequences, including amphibious teleost fish (mudskipper), cave-restricted or eyeless fish (Sinocyclocheilus anshuiensis), tetraploid fishes (Amazon molly and Sinocyclocheilus spp.) and migratory fishes (rainbow trout and salmon), which have been derived from whole genome sequencing. This allows us to determine the presence or absence of aanat isoforms, as well as sequence differences (both coding and non-coding) across species. In this report we have analyzed the regulatory regions of aanat genes, compared the protein sequence differences in three AANAT isoforms (AANAT1a, AANAT1b, and AANAT2), constructed phylogenetic trees and completed synteny analysis. Finally, we discussed the molecular and functional evolution of AANAT in fish.
2. Results
2.1. Variation of Aanat Copy Number in Different Species
A total of 84 aanat sequences were derived from 37 vertebrate species; these sequences and their encoded proteins were used for our data analysis. Sequences for 25 species, including seven mammal species, one bird species, two reptile species, one amphibian species and 14 fish species, were downloaded from the NCBI and Ensembl databases (related accession numbers are given in Table 1). Sequences extracted from another 12 fish species whose unpublished genomes were sequenced by us, or our collaborators, were included (see details in Table 2).

Table 1.
The accession numbers for known aanat sequences.
| Class | Common Name | Species Name | aanat | Location | Accession Number | Source | |
|---|---|---|---|---|---|---|---|
| Nucleotide Sequence | Protein Sequence | ||||||
| Mammalis | Human | Homo sapiens | 1 | chromosome: 17 | NP_001160051.1 | EAW89400.1 | NCBI |
| House mouse | Mus musculus | 1 | chromosome: 11 | NM_009591.3 | EDL34585.1 | NCBI | |
| Chimpanzee | Pan troglodytes | 1 | chromosome: 17 | AY665273.2 | AAV74311.2 | NCBI | |
| Sheep | Ovis aries | 1 | chromosome: 11 | NM_001009461.1 | NP_001009461.1 | NCBI | |
| Minke whale | Balaenoptera acutorostrata scammoni | 1 | chromosome: Un | XM_007185968.1 | XP_007186030.1 | NCBI | |
| Platypus | Ornithorhynchus anatinus | 1 | Ultra430:237283–238471: −1 | ENSOANG00000011565 | ENSOANT00000018326 | Ensembl | |
| Panda | Ailuropoda melanoleuca | 1 | GL192703.1:1467291–1468587: 1 | ENSAMEG00000010321 | ENSAMET00000011309 | Ensembl | |
| Aves Reptilia | Rock pigeon | Columba livia | 1 | chromosome: Un | XM_005503150 | XP_005503207.1 | NCBI |
| Green sea turtle | Chelonia mydas | 1 | chromosome: Un | XM_007072534.1 | EMP23774 | NCBI | |
| Anole lizard | Anolis carolinensis | 1 | chromosome: 2 | XM_003217189.2 | XP_003217237.1 | NCBI | |
| Amphibia | Western clawed frog | Xenopus laevis | 1 | chromosome: Un | XM_002935933.2 | XP_002935979.1 | NCBI |
| Actinopterygii | Zebrafish | Danio rerio | 1 | chromosome: 6 | BC059448.1 | AAH59448.1 | NCBI |
| 2 | chromosome: 3 | NM_131411.2 | NP_571486.1 | NCBI | |||
| Mexican tetra | Astyanax mexicanus | 1 | KB882152.1:1429796–1431775: −1 | XM_007252167.1 | XP_007252229.1 | NCBI | |
| 2 | KB872051.1:338907-348651: −1 | ENSAMXG00000016495 | ENSAMXT00000016988 | Ensembl | |||
| Gilthead seabream | Sparus aurata | 1 | – | AY533402.1 | AAT02159.1 | NCBI | |
| 2 | – | AY533403.2 | AAT02160.2 | NCBI | |||
| European seabass | Dicentrarchus labrax | 1a | – | EU378922.1 | ACB13284.1 | NCBI | |
| 1b | – | EU378923.1 | ACB13285.1 | NCBI | |||
| Atlantic cod | Gadus morhua | 1 | GeneScaffold 1449:283672–285101: −1 | ENSGMOG00000010804 | ENSGMOT00000011867 | Ensembl | |
| 2 | GeneScaffold 1876:387650–388957: −1 | ENSGMOG00000004277 | ENSGMOT00000004660 | Ensembl | |||
| Medaka | Oryzias latipes | 1a | chromosome: 1 | NM_001104832.1 | NP_001098302.1 | NCBI | |
| 1b | chromosome: 8 | NM_001104860.1 | NP_001098330.1 | NCBI | |||
| 2 | chromosome: 8 | AB248276.1 | BAE78762.1 | NCBI | |||
| Fugu | Takifugu rubripes | 1a | scaffold 2391:5485–6279: −1 | ENSTRUG00000017502 | ENSTRUT00000045022 | Ensembl | |
| 1b | scaffold 65:466946-468376: −1 | ENSTRUG00000009927 | ENSTRUT00000025045 | Ensembl | |||
| 2 | scaffold15:451903–452747: −1 | ENSTRUG00000011652 | ENSTRUT00000029563 | Ensembl | |||
| Tilapia | Oreochromis niloticus | 1a | GL831193.1:3303726–3305445: 1 | ENSONIG00000018253 | ENSONIT00000023007 | Ensembl | |
| 1b | Un random:3751825–3761582: 1 | ENSTNIG00000004312 | ENSTNIT00000007096 | Ensembl | |||
| 2 | 3:12195390–12196528: −1 | ENSTNIG00000018581 | ENSONIT00000001810 | Ensembl | |||
| Tetraodon | Tetraodon nigroviridis | 1a | Un random:26969504–26970941: −1 | ENSTNIG00000017516 | ENSTNIT00000020893 | Ensembl | |
| 1b | Un random:3751825–3761582: 1 | ENSTNIG00000004312 | ENSTNIT00000007096 | Ensembl | |||
| 2 | 3:12195390–12196528: −1 | ENSTNIG00000018581 | ENSTNIT00000021990 | Ensembl | |||
| Amazon molly 1 | Poecilia formosa | 1a | KI519952.1:480412–483140: −1 | ENSPFOG00000012646 | ENSPFOT00000012663 | Ensembl | |
| 1b | KI519724.1:1037528–1042207: 1 | ENSPFOG00000003689 | ENSPFOT00000003582 | Ensembl | |||
| 2a | KI519725.1:742206-743398: −1 | ENSPFOG00000010360 | ENSPFOT00000010350 | Ensembl | |||
| 2b | KI519725.1:771388–772351:-1 | ENSPFOG00000010492 | ENSPFOT00000010477 | Ensembl | |||
| Platyfish | Xiphophorus maculatus | 1a | JH556678.1:1270978–1287717: 1 | ENSXMAG00000006015 | ENSXMAT00000006036 | Ensembl | |
| 1b | JH557102.1:45038–49228: 1 | ENSXMAG00000008291 | ENSXMAT00000008323 | Ensembl | |||
| 2 | JH556906.1:38852–39807: 1 | ENSXMAG00000000128 | ENSXMAT00000000129 | Ensembl | |||
| Stickleback 2 | Gasterosteus aculeatus | 1a | groupIX:16205994–16208003:-1 | ENSGACG00000019276 | ENSGACT00000025530 | Ensembl | |
| 1b | groupXI:3766738–3767762: −1 | ENSGACG00000007070 | ENSGACT00000009381 | Ensembl | |||
| 2 | groupXI:13702853–13703781: −1 | ENSGACG00000013957 | ENSGACT00000018464 | Ensembl | |||
| Chondrichthyes arcopterygii | Elephant shark | Callorhinchus milii | 1 | chromosome: Un | NM_001292772.1 | NP_001279701.1 | NCBI |
| Coelacanth | Latimeria chalumnae | 1 | chromosome: Un | XM_005993207.1 | XP_005993269.1 | NCBI | |
