3. Discussion
HSC is composed of 11 crude drugs and several of them have been reported to have protective effects against radiation. For example, ginseng appeared to be a promising radio-protector for therapeutic or preventive protocols capable of attenuating the deleterious effects of radiation on normal human tissue, especially for cancer patients undergoing radiotherapy [
14]. Catalpol (a main bioactive component in the roots of
Rehmannia glutinosa) decreased plasma malondialdehyde (MDA) intestinal 8-hydroxydeoxyguanosine (8-OHdG) levels and increased plasma endogenous antioxidants and peripheral white blood cells and platelets
in vivo, which suggested that catalpol possessed notable radio-protective activity by reducing reactive oxygen species (ROS) [
17]. The components of
Cornus officinalis showed significant free radical-scavenging activity and inhibitory effects on melanogenesis induced by radiation [
11].
Millettia dielsiana had an anti-inflammatory effect, decreasing NO production [
8]. The extracts of
Fructus crataegi were reported to have a radio-protective effect with antioxidant activity [
12,
13] and protected lymphocytes from the effects of radiation [
10].
Citri reticulatae pericarpium possessed various pharmacological effects involved in antioxidant ability against hydroxyl-induced DNA damage [
9].
Anemarrhenae Rhizoma showed various bioactivities, such as anti-tumor, anti-oxidation, anti-microbial, anti-virus, anti-inflammation, anti-osteoporosis, anti-skin aging and damage effects, as well as other activities [
18]. The combination of different types of medical herbs above in HSC can benefit from each other with different roles in the formula, and ultimately gain the goal of enhancing efficacy, which caters to the core thinking of traditional Chinese medicine theory.
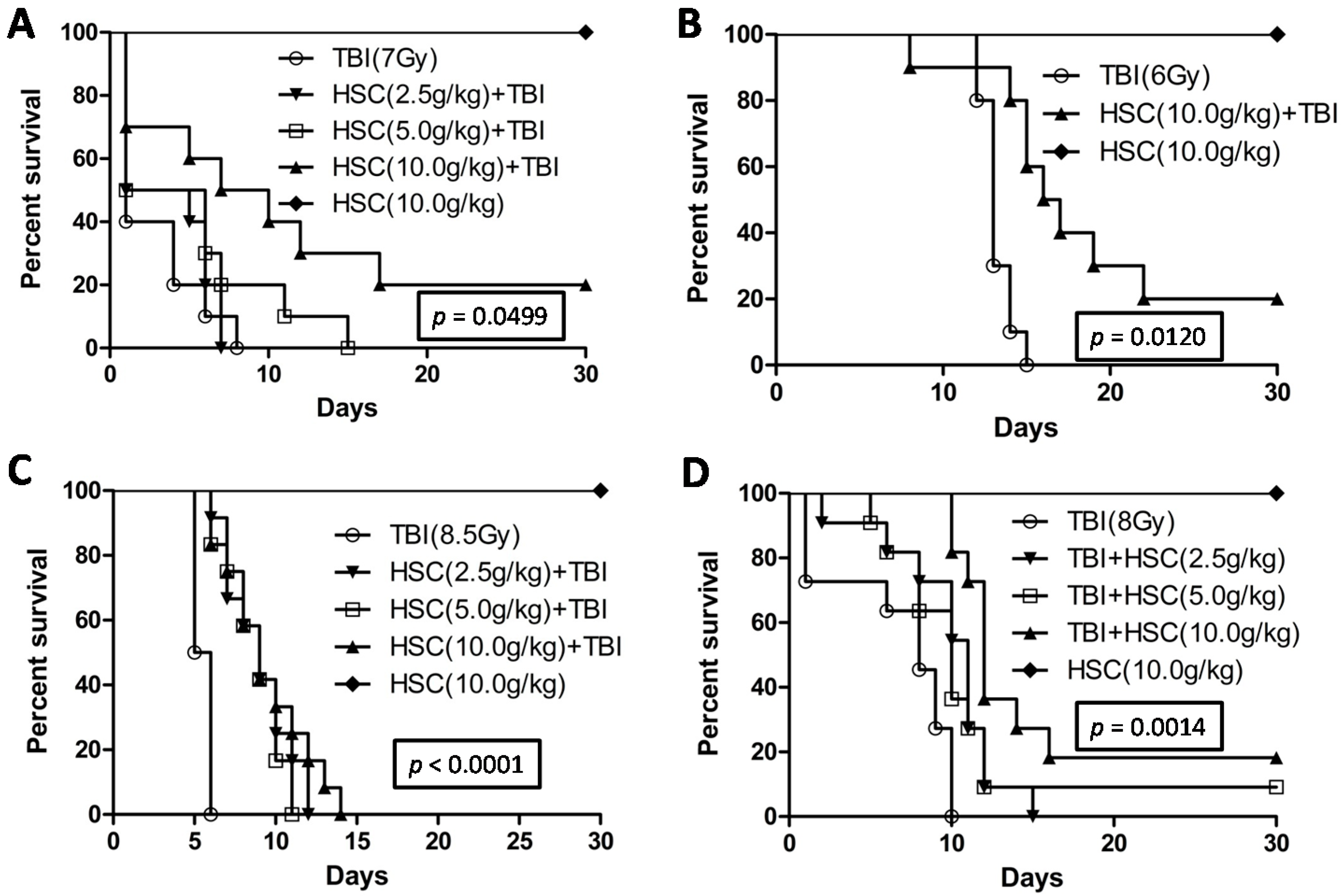
The present study revealed that pre-administration of HSC reduced the radiation sickness characteristics and improved the 30-day survival of the irradiated rats and mice (
Figure 1). In addition, the administration of HSC also increased the 30-day survival of mice after exposure to lethal TBI (
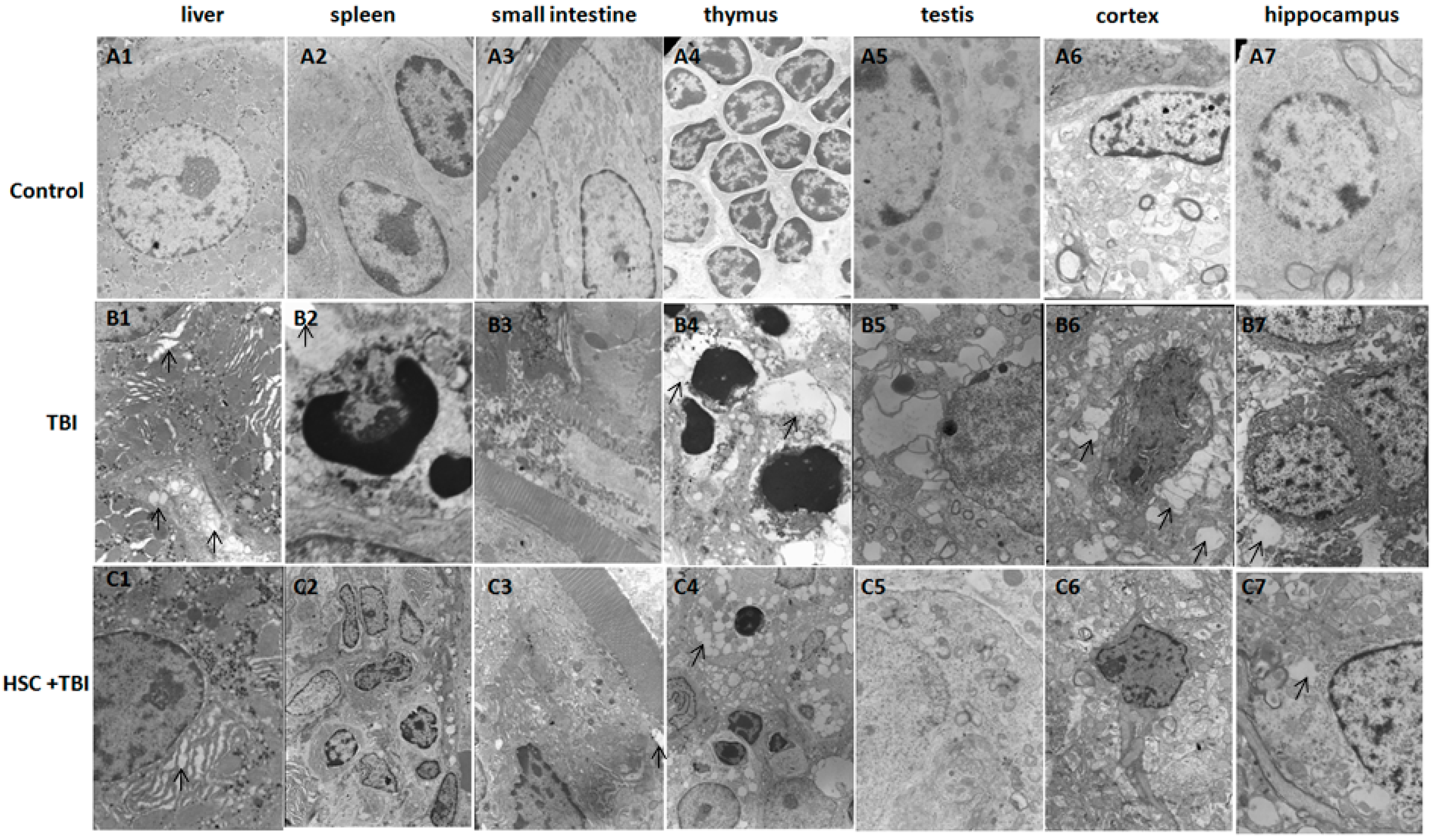
Figure 1D). Our study clearly demonstrated that acute, lethal TBI of different organs was associated with significant ultrastructural changes indicative of cell damage. It also indicated that the administration of HSC was able to minimize these changes (
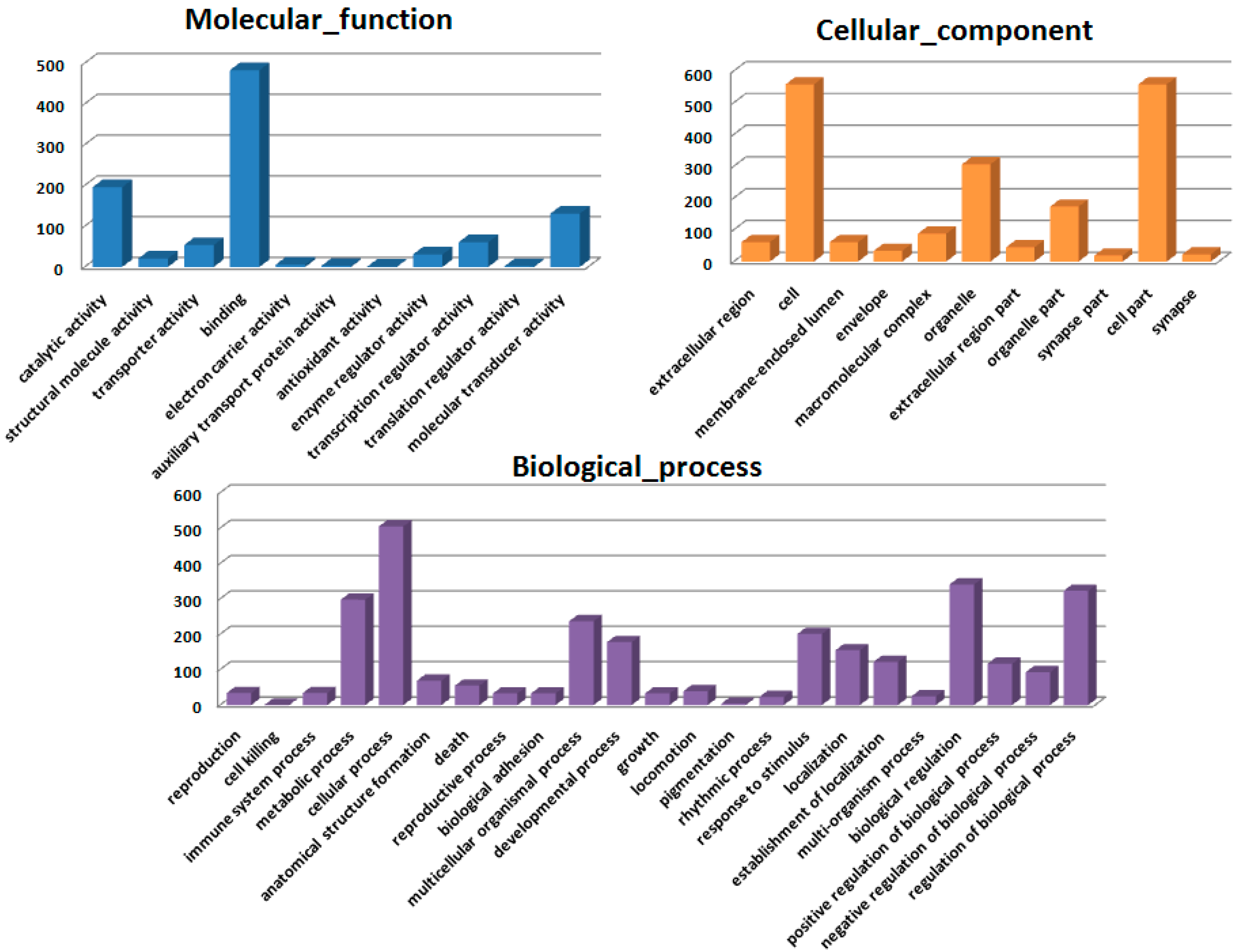
