Virulence Factors of Erwinia amylovora: A Review
Abstract
:1. Introduction
2. Virulence Factors
2.1. Exopolysaccharides (EPS) Amylovoran and Levan
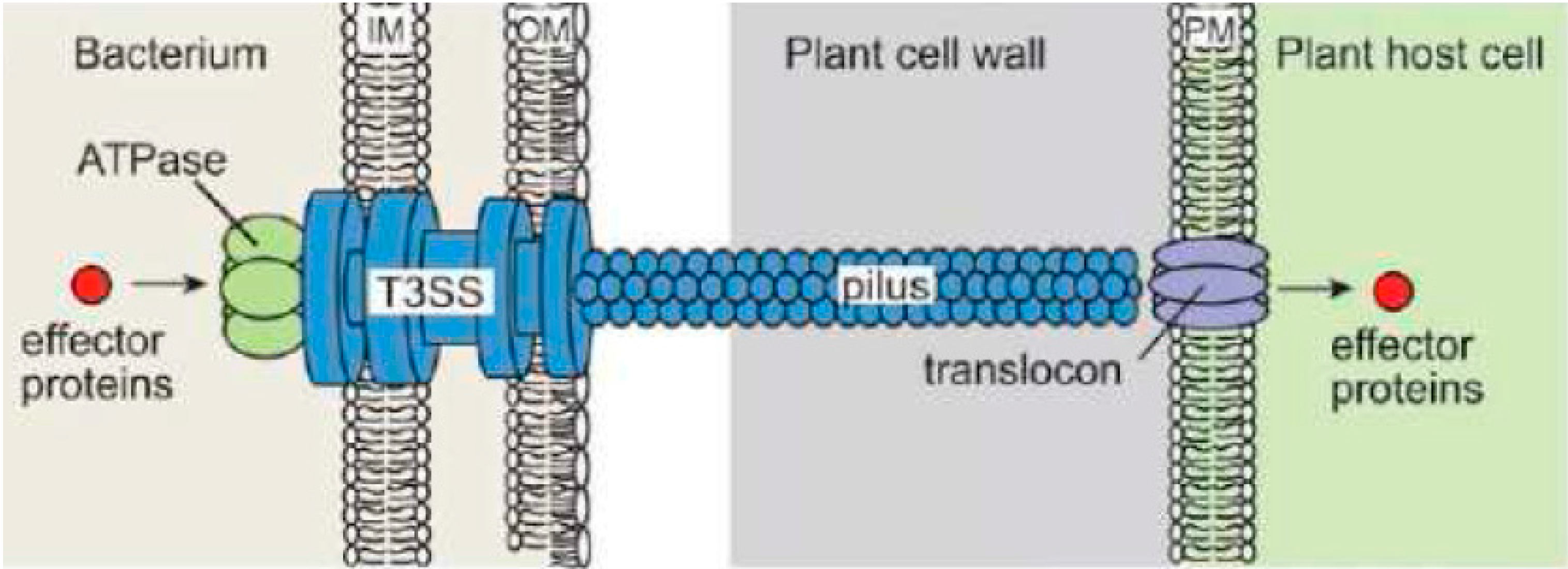
2.2. Type III Secretion System (T3SS)
2.3. Curli
2.4. Biofilm Formation
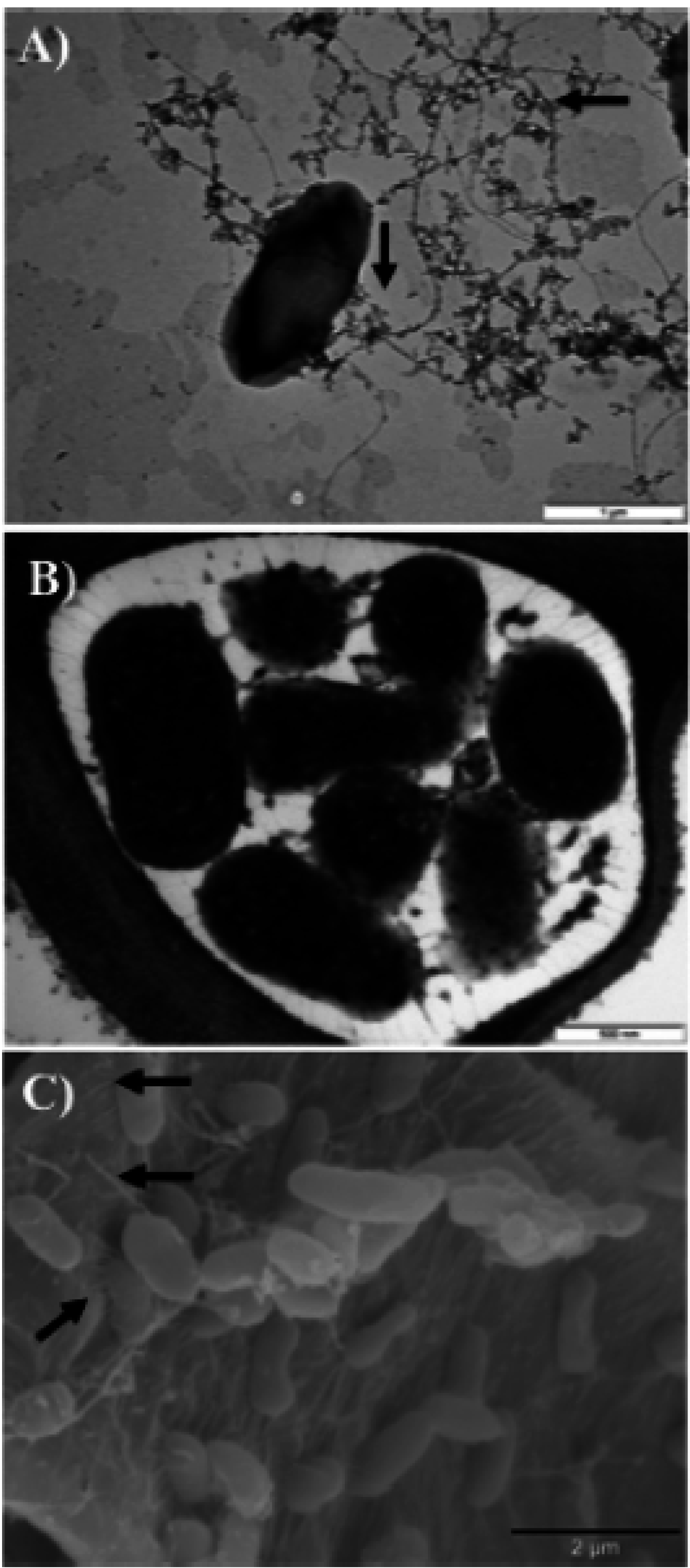
2.5. Motility
2.6. Lipopolysaccharide (LPS)
2.7. Metalloprotease PrtA
2.8. Iron-Scavenging Siderophore Desferrioxamine
2.9. Multidrug Efflux Pump AcrAB
2.10. Other Virulence Factors
3. Genetics
4. Regulation of Virulence Factors
4.1. Two Component Transduction Systems
4.2. Small RNA
4.3. Quorum Sensing
4.4. c-di-GMP
4.5. ppGpp
4.6. Type 3 Effectors
4.7. Hrp X/Y, HrpS and HrpL Cascade Leading to Activation of hrpT3SS
4.8. Regulatory Cascade for Amylovoran Synthesis
5. Discussion
Acknowledgments
Conflicts of Interest
References
- Griffith, C.S.; Sutton, T.B.; Peterson, P.D. Fire Blight: The Foundation of Phytobacteriology; APS Press: St. Paul, MN, USA, 2003. [Google Scholar]
- Hauben, L.; Swings, J. Genus XIII Erwinia. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Brenner, D.J., Krieg, N.R., Staley, J.T., Garrity, G.M., Eds.; Springer: New York, NY, USA, 2005; Volume 2B, pp. 670–679. [Google Scholar]
- Bonn, W.G.; van der Zwet, T. Distribution and economic importance of fire blight. In Fire Blight: The Disease and Its Causative Agent, Erwinia Amylovora; Vanneste, J.L., Ed.; CAB International: Wallingford, CT, USA, 2000; pp. 37–53. [Google Scholar]
- Thomson, S.V. Epidemiology of fire blight. In Fire Blight: The Disease and Its Causative Agent, Erwinia Amylovora; Vanneste, J.L., Ed.; CAB International: Wallingford, UK, 2000; pp. 9–36. [Google Scholar]
- Born, Y.; Fieseler, L.; Klumpp, J.; Eugster, M.R.; Zurfluh, K.; Duffy, B.; Loessner, M.J. The tail-associated depolymerase of Erwinia amylovora phage L1 mediates host cell adsorption and enzymatic capsule removal, which can enhance infection by other phage. Environ. Microbiol. 2014, 16, 2168–2180. [Google Scholar] [CrossRef] [PubMed]