1 Tetraploid; 2 As a genome reference sequence for synteny analysis.

Table 2.
The copy number of aanat genes from the fishes sequenced by our lab or collaborators.
| Class | Common Name | Species Name | Total Number | aanat1a | aanat1b | aanat2 |
|---|---|---|---|---|---|---|
| Actinopterygii | Giant-fin mudskipper/PM 1 | Periophthalmus magnuspinnatus | 2 | – | 1 | 1 |
| Blue-spotted mudskipper/BP 1 | Boleophthalmus pectinirosris | 3 | 1 | 1 | 1 | |
| Malabar grouper | Epinephelus malabaricus | 3 | 1 | 1 | 1 | |
| Half-smooth tongue sole | Cynoglossus semilaevis | 3 | 1 | 1 | 1 | |
| Hedgehog seahorse | Hippocampus spinosissimus | 3 | 1 | 1 | 1 | |
| Atlantic salmon 2 | Salmo salar | 5 | 1 | 2 | 2 | |
| Rainbow trout 2 | Oncorhynchus mykiss | 5 | 1 | 2 | 2 | |
| Northern pike | Esox lucius | 3 | 1 | 1 | 1 | |
| Golden-line fishes (SIA) 2 | Sinocyclocheilus anshuiensis | 3 | 1 | – | 2 | |
| (SIG) 2 | Sinocyclocheilus graham | 3 | 1 | – | 2 | |
| (SIR) 2 | Sinocyclooheilus rhinocerous | 3 | 1 | – | 2 | |
| Asian arowana | Scleropages formosus | 3 | 1 | 1 | 1 |
1 Amphibious fishes; 2 Tetraploid fishes.
In mammals, amphibians, reptiles, birds, sarcopterygii, and chondrichthyes (although there were two aanat genes, one non-vertebrate aanat, and one vertebrate aanat, found in the elephant shark, in this paper we focused only on vertebrate type), only one aanat gene was identified in their genomes. However, in teleosts, there are several copies of aanat genes. For common diploid teleosts, there are two or three different aanat genes. In detail (Table 1 and Table 2), for species like zebrafish (Danio rerio) possessing two aanat genes, there is one aanat1 expressed in the retina, and one aanat2 expressed in the pineal gland. For those like medaka (Oryzias latipes) with three aanat genes, there are two aanat1s, which have subtypes aanat1a and aanat1b expressed in the retina, and one aanat2 in the pineal gland. Interestingly, for giant-fin mudskipper (PM) and Atlantic cod, there are aanat1b and aanat2 with aanat1a missing. For a typical tetraploid teleost, like Atlantic salmon (Salmo salar) which experienced the salmonid-specific genome duplication, its genome has five copies of aanat genes, having lost one copy of its aanat1a. Likewise, in another tetraploid group, the golden-line fishes (SIA, Sinocyclocheilus anshuiensis; SIG, Sinocyclocheilus graham; SIR, Sinocycloheilus rhinocerous), there are three aanat isoforms (compared to the expected six), due to loss of one aanat1a and both aanat1bs.
2.2. The Phylogenetic Relationships among aanat Genes
Using the lamprey (Petromyzon marinus) aanat as the out-group, three different reconstruction methods of phylogenetic trees generated the same tree topology with slight differences. According to the conserved blocks of aanat coding sequence (CDS), we generated the radial Bayesian tree in Figure 2. The left of Figure 3A is the aanat1 rectangular Bayesian tree and the left of Figure 3B is the aanat2 phylogenetic tree. According to the phylogenetic tree, the aanat gene divides into two main groups (aanat1 and aanat2). Specifically, teleost aanat1 is much closer to the tetrapod aanat, while aanat2 is more distant and seems to be teleost unique. Teleost aanat1 divides into two subgroups (aanat1a and aanat1b) and separates from tetrapod aanat. Interestingly, in the ancient fish Asian arowana (Scleropages formosus) aanat1a locates out of the entire teleost aanat1 clade. In another ancient fish coelacanth (Latimeria chalumnae) the single aanat gene locates within the reptile clade, represented here by the Anole lizard (Anolis carolinensis). Mexican tetra (Astyanax mexicanus), three kinds of golden-line fishes (SIA, SIG, and SIR) and zebrafish belong to Ostariophysi, and in contrast to other teleosts possess only a single copy of the aanat1 gene; this gene seems to be more closely related to aanat1a in the phylogenetic analysis shown in Figure 2, but related to aanat1b in the left panel of Figure 3a. Ultimately, synteny analysis (Figure 3a) and protein sequence alignment (Figure 4) confirm that the Ostariophysi aanat1 corresponds to aanat1a. Within these same five Ostariophysi species, their aanat2 seems to be divided from other teleost aanat2 and to form a new clade. Protacanthopterygii (Atlantic salmon; rainbow trout, Oncorhynchus mykiss) aanat1b as well as aanat2 diverge and form two different clades, consistent with the Salmonidae experiencing an additional unique whole genome duplication.

Figure 2.