Figure 2), confirming the protective effect of HSC on radiation-induced tissue injury. Gene expression profiles revealed a dramatic effect of HSC on alterations of gene expression caused by lethal TBI. Pretreatment with HSC prevented the differential expression of 66% (1398 genes) of 2126 genes differentially expressed in response to radiation. We focused on these genes regulated by radiation and the alterations of expression were abolished or attenuated by HSC pretreatment, as these genes might be regulated by HSC and involved in the protective effects on radiation. GO ontology analysis indicated that these genes were mainly involved in the highly enriched terms such as binding and catalytic activity for molecular function, cell parts, and cells for cellular components, cellular processes, and biological regulations for biological processes. Pathway enrichment analysis indicated that these genes were mainly involved in a total of 32 pathways (
Table 1). Most of these pathways were reported to be induced by radiation in the previous studies. The pathways include olfactory transduction [
19], cytokine-cytokine receptor interaction [
19], neuroactive ligand-receptor interaction [
20], pathways in cancer [
21,
22,
23], MAPK signaling pathways [
24], PPAR signaling pathways [
25], GnRH signaling pathways [
26], Notch signaling pathways [
17,
27,
28], calcium signaling pathways [
22], Wnt signaling pathways [
29,
30], and Jak-STAT signaling pathways [
31], suggesting that HSC has radio-protective effects mainly or partially through the modulation of these pathways. For example, the Wnt pathway was activated by ionizing radiation [
30]. Here, all the gene expressions involved in the Wnt signaling pathway (
Table 5) were evidently up-regulated by radiation and these up-regulations were abolished or attenuated by pretreatment with HSC. Changes in Notch signaling were previously identified under the action of ionizing radiation [
28]. Here, five genes were involved in the Notch signaling pathway (
Table 6). Four of these gene expressions were evidently up-regulated by radiation and these up-regulations abolished or were attenuated by pretreatment with HSC. In addition, metabolomic studies demonstrated that HSC could restore the metabolic pathways perturbed by radiation, such as fatty acid metabolism, purine metabolism, tryptophan metabolism, and phenylalanine metabolism [
15]. Here, our analysis indicated that the pretreatment of rats with HSC modulated radiation-induced fatty acid metabolism, purine metabolism, glutathione metabolism, starch and sucrose metabolism, and glutathione metabolism, suggesting that HSC might provide protective effects partially through the modulation of these metabolic pathways.