- Vanneste, J.L. Fire blight: The disease and its causative agent. In Erwinia Amylovora; CABI Publishing: Oxfordshire, UK, 2000. [Google Scholar]
- Van der Zwet, T.; Orolaza-Halbrendt, N.; Zeller, W. Fire Blight: History, Biology, and Management; APS Press: St. Paul, MN, USA, 2012. [Google Scholar]
- Santander, R.D.; Catala-Senent, J.F.; Marco-Noales, E.; Biosca, E.G. In planta recovery of Erwinia amylovora viable but nonculturable cells. Trees 2012, 26, 75–82. [Google Scholar] [CrossRef]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [PubMed]
- McManus, P.S.; Stockwell, V.O.; Sundin, G.W.; Jones, A.L. Antibiotic use in plant agriculture. Annu. Rev. Phytopathol. 2002, 40, 443–465. [Google Scholar] [CrossRef] [PubMed]
- Duffy, B.; Schärer, H.-J.; Bünter, M.; Klay, A.; Holliger, E. Regulatory measures against Erwinia amylovora in Switzerland. EPPO Bull. 2005, 35, 239–244. [Google Scholar] [CrossRef]
- Stockwell, V.O.; Duffy, B. Use of antibiotics in plant agriculture. Rev. Sci. Tech. 2012, 31, 199–210. [Google Scholar] [PubMed]
- Johnson, K.B.; Stockwell, V.O. Management of fire blight: A case study in microbial ecology. Annu. Rev. Phytopathol. 1998, 36, 227–248. [Google Scholar] [CrossRef] [PubMed]
- Norelli, J.L.; Jones, A.L.; Aldwinckle, H.S. Fire blight management in the twenty-first century: Using new technologies that enhance host resistance in apple. Plant Dis. 2003, 87, 756–765. [Google Scholar] [CrossRef]
- Zeng, Q.; Sundin, G.W. Genome-wide identification of Hfq-regulated small RNAs in the fire blight pathogen Erwinia amylovora discovered small RNAs with virulence regulatory function. BMC Genomics 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Thomson, S.V. The role of the stigma in fire blight infections. Phytopathology 1986, 76, 476–482. [Google Scholar] [CrossRef]
- Dellagi, A.; Brisset, M.N.; Paulin, J.P.; Expert, D. Dual role of desferrioxamine in Erwinia amylovora pathogenicity. Mol. Plant Microbe Interact. 1998, 11, 734–742. [Google Scholar] [CrossRef] [PubMed]