The radial Bayesian phylogenetic tree based on the conserved blocks of all aanat CDS sequences. The analysis was performed using the MrBayse 3.2 software. Evolutionary model selection was calculated using the MrMTgui program (the best nucleotide substitution model was GTR + I + G). The phylogenetic tree is rooted with lamprey (Petromyzon marinus) aanat. Although the PhyML and Mega phylogenetic trees are not shown here, they present the same tree topology. Four aanat subgroups are shown with different colors (Blue, tetrapod aanat; Red, teleost aanat1a; Magenta, teleost aanat1b; Lime, teleost aanat2).
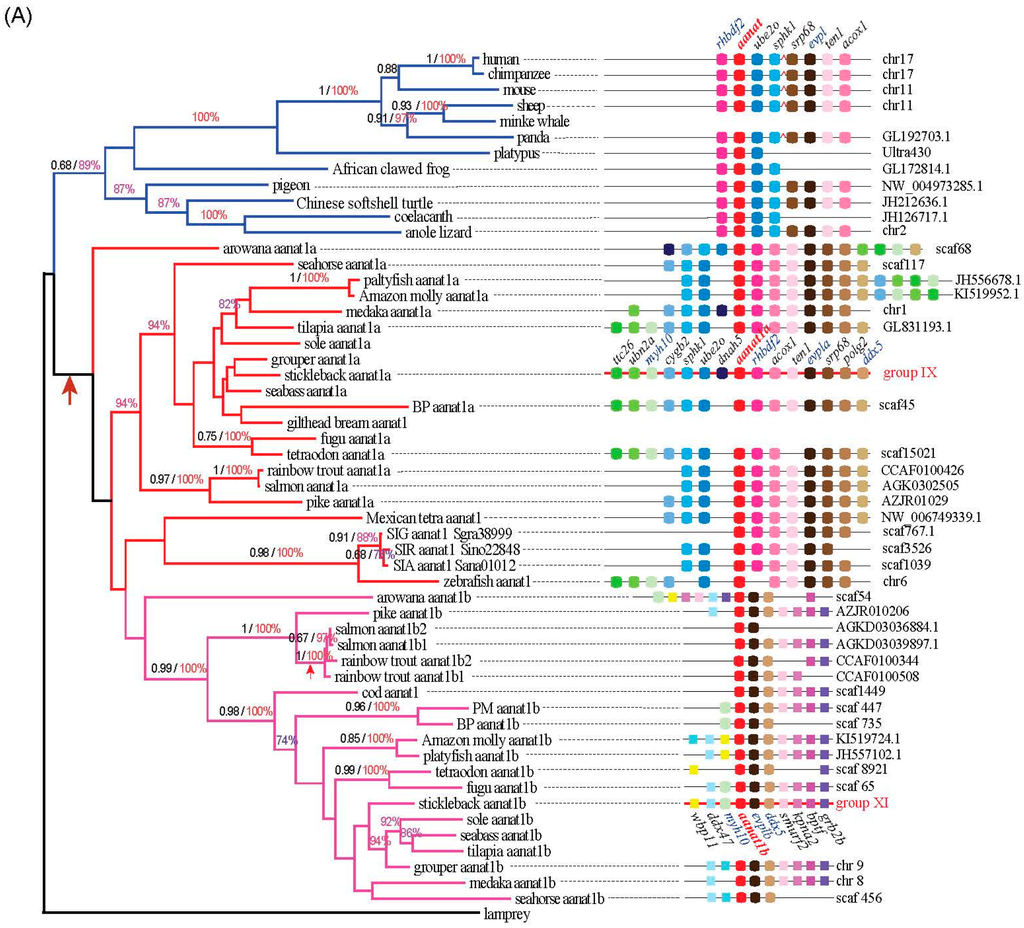
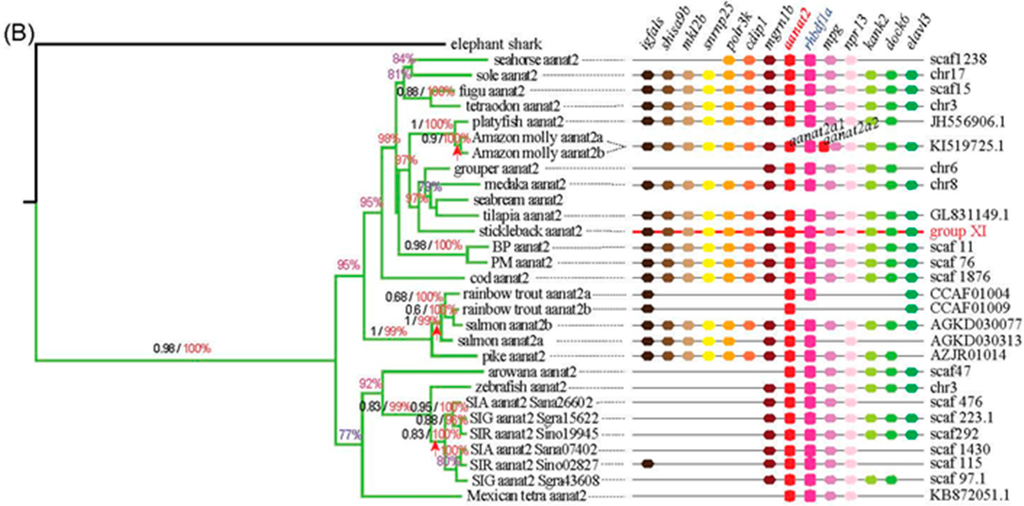
Figure 3.
Phylogenetic trees and synteny of aanat1 (A) and aanat2 (B). The left of (A,B) are rectangular Bayesian trees of aanat1 and aanat2, respectively. Numbers on branches from left to right are bootstrap supports (black) obtained in the phyML reconstruction and the Bayesian posterior probabilities (colored). The symbol of arrow represents the whole genome duplication. Note that bootstrap support under 0.6 and posterior probabilities under 70% are not shown. The color of branches is similar to Figure 2; The right of (A,B) are synteny of aanat1 and aanat2, respectively. Distances between genes or gene length are not drawn to scale. In mammals, four genes (srp68, evpl, ten1, acox1) are located 400-kb upstream of aanat within several genes intervening (shown with a red broken line). Stickleback aanat1a, shown with a red backbone, is used as the reference sequence.