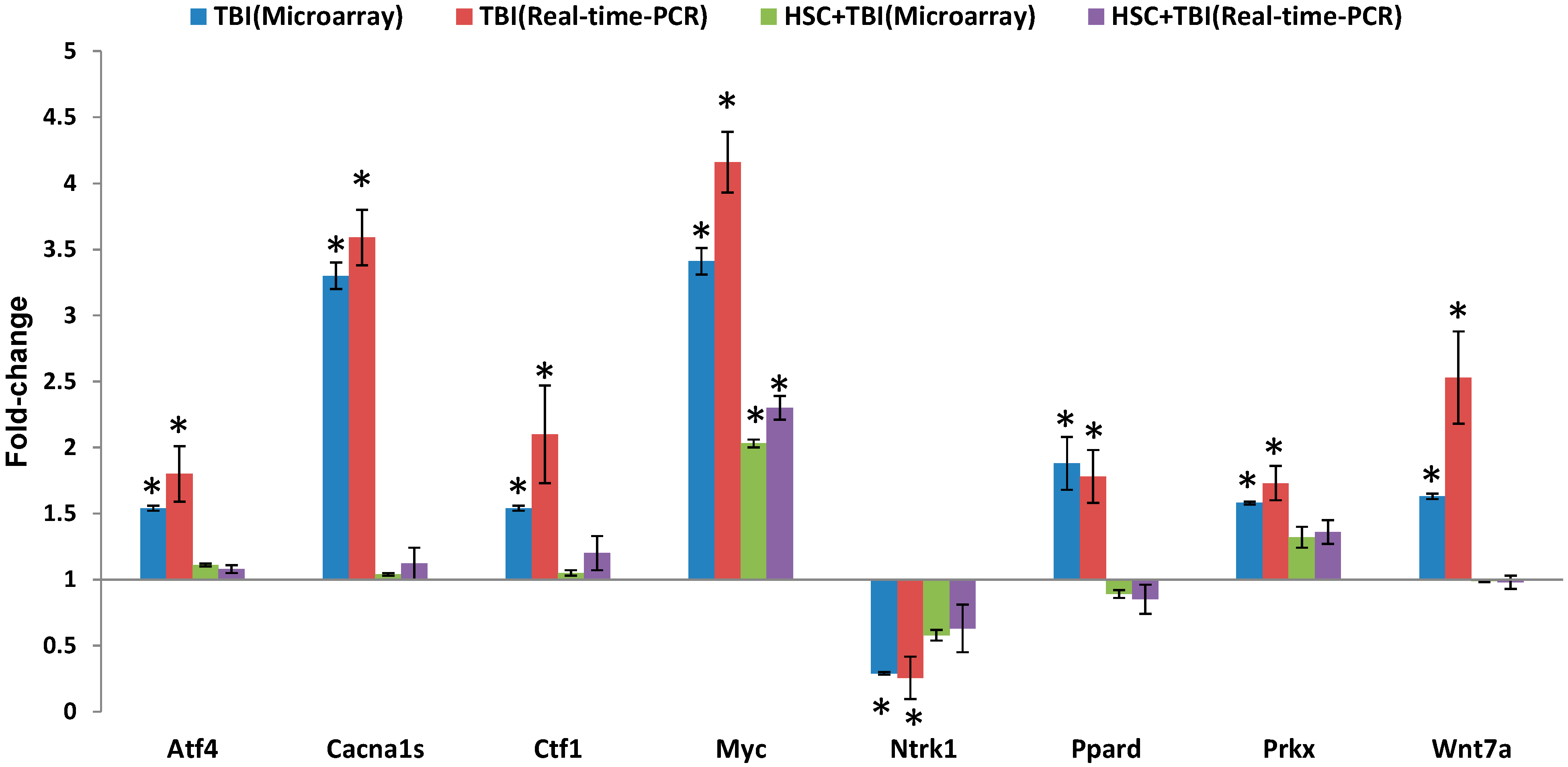
Since ionizing radiation induces the simultaneous compensatory activation of multiple MAPK pathways [
24], we focused on these pathways. These pathways played critical roles in controlling cell survival and repopulation effects following irradiation, in a cell-type-dependent manner [
24]. The 10 genes presented in
Table 2 were involved in the MAPK pathways. Eight of the gene expressions were evidently up-regulated by radiation and these up-regulations were abolished or attenuated by pretreatment with HSC. These radiation-induced genes included myelocytomatosis oncogene (Myc), phospholipase A2, group IIA (Pla2g2a or sPLA2), mitogen-activated protein kinase 4 (Map4k4), protein kinase X-linked (Prkx), voltage-dependent calcium channel R type α 1E subunit (Cacna1e), voltage-dependent calcium channel L type α 1S subunit (Cacna1s), TAO kinase 2 (Taok2), and activating transcription factor 4 (Atf4). In contrast, the expressions of neurotrophic tyrosine kinase receptor type 1 (Ntrk1) and apoptosis signal-regulating kinase 1 (Ask1) were down-regulated by radiation and these down-regulations were abolished or enhanced by pretreatment with HSC. Interestingly, activating transcription factor 4 (Atf4) was a member of the ATF/CREB (activating transcription factor/cyclic AMP response element binding protein) family of basic region-leucine zipper (bZip) transcription factors. Atf4 regulated the expression of genes involved in oxidative stress, amino acid synthesis, differentiation, metastasis, and angiogenesis [
32]. As described by the earlier reports [
33,
34], our data indicated that TBI increased the expression of Atf4. Myc was involved in a wide range of cellular processes including cell-cycle control, metabolism, signal transduction, self-renewal, maintenance of pluripotency, and control of cell fate decisions [
35]. Here, our data indicated that the radiation-induced increase in Myc was attenuated by pretreatment with HSC. This was consistent with previous reports that irradiation significantly increased Myc [