- Malnoy, M.; Martens, S.; Norelli, J.L.; Barny, M.A.; Sundin, G.W.; Smits, T.H.; Duffy, B. Fire blight: Applied genomic insights of the pathogen and host. Annu. Rev. Phytopathol. 2012, 50, 475–494. [Google Scholar] [CrossRef] [PubMed]
- Koczan, J.M.; Lenneman, B.R.; McGrath, M.J.; Sundin, G.W. Cell surface attachment structures contribute to biofilm formation and xylem colonization by Erwinia amylovora. Appl. Environ. Microbiol. 2011, 77, 7031–7039. [Google Scholar] [CrossRef] [PubMed]
- Mann, R.A.; Smits, T.H.; Bühlmann, A.; Blom, J.; Goesmann, A.; Frey, J.E.; Plummer, K.M.; Beer, S.V.; Luck, J.; Duffy, B.; et al. Comparative genomics of 12 strains of Erwinia amylovora identifies a pan-genome with a large conserved core. PLoS ONE 2013, 8, e55644. [Google Scholar] [CrossRef] [PubMed]
- Koczan, J.M.; McGrath, M.J.; Zhao, Y.; Sundin, G.W. Contribution of Erwinia amylovora exopolysaccharides amylovoran and levan to biofilm formation: Implications in pathogenicity. Phytopathology 2009, 99, 1237–1244. [Google Scholar] [CrossRef] [PubMed]
- Burse, A.; Weingart, H.; Ullrich, M.S. NorM, an Erwinia amylovora multidrug efflux pump involved in in vitro competition with other epiphytic bacteria. Appl. Environ. Microbiol. 2004, 70, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Ordax, M.; Marco-Noales, E.; Lopez, M.M.; Biosca, E.G. Exopolysaccharides favor the survival of Erwinia amylovora under copper stress through different strategies. Res. Microbiol. 2010, 161, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Vrancken, K.; Holtappels, M.; Schoofs, H.; Deckers, T.; Valcke, R. Pathogenicity and infection strategies of the fire blight pathogen Erwinia amylovora in Rosaceae: State of the art. Microbiology 2013, 159, 823–832. [Google Scholar] [CrossRef] [PubMed]