2.3. Synteny
All aanat genes share a conserved suite of genes bounding them on their side, although some species may display gene loss and location variation (see details in the right of Figure 3A,B). Seven genes (rhbdf2, ube2o, sphk1, srp68, evpl, ten1, and acox1) are neighboring the tetrapod aanat gene. In mammals, four genes (srp68, evpl, ten1, and acox1) are located 400-kb upstream of aanat within several intervening genes. All these neighboring genes can be identified in fish genomes with minor variations in location. Rhbdf can be found adjacent to aanat1a and aana2, but not aanat1b. Evpl can be found in tetrapod aanat and teleost aanat1 regions. Three genes (myh10, myh11b, and ddx47) and six genes (evp1b, ddx5, smurf2, kpna2, bptf, and grb2b) are located upstream and downstream, respectively, of fish aanat1b genes. Two genes (ddx5 and evpl/evpl1b) appear near the two isoforms of aanat1. Seven genes (igfals, shisa9b, mkl2b, snrnp25, polrk3k, cdip1, and mgrn1b) are located upstream of fish aanat2; six genes (rhbdf1a, mpg, nprl3, kank2, dock6, and elavl3) are positioned downstream of fish aanat2. The gene context of the two copies of Amazon molly aanat2 shows differences because these two copies of aanat2 are close to each other and locate within the same scaffold. Ostariophysi, compared with other teleosts, has lost many genes near aanats. All data illustrate that the synteny regions were conserved across species with slight differences.
2.4. The Regulatory Elements of aanat Genes
In mammals, such as human and mouse, there are fewer PCEs associated with aanats compared to fishes (Figure 5 and Supplementary Figure S1). This is correlated with a significantly reduced role for photoreception in control of aanat expression in mammalian pinealocytes compared with their teleost counterparts. The total number of E-box and E-box-like elements are seven in human and eight in mouse. In contrast, the least is one in Mexican tetra aanat1 and Atlantic salmon aanat1b2, and the most is 14 in fugu aanat2 because of the short tandem repeats. The average number of E-box and E-box-like elements for teleost aanat2 is much more than that of teleost aanat1. This suggests that E-box and E-box-like elements are the likely key regulators of pineal-specific aanat expression.

Figure 4.
Alignment of AANAT protein sequences. AANAT proteins from different species were aligned with sheep AANAT and the structural template 1CJW. The analysis was completed with MAFFT and colorized using TEXshade. Secondary structures include alpha helix (α), beta strand (β), and 310 helix (η). Important residues, including substrate binding residues (§, red boxes), catalytic residues (‡) and phosphorylation sites (up arrow, pink boxes), are marked for comparison. Residues differentiating between AANAT1 and AANAT2 are marked by a dagger (†). Numbering is relative to sheep AANAT. The color code for the conservation track ranges from red for the most conserved, to blue for the least conserved, as per TEXshade.
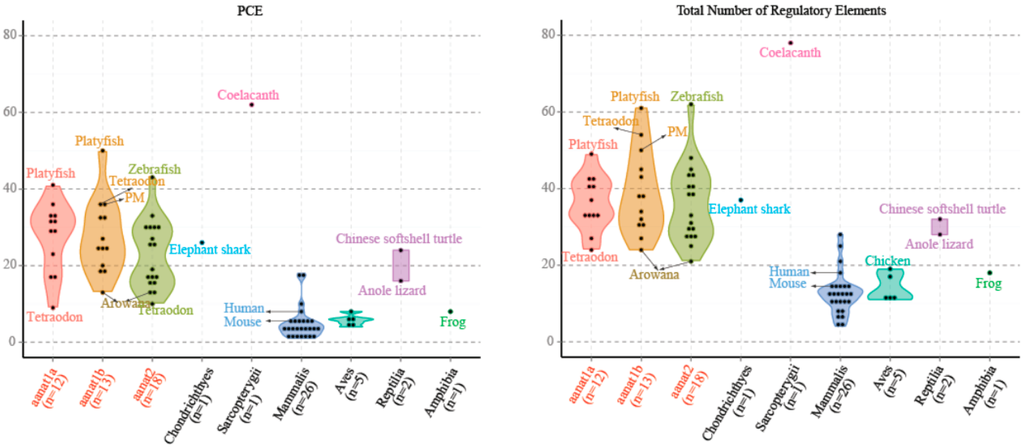
Figure 5.
The numbers of PCE (Photoreceptor Conserved Elements) and the total numbers of regulatory elements (PCE, E-box, E-box-like, and PIPES, as defined in the Introduction) for different classes of aanat. We count aanat regulatory elements from 54 species, including those species mentioned in Supplementary Figure S1. The average number of aanat regulatory elements in fishes is larger than that of non-fish species.
2.5. The Structure of AANAT Proteins
Representative AANAT proteins are aligned in Figure 4. The similarity and identity of these AANAT proteins are summarized in Supplementary Table S1. Previous research [12,30] has shown that AANAT possesses a catalytic funnel to bond with the reactive groups (i.e., the amine and AcCoA). Catalytic histidines (His120, His122) at the bottom of this pocket form a proton channel that deprotonates the protonated amine group to initiate the acetyl transfer reaction. The thioester bond of AcCoA comes under nucleophilic attack through the deprotonation. A conserved tyrosine (Tyr168) promotes protonation of the CoA thiolate-leaving group.
As shown in this alignment, amino acid residues 130 and 153 clearly differentiate between AANAT1 and AANAT2, regardless of species. Position of 130-amino acid of AANAT1 is Phe (F) compare with the Cys130 of AANAT2. The position of 153-amino acid of AANAT1 is Val (V) compared with the Leu153 of AANAT2. According to the template 1CJW [31], 153-amino acid locates close to the β-turn and the 130-amino acid participates in forming a turn. Therefore, F/C variation may play the biggest difference of these two kinds of AANATs. Some other variation sites also are pointed out in Figure 4. Altogether, our alignments show the highly conserved nature of AANAT proteins in all vertebrates.
3. Discussion
This study surveys a range of facets of the aanat gene and provides a global view about the diversity of AANAT in fish, not only from the DNA level but also from the protein level. We compared not only the differences in the regulatory regions, but also protein structural differences.
3.1. The Potential Reasons of Copy Number Variation among Vertebrates
Jack Falcon and his co-workers [14] clarified that 500 million years ago vertebrate aanat (VT-aanat) evolved from the invertebrate aanat (NV-aanat) isoform after the vertebrates split from the cephalochordates. NV-aanat is suggested to act as a detoxifier to prevent toxic reactions from endogenous and exogenous aralkylamines and polyamines [14]. The evolution of VT-aanat was facilitated by a duplication of NV-aanat early during vertebrate evolution. One duplicated copy was neofunctionalized to function primarily in the acetylation of serotonin in the synthesis of melatonin. Eventually the other copy, which would be recognized as the NV-aanat form, was lost from the vertebrate lineage.