36,
37]. Prkx was a member of an ancient family of cAMP-dependent serine/threonine kinases distinct from the classical PKA, PKB/Akt, PKC, SGK, and PKG families [
38]. As described by the earlier reports [
39,
40], Prkx protein kinase was up-regulated by radiation. Ntrk1 was a member of the neurotrophic tyrosine kinase receptor (NTKR) family. The presence of Ntrk1 led to cell differentiation and might play a role in specifying sensory neuron subtypes [
41,
42]. In UV-irradiated normal skin, there was a significant reduction in Trk A/Ntrk1 tyrosine kinase receptor immunostaining after UV irradiation [
43]. Though our data strongly supported the radiation-induced down-regulation of Ntrk1 [
43], there were also reports on UV-induced up-regulation of both nerve growth factor NGF and its high-affinity receptor Ntrk1 [
44]. cPLA2 was a member of the PLA2 enzyme super-family, which included secretory PLA2 (sPLA2), cytosolic PLA2 (cPLA2), and other members. cPLA2, which activated AA hydrolysis, existed in three isoforms: α, β, and γ. cPLA2-α was known to be a major component of the arachidonate-releasing signal transduction pathway [
45,
46]. Low-level laser irradiation significantly inhibited phospholipase cPLA2-α mRNA expression, which was increased in response to mechanical stress [
45]. Here, our data showed that the radiation-induced increase in sPLA2 was attenuated by pretreatment with HSC. Taken together, these results indicated that the pretreatment of rats with HSC modulated multiple lethal TBI-induced MAPK pathways, suggesting that HSC might provide the protective effects partially through the modulation of MAPK pathways. These findings will help us better understand the protective effects of HSC on lethal TBI damage and provide new insights into the molecular mechanism underlying the radio-protective role of HSC in rats and create a basis for furthering our knowledge about radiation-induced molecular and cellular pathways.