It has been proven that two whole-genome duplications (WGD) occurred in the common ancestor of early vertebrates before ray-fined fishes split with tetrapods [32,33,34]. More specifically, the first round of WGD occurred before the agnatha-gnatostoma split and the second occurred before the chrondrichthyes-osteichthyes split. Subsequently, a third round of teleost-specific WGD occurred. Some fish families (Acipenseridae, Catostomidae, Cobitidae, Cyprininae, and Salmonidae) experienced a fourth round [35]. The teleost-specific WGD is thought of as the main evolutionary source of new genetic traits to support the expansion of this group. According to our analysis, we can confirm that the main reasons for copy number variation of aanat genes in different vertebrates are WGD and gene loss. The ancestral VT-aanat formed two copies of aanat gene (aanat1 and aanat2) after undergoing WGD. The tetrapods and the ancient fishes, including the Actinistia coelacanth, lost aanat2 after they split with the bony fish, meanwhile the Holocephalan elephant shark lost aanat1. Because of another special WGD in teleosts, the teleost genomes may contain double copies of genes compared to their tetrapod counterparts. Hence, the teleost aanat1 splits into two forms: aanat1a and aanat1b. Meanwhile, according to our synteny analysis, the two copies of aanat2 may lose a copy and eventually form only one copy of aanat2 gene in the teleost genomes.
The tetrapod and the ancient fish aanats are similar to the bony fish aanat1a in the gene sequence (details shown in Figure 5 and Supplementary Table S1). The phylogenetic analysis also supports the view.
For tetraploid teleosts like rainbow trout [22,36], the salmonid-specific genome duplication leads to appearance of five copies of aanat genes in the genome. This phenomenon was also observed in Atlantic salmon. In Amazon molly, another tetraploid species, one copy of aanat1a and one copy of aanat1b are lost, generating four aanat genes in its genome. However, synteny analysis of Amazon molly indicates that the existence of two aanat2 is due to a tandem duplication, not because of genome duplication. The copy number between diploid and tetraploid is not always the one-to-two correspondence due to selective gene loss.
With the development of new sequencing technology, the quality of sequenced whole genomes has been vastly improved. We therefore can gain more complete information about aanat gene in each genome. For example, we originally thought that there were only two copies of aanat genes existing in European seabass (Dicentrarchus labrax), which were aanat1a and aanat1b. However, now aanat2 has been also identified using the newest version of European seabass reference genome [37]. More accurate and complete information about the aanat genes may be obtained in the future, and the evolution of aanat in fish can be better illustrated.
3.2. Adaptive Evolution of aanat
It is interesting that teleosts have retained multiple isoforms of AANAT where chondrichthyes and tetrapods have retained only one. This implies that there is some selective advantage to having multiple forms. In addition to having a spatial specialization, with AANAT1 being expressed primarily in the retina and AANAT2 being expressed primarily in the pineal gland, there are also instances where different isoforms are temporally regulated within a tissue. For example, two isoforms of aanat1 (ssaanat1a and ssaanat1b) from the metamorphic flatfish Senegalese sole retina have been identified [38]. The timing of expression of these two aanat1s differs during sole development. In the early stages, ssaanat1a increases, reaching a peak at 4 dpf (days post fertilization) with a clear daily rhythm; after that its expression declines, and becomes arrhythmic. In contrast, ssaanat1b increases after metamorphosis, with a day/night rhythm in the adult. These data suggest different roles for the two ssaanat1 during metamorphosis.
An instance where loss of an AANAT1 isoform may have functional consequences may be found in amphibious mudskippers [39]. Fully aquatic fish would be expected to have myopic vision out of water, but amphibious mudskippers apparently do not have this problem. AANAT1a in the retina can acetylate dopamine [26], and low-levels of dopamine during development may result in myopia [40]. As a consequence You et al. [39] speculated that loss of aanat1a in the genome of terrestrial giant-fin mudskipper compared with water-living blue-spotted mudskipper, may be adaptive for its aerial vision. Mexican tetra (Astyanax mexicanus), three kinds of golden-line fishes (SIA, SIG, and SIR) and zebrafish belong to Ostariophysi, and in contrast to other teleosts possess only a single copy of the aanat1 gene; this gene seems to be more closely related to aanat1a in the phylogenetic analysis shown in Figure 2, but related to aanat1b in the left panel of Figure 3a. Ultimately, synteny analysis (Figure 3a) and protein sequence alignment (Figure 5) confirm that the Ostariophysi aanat1 corresponds to aanat1a. Within these same five Ostariophysi species, their aanat2 seems to be divided from other teleost aanat2 and to form a new clade. However, the functional significance of these two isoforms of aanat1 in teleosts is still under investigation.
The principle function of teleost aanat2 appears to be melatonin synthesis. Like tetrapod aanat, teleost aanat2 is primarily expressed in the pineal gland. The kinetics of teleost aanat2 is similar to that of tetrapod aanat in terms of substrate selectivity [22]. Intriguingly, it is teleost aanat1 that is more closely phylogenetically related to tetrapod aanat. Apparently this is related to the duplicated copies of aanat present in the common ancestor of teleosts and tetrapods. Following divergence of these lineages, tetrapod aanat retained or acquired characteristics of what would become teleost aanat2. Meanwhile, teleost aanat1 evolved to gain traits specific for functions related to the retina. A better understanding of the ancestral forms of aanat present at the point of divergence would help decipher this dilemma.
4. Experimental Section
4.1. Sequence Collection
For our research, aanat nucleotide and amino acid sequences were collected from 37 species for comparison. For details, those reported aanat sequences, for example human (Homo sapiens) and zebrafish (Danio rerio), were downloaded from NCBI and Ensembl (Table 1). AANAT sequences of Malabar grouper (Epinephelus malabaricus), half-smooth tongue sole (Cynoglossus semilaevis), giant-fin mudskipper or PM (Periophthalmus magnuspinnatus), blue-spotted mudskipper or BP (Boleophthalmus pectinirosris), seahorse (Hippocampus spinosissimus), Atlantic salmon (Salmo salar), rainbow trout (Oncorhynchus mykiss), pike (Esox lucius), golded-line fishes (SIA, Sinocyclocheilus anshuiensis; SIG, Sinocyclocheilus graham; SIR, Sinocyclooheilus rhinocerous), Asian arowana (Scleropages formosus), and Mexican tetra (Astyanax mexicanus) were extracted from the genome databases, generated by us and our collaborators, through BLAST [41] and Gene Wise [42].
4.2. Sequence Alignment and Phylogenetic Reconstruction
We conducted phylogenetic analyses using these collected sequences. After finishing the alignment of aanat gene sequences using MAFFT software [43], we chose conserved blocks by the online Gblocks [44]. Subsequently, MrMTgui program [45] was applied to complete the nucleotide substitution model test (the best model based on the Akaike Information Criterion, AIC, was GTR + I + G). Finally, we analyzed the sequences using MrBayes 3.2 [46] through the Bayesian tactics with the Metropolis-coupled Markov chain Monte Carlo algorithm [47]. By using four chains run, setting four million generations for Ngen and 100 for Samplefreq, the best phylogenetic tree was explored. The first 5000 trees were discarded (Burnin) before calculating summary statistics that allowed identification of the point when the chains became fixable. A consensus tree was constructed with the remaining trees. Branch support was evaluated using Bayesian posterior probabilities.
In addition, PhyML [48] and MEGA 6 [49] were used to rebuild AANAT protein phylogenetic trees, with a local Prottest [50] program to analyze the amino acid substitution model (the best substitution model was JTT + G + F) and the 1000 bootstrap replications. The best tree was explored using PhyML and MEGA 6 with manual alignment corrections.
Meanwhile, we also selected the tightly conserved mitochondrial gene cytb of the examined species to build the species phylogenetic tree using the same methods. Comparing the species phylogenetic tree and the AANAT tree, we evaluated the relatedness between species evolution and aanat gene evolution.
4.3. Identification of Conserved Synteny
To assess the degree of aanat gene conservation, we searched several genes upstream and downstream of each aanat paralog. We collected genome data from Ensembl dataset and our lab as mentioned in Section 2.1. The stickleback (Gasterosteus aculeatus) genome was used as the reference sequence for searching aanat upstream and downstream. The genome assemblies of different species were searched using the BLAST and the best hit was selected using a Perl script.
4.4. Analysis of Regulatory Regions and the Differences between AANAT Proteins
We obtained the complete genomic sequences of including 4 kb upstream and downstream of 84 aanat genes. Four typical regulatory elements, including E-box, E-box like element, PCE and cis-acting regulatory element, were searched for in each sequence including the upstream and downstream regions using BLAST program. The similarity of target was required to be 100% with the sequences of four regulatory elements (described in Introduction Section).
We also downloaded a common AANAT protein model of sheep (Ovis aries, 1CJW) from the Protein Data Bank (PDB) for comparing the difference between AANAT1 and AANT2. MAFFT software was used to complete alignment. The result of alignment was colorized using TEXshade [51].
5. Conclusions
The diversity of aanat in fish is generated by whole genome duplication and gene loss. During evolution, due to different external circumstances, aanat1 and aanat2 separated, were expressed in different organs and took on different roles. In this paper, we have investigated and refined this view based on additional sequences from various fish species not previously available. At the same time, we also use an evolutionary view to reveal many similarities or identities between AANAT1 protein and AANAT2 protein. Through these analyses, to some extent, the evolutionary features of aanat also reveal aspects of the relationship between the pineal gland and the environmental circumstance.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/17/1/51/s1.
Acknowledgments
The work was supported by China 863 Project (No. 2012AA10A407), Special Project on the Integration of Industry, Education and Research of Guangdong Province (No. 2013B090800017), Shenzhen Special Program for Future Industrial Development (No. JSGG20141020113728803), Shenzhen Dapeng Special Program for Industrial Development (No. QT20140101), Shenzhen Scientific R & D Grant (No. CXB201108250095A), and the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development at the National Institutes of Health, Bethesda, MD, USA.
Author Contributions
Qiong Shi and Jia Li conceived and designed the experiments. Jia Li performed the experiments. Jia Li, Steven L. Coon and Qiong Shi prepared the manuscript. Xinxin You, Chao Bian and Hui Yu participated in discussions and provided suggestions.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
acox1: peroxisomal acyl-coenzyme A oxidase 1; bptf: nucleosome-remodeling factor subunit BPTF; cdip1: cell death-inducing p53 target 1; cygb2: cytoglobin 2; ddx47: DEAD (Asp-Glu-Ala-Asp) box polypeptide 47; ddx5: DEAD (Asp-Glu-Ala-Asp) box polypeptide 5; dock6: dedicator of cytokinesis 6; elavl3: ELAV like neuron-specific RNA binding protein 3; evpl: envoplakin; evpla: envoplakin a; evplb: envoplakin b; grb2b: growth factor receptor-bound protein 2b; igfals: insulin-like growth factor binding protein, acid labile subunit; kank2: KN motif and ankyrin repeat domains 2; kpna2: karyopherin alpha 2; mgrn1b: MGRN1 mahogunin ring finger 1, E3 ubiquitin protein ligase; mkl2b: MKL/myocardin-like 2b; mpg: DNA-3-methyladenine glycosylase; myh10: myosin, heavy chain 10, non-muscle; nprl3: nitrogen permease regulator-like 3; polg2: polymerase (DNA directed), gamma 2; polr3k: polymerase (RNA) III (DNA directed) polypeptide K; rhbdf1a: rhomboid 5 homolog 1a; rhbdf2: inactive rhomboid protein 2; shisa9b: shisa family member 9b; smurf2: SMAD specific E3 ubiquitin protein ligase 2; snrnp25: small nuclear ribonucleoprotein 25; sphk1: sphingosine kinase 1; srp68: signal recognition particle 68; ten1: TEN1 CST complex subunit; ttc26: tetratricopeptide repeat domain 26; ube2o: ubiquitin-conjugating enzyme E2 O; ubn2a: ubinuclein 2a; wbp11: WW domain-binding protein 11.