4. Materials and Methods
4.1. Animals
Male ICR mice (20–22 g, 6–8 weeks old) and male Wistar rats (200–220 g) were purchased from SLAC Laboratory Animal Co., Ltd. (SLAC, Shanghai, China) and divided randomly into several groups. Animals were kept under standard laboratory conditions of temperature, pressure, and humidity. Food and water were sterilized by 60Co γ-irradiation and high pressure, respectively. All animal procedures were performed according to protocols approved by Institutional Animal Care and Use Committees of the Second Military Medical University (Shanghai, China). Research was conducted according to the Guide for the Care and Use of Laboratory Animals prepared by the Centre of Laboratory Animals of the Second Military Medical University. All efforts were made to minimize animal suffering.
4.2. Radiation and Administration
The animals were randomly assigned to one of the six following treatment groups (10–12 animals per group): normal control, HSC-High, radiation and HSC-Low, Medium, and High dose (2.5, 5, 10 g crude drug/kg body weight/day) + radiation. HSC was purchased from Shanxi Xiangju Pharmaceutical Co., Ltd. (Shanxi, China). Different doses of HSC dissolved in double-distilled water were administered intragastrically to the male animals for three consecutive days before or after irradiation. All animals, except the normal control group, were placed in specially designed, well-ventilated acrylic containers and subjected to total-body irradiation (TBI). Radiation was delivered by the 60Co source (Radiation facility, the Second Military Medical University, Shanghai, China). Radiation doses were 6, 6.5, 7, 8, and 8.5 Gy at a rate 2 Gy/min. The animals were monitored daily for the development of symptoms of radiation sickness and mortality.
4.3. Survival Assays
Wistar rats (
n = 10 rats/group) were administered HSC before exposure to TBI with 6.0 and 7.0 Gy. ICR mice (
n = 11–12 mice/group) were administered HSC after or before exposure to TBI with 8.0 or 8.5 Gy, respectively. We observed animals twice or three times daily for a period of 30 days to determine survival rates, and moribund animals were euthanized according to humane endpoints. The clinical criteria of moribund is being in the state of dying with no expectation of recovery, where animals display a combination of the following: lowered body temperature, continuous shaking, hunched back, impaired or slow motion, and inability to maintain sternal recumbency [
47]. Moribund animals were placed in a separate cage with CO
2 until no breathing was observed, followed by a cervical dislocation as a secondary confirmatory method of euthanasia. Any surviving animals at the end of the study were also subjected to euthanasia by the application of CO
2 followed by cervical dislocation.
Survival of rats or mice from all the different groups was monitored from the day of onset of the experiment until the 30th day. The probabilities of the survival of all different groups were plotted as Kaplan-Meier survival curves until the 30th day. Significant differences in the survival curves among the different groups were evaluated using the log rank test (Mantel-Cox test) for multiple groups.
4.4. Sample Collection
Twenty-four hours following irradiation, the rats were subjected to anesthesia and then sacrificed by cervical dislocation. Thymus, spleen, liver, small intestine, testis, hippocampus, and cortex were dissected out from each animal. The specimens of the all groups were processed for ultrastructural examination. In addition, liver tissues of the all groups were processed for the microarray experiment.