References
- Iuvone, P.M.; Tosini, G.; Pozdeyev, N.; Haque, R.; Klein, D.C.; Chaurasia, S.S. Circadian clocks, clock networks, arylalkylamine n-acetyltransferase, and melatonin in the retina. Prog. Retin. Eye Res. 2005, 24, 433–456. [Google Scholar] [CrossRef] [PubMed]
- Li, D.Y.; Smith, D.G.; Hardeland, R.; Yang, M.Y.; Xu, H.L.; Zhang, L.; Yin, H.D.; Zhu, Q. Melatonin receptor genes in vertebrates. Int. J. Mol. Sci. 2013, 14, 11208–11223. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.Y.; Hong, W.S.; Zhu, W.B.; Shi, Q.; You, X.X.; Chen, S.X. Cloning and expression of melatonin receptors in the mudskipper boleophthalmus pectinirostris: Their role in synchronizing its semilunar spawning rhythm. Gen. Comp. Endocrinol. 2014, 195, 138–150. [Google Scholar] [CrossRef] [PubMed]
- Pinillos, M.L.; De Pedro, N.; Alonso-Gomez, A.L.; Alonso-Bedate, M.; Delgado, M.J. Food intake inhibition by melatonin in goldfish (Carassius auratus). Physiol. Behav. 2001, 72, 629–634. [Google Scholar] [CrossRef]
- Lopez Patino, M.A.; Guijarro, A.I.; Alonso-Gomez, A.L.; Delgado, M.J. Characterization of two different melatonin binding sites in peripheral tissues of the teleost tinca tinca. Gen. Comp. Endocrinol. 2012, 175, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Velarde, E.; Alonso-Gomez, A.L.; Azpeleta, C.; Isorna, E.; Delgado, M.J. Melatonin attenuates the acetylcholine-induced contraction in isolated intestine of a teleost fish. J. Comp. Physiol. B 2009, 179, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Lima-Cabello, E.; Diaz-Casado, M.E.; Guerrero, J.A.; Otalora, B.B.; Escames, G.; Lopez, L.C.; Reiter, R.J.; Acuna-Castroviejo, D. A review of the melatonin functions in zebrafish physiology. J. Pineal Res. 2014, 57, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hassell, K.J.; Reiter, R.J.; Robertson, N.J. Melatonin and its role in neurodevelopment during the perinatal period: A review. Fetal Matern. Med. Rev. 2013, 24, 76–107. [Google Scholar] [CrossRef]
- Lombardo, F.; Gioacchini, G.; Fabbrocini, A.; Candelma, M.; D’Adamo, R.; Giorgini, E.; Carnevali, O. Melatonin-mediated effects on killifish reproductive axis. Comp. Biochem Physiol. Part A 2014, 172, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Appelbaum, L.; Wang, G.X.; Maro, G.S.; Mori, R.; Tovin, A.; Marin, W.; Yokogawa, T.; Kawakami, K.; Smith, S.J.; Gothilf, Y.; et al. Sleep-wake regulation and hypocretin-melatonin interaction in zebrafish. Proc. Natl. Acad. Sci. USA 2009, 106, 21942–21947. [Google Scholar] [CrossRef] [PubMed]
- Klein, D.C.; Coon, S.L.; Roseboom, P.H.; Weller, J.L.; Bernard, M.; Gastel, J.A.; Zatz, M.; Iuvone, P.M.; Rodriguez, I.R.; Begay, V.; et al. The melatonin rhythm-generating enzyme: Molecular regulation of serotonin n-acetyltransferase in the pineal gland. Recent Prog. Horm. Res. 1997, 52, 307–357. [Google Scholar] [PubMed]
- Klein, D.C. Arylalkylamine n-acetyltransferase: “The timezyme”. J. Biol. Chem. 2007, 282, 4233–4237. [Google Scholar] [CrossRef] [PubMed]
- Cazamea-Catalan, D.; Besseau, L.; Falcon, J.; Magnanou, E. The timing of timezyme diversification in vertebrates. PLoS ONE 2014, 9, e112380. [Google Scholar] [CrossRef] [PubMed]
- Falcon, J.; Coon, S.L.; Besseau, L.; Cazamea-Catalan, D.; Fuentes, M.; Magnanou, E.; Paulin, C.H.; Boeuf, G.; Sauzet, S.; Jorgensen, E.H.; et al. Drastic neofunctionalization associated with evolution of the timezyme aanat 500 mya. Proc. Natl. Acad. Sci. USA 2014, 111, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Pavlicek, J.; Sauzet, S.; Besseau, L.; Coon, S.L.; Weller, J.L.; Boeuf, G.; Gaildrat, P.; Omelchenko, M.V.; Koonin, E.V.; Falcon, J.; et al. Evolution of aanat: Expansion of the gene family in the cephalochordate amphioxus. BMC Evol. Biol. 2010, 10, 154. [Google Scholar] [CrossRef] [PubMed]
- Appelbaum, L.; Vallone, D.; Anzulovich, A.; Ziv, L.; Tom, M.; Foulkes, N.S.; Gothilf, Y. Zebrafish arylalkylamine-n-acetyltransferase genes-targets for regulation of the circadian clock. J. Mol. Endocrinol. 2006, 36, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Appelbaum, L.; Toyama, R.; Dawid, I.B.; Klein, D.C.; Baler, R.; Gothilf, Y. Zebrafish serotonin-n-acetyltransferase-2 gene regulation: Pineal-restrictive downstream module contains a functional e-box and three photoreceptor conserved elements. Mol. Endocrinol. 2004, 18, 1210–1221. [Google Scholar] [CrossRef] [PubMed]
- Tosini, G.; Chaurasia, S.S.; Michael Iuvone, P. Regulation of arylalkylamine n-acetyltransferase (aanat) in the retina. Chronobiol. Int. 2006, 23, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, S.; Gastel, J.A.; Weller, J.L.; Schwartz, C.; Jaffe, H.; Namboodiri, M.A.; Coon, S.L.; Hickman, A.B.; Rollag, M.; Obsil, T.; et al. Role of a pineal camp-operated arylalkylamine n-acetyltransferase/14-3-3-binding switch in melatonin synthesis. Proc. Natl. Acad. Sci. USA 2001, 98, 8083–8088. [Google Scholar] [CrossRef] [PubMed]
- Asaoka, Y.; Mano, H.; Kojima, D.; Fukada, Y. Pineal expression-promoting element (pipe), a cis-acting element, directs pineal-specific gene expression in zebrafish. Proc. Natl. Acad. Sci. USA 2002, 99, 15456–15461. [Google Scholar] [CrossRef] [PubMed]
- Falcon, J.; Galarneau, K.M.; Weller, J.L.; Ron, B.; Chen, G.; Coon, S.L.; Klein, D.C. Regulation of arylalkylamine n-acetyltransferase-2 (aanat2, ec 2.3.1.87) in the fish pineal organ: Evidence for a role of proteasomal proteolysis. Endocrinology 2001, 142, 1804–1813. [Google Scholar] [CrossRef] [PubMed]
- Cazamea-Catalan, D.; Magnanou, E.; Helland, R.; Besseau, L.; Boeuf, G.; Falcon, J.; Jorgensen, E.H. Unique arylalkylamine n-acetyltransferase-2 polymorphism in salmonids and profound variations in thermal stability and catalytic efficiency conferred by two residues. J. Exp. Biol. 2013, 216, 1938–1948. [Google Scholar] [CrossRef] [PubMed]