4.5. Transmission Electron Microscopy
Transmission electron microscopy was performed essentially as described previously [
48]. Biopsy samples were cut and fixed in 2.5% glutaraldehyde in 0.1 M sodium cacodylate, then post-fixed with 1% osmium tetroxide. The specimens were dehydrated through a graded series of ethanol and then embedded in labeled capsules with freshly prepared resin and left to polymerize. Ultrathin sections were cut on a Reichert Ultracut UCT (Leica, Wetzlar, Germany), stained with uranyl acetate and lead citrate and examined by a JEOL 1200EX transmission electron microscopy (Peabody, MA, USA).
4.6. Gene Expression Microarray and Data Analysis
The microarray experiments were performed as described previously [
49]. The 4 × 44 K Whole Rat Genome Oligo Microarray (Agilent Technologies, Santa Clara, CA, USA) was hybridized with Cy3-labeled cRNA using Gene Expression Hybridization Kit (Agilent Technologies) in Hybridization Oven (Agilent Technologies), according to the manufacturer’s instructions. Raw data were obtained by Feature Extraction software 10.7 (Agilent Technologies) and normalized by Quantile algorithm, Gene Spring Software 11.0 (Agilent Technologies). The microarray experiments were conducted at the National Engineering Center for Biochip in Shanghai, China, according to the procedures in the Agilent technical manual. After normalization, genes in the treatment groups with at least 1.5-fold change in expression were considered as up-regulated or down-regulated in comparison to non-treated groups (control). To determine significant proportions of differentially expressed genes within functional groups, the hypergeometric probability
p was calculated.
p < 0.05 was considered significant.
Microarray data analysis was performed using the SBC Analysis system, which is available on the website:
http://sas.ebioservice.com/protal/root/molnet_shbh/index.jsp. The username and password to access this website are available upon request. A general description of the SBC analysis system can be found on the website:
http://www.ebioservice.com/eng/index.asp. The microarray data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database under the accession number GSE57692.
4.7. Quantitative Real-Time (qRT)-PCR Array Validation
qRT-PCR was performed essentially as described previously [
49]. In total, eight genes were chosen for RT-PCR validation. PCR primers (
Table 7) were designed to either span or flank introns by using the ProbeFinder version 2.50 (
http://www.roche-applied-science.com). Data are presented as mean ± SD.
Table 7.
Oligonucleotides primers used in this study for quantitative real-time-PCR analysis. The primer sequences were designed by using the ProbeFinder version 2.49 (
http://www.roche-applied-science.com).
Table 7.
Oligonucleotides primers used in this study for quantitative real-time-PCR analysis. The primer sequences were designed by using the ProbeFinder version 2.49 (http://www.roche-applied-science.com).
| Gene ID | Symbol | Primer | Sequence |
|---|
| 79255 | Atf4 | Forward | tcagacaccggcaaggag |
| Reverse | gtggccaaaagctcatctg |
| 682930 | Cacna1s | Forward | gcgtcgtggagtggaaac |
| Reverse | ctctggcatgggcaggta |
| 24577 | Myc | Forward | gaatttttgtctatttggggaca |
| Reverse | gcatcgtcgtgactgtcg |
| 114850 | Wnt7a | Forward | ccctcatgaacttacacaataacg |
| Reverse | tggcacttacactccagtttcat |
| 501563 | Prkx | Forward | gcctgggcaacatgaaga |
| Reverse | tccacacctcggaaccac |
| 29201 | Ctf1 | Forward | tcctgaccaaatatgcagacc |
| Reverse | agggctctccctgttgct |
| 25682 | Ppard | Forward | ggaccagagcacacccttc |
| Reverse | gaggaaggggaggaattctg |
| 59109 | Ntrk1 | Forward | cagcttctggctgtggcta |
| Reverse | aagtgcaggctggctaggta |
| 81822 | β-Actin | Forward | cccgcgagtacaaccttct |
| Reverse | cgtcatccatggcgaact |
4.8. Statistical Analysis
Data were presented as mean ± SD. A one-way analysis of variance (ANOVA) was used to examine differences among untreated (Control) and treated (Radiation, HSC + Radiation) groups. Differences were considered significant when p < 0.05 or p < 0.01.