- Falcon, J.; Migaud, H.; Munoz-Cueto, J.A.; Carrillo, M. Current knowledge on the melatonin system in teleost fish. Gen. Comp. Endocrinol. 2010, 165, 469–482. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, A.V.; Mosser, E.A.; Oikonomou, G.; Prober, D.A. Melatonin is required for the circadian regulation of sleep. Neuron 2015, 85, 1193–1199. [Google Scholar] [CrossRef] [PubMed]
- Paulin, C.H.; Cazamea-Catalan, D.; Zilberman-Peled, B.; Herrera-Perez, P.; Sauzet, S.; Magnanou, E.; Fuentes, M.; Gothilf, Y.; Munoz-Cueto, J.A.; Falcon, J.; et al. Subfunctionalization of arylalkylamine n-acetyltransferases in the sea bass dicentrarchus labrax: Two-ones for one two. J. Pineal Res. 2015, 59, 354–364. [Google Scholar] [CrossRef] [PubMed]
- Zilberman-Peled, B.; Ron, B.; Gross, A.; Finberg, J.P.; Gothilf, Y. A possible new role for fish retinal serotonin-n-acetyltransferase-1 (aanat1): Dopamine metabolism. Brain Res. 2006, 1073–1074, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Nisembaum, L.G.; Tinoco, A.B.; Moure, A.L.; Alonso Gomez, A.L.; Delgado, M.J.; Valenciano, A.I. The arylalkylamine-n-acetyltransferase (aanat) acetylates dopamine in the digestive tract of goldfish: A role in intestinal motility. Neurochem. Int. 2013, 62, 873–880. [Google Scholar] [CrossRef] [PubMed]
- Velarde, E.; Cerda-Reverter, J.M.; Alonso-Gomez, A.L.; Sanchez, E.; Isorna, E.; Delgado, M.J. Melatonin-synthesizing enzymes in pineal, retina, liver, and gut of the goldfish (carassius): mRNA expression pattern and regulation of daily rhythms by lighting conditions. Chronobiol. Int. 2010, 27, 1178–1201. [Google Scholar] [CrossRef] [PubMed]
- Coon, S.L.; Klein, D.C. Evolution of arylalkylamine n-acetyltransferase: Emergence and divergence. Mol. Cell. Endocrinol. 2006, 252, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Scheibner, K.A.; De Angelis, J.; Burley, S.K.; Cole, P.A. Investigation of the roles of catalytic residues in serotonin n-acetyltransferase. J. Biol. Chem. 2002, 277, 18118–18126. [Google Scholar] [CrossRef] [PubMed]
- Hickman, A.B.; Klein, D.C.; Dyda, F. Melatonin biosynthesis: The structure of serotonin n-acetyltransferase at 2.5 a resolution suggests a catalytic mechanism. Mol. Cell 1999, 3, 23–32. [Google Scholar] [CrossRef]
- Guyomard, R.; Boussaha, M.; Krieg, F.; Hervet, C.; Quillet, E. A synthetic rainbow trout linkage map provides new insights into the salmonid whole genome duplication and the conservation of synteny among teleosts. BMC Genet. 2012, 13, 15. [Google Scholar] [CrossRef] [PubMed]
- Glasauer, S.M.; Neuhauss, S.C. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genom. 2014, 289, 1045–1060. [Google Scholar] [CrossRef] [PubMed]
- Zilberman-Peled, B.; Bransburg-Zabary, S.; Klein, D.C.; Gothilf, Y. Molecular evolution of multiple arylalkylamine n-acetyltransferase (aanat) in fish. Mar. Drugs 2011, 9, 906–921. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.M.; Brunet, F.; Petit, J.L.; Stange-Thomann, N.; Mauceli, E.; Bouneau, L.; Fischer, C.; Ozouf-Costaz, C.; Bernot, A.; et al. Genome duplication in the teleost fish tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 2004, 431, 946–957. [Google Scholar] [CrossRef] [PubMed]
- Berthelot, C.; Brunet, F.; Chalopin, D.; Juanchich, A.; Bernard, M.; Noel, B.; Bento, P.; Da Silva, C.; Labadie, K.; Alberti, A.; et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat. Commun. 2014, 5, 3657. [Google Scholar] [CrossRef] [PubMed]
- Shotgun Assembly Sequences: Genome (WGS) and Transcriptome (TSA). Available online: http://www.Ncbi.Nlm.Nih.Gov/traces/wgs/?val=CBXY01 (accessed on 5 March 2015).
- Isorna, E.; Aliaga-Guerrero, M.; M’Rabet, A.E.; Servili, A.; Falcon, J.; Munoz-Cueto, J.A. Identification of two arylalkylamine n-acetyltranferase 1 genes with different developmental expression profiles in the flatfish solea senegalensis. J. Pineal Res. 2011, 51, 434–444. [Google Scholar] [CrossRef]
- You, X.; Bian, C.; Zan, Q.; Xu, X.; Liu, X.; Chen, J.; Wang, J.; Qiu, Y.; Li, W.; Zhang, X.; et al. Mudskipper genomes provide insights into the terrestrial adaptation of amphibious fishes. Nat. Commun. 2014, 5, 5594. [Google Scholar] [CrossRef] [PubMed]
- Feldkaemper, M.; Schaeffel, F. An updated view on the role of dopamine in myopia. Exp. Eye Res. 2013, 114, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Mount, D.W. Using the basic local alignment search tool (blast). CSH Protoc. 2004, 2007. [Google Scholar] [CrossRef] [PubMed]
- Birney, E.; Durbin, R. Using genewise in the drosophila annotation experiment. Genom. Res. 2000, 10, 547–548. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. Mafft: A novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Available online: http://phylogeny.Lirmm.Fr/ (accessed on 19 April 2008).
- Mrmtgui. V 1.0; Paulo Nuin: Boston, MA, USA, 2007.
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Huggins, P.M.; Li, W.; Haws, D.; Friedrich, T.; Liu, J.; Yoshida, R. Bayes estimators for phylogenetic reconstruction. Syst. Biol. 2011, 60, 528–540. [Google Scholar] [CrossRef] [PubMed]
- Criscuolo, A. Morephyml: Improving the phylogenetic tree space exploration with phyml 3. Mol. Phylogenet. Evol. 2011, 61, 944–948. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. Mega6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. Prottest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [PubMed]
- Beitz, E. Texshade: Shading and labeling of multiple sequence alignments using latex2 epsilon. Bioinformatics 2000, 16, 135–139. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).