- Bellemann, P.; Bereswill, S.; Berger, S.; Geider, K. Visualization of capsule formation by Erwinia amylovora and assays to determine amylovoran synthesis. Int. J. Biol. Macromol. 1994, 16, 290–296. [Google Scholar] [CrossRef]
- Ramey, B.E.; Koutsoudis, M.; von Bodman, S.B.; Fuqua, C. Biofilm formation in plant-microbe associations. Curr. Opin. Microbiol. 2004, 7, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Maes, M.; Orye, K.; Bobev, S.; Devreese, B.; van Beeumen, J.; de Bruyn, A.; Busson, R.; Herdewijn, P.; Morreel, K.; Messens, E. Influence of amylovoran production on virulence of Erwinia amylovora and a different amylovoran structure in E. amylovora isolates from Rubus. Eur. J. Plant Pathol. 2001, 107, 839–844. [Google Scholar] [CrossRef]
- Nimtz, M.; Mort, A.; Domke, T.; Wray, V.; Zhang, Y.; Qiu, F.; Coplin, D.; Geider, K. Structure of amylovoran, the capsular exopolysaccharide from the fire blight pathogen Erwinia amylovora. Carbohydr. Res. 1996, 287, 59–76. [Google Scholar] [CrossRef]
- Schollmeyer, M.; Langlotz, C.; Huber, A.; Coplin, D.L.; Geider, K. Variations in the molecular masses of the capsular exopolysaccharides amylovoran, pyrifolan and stewartan. Int. J. Biol. Macromol. 2012, 50, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Bellemann, P.; Geider, K. Localization of transposon insertions in pathogenicity mutants of Erwinia amylovora and their biochemical characterization. J. Gen. Microbiol. 1992, 138, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Bugert, P.; Geider, K. Characterization of the amsI gene product as a low molecular weight acid phosphatase controlling exopolysaccharide synthesis of Erwinia amylovora. FEBS Lett. 1997, 400, 252–256. [Google Scholar] [CrossRef]
- Eastgate, J.A. Erwinia amylovora: The molecular basis of fire blight disease. Mol. Plant Pathol. 2000, 1, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Langlotz, C.; Schollmeyer, M.; Coplin, D.; Nimtz, M.; Geider, K. Biosynthesis of the repeating units of the exopolysaccharides amylovoran from Erwinia amylovora and steewartan from Pantoea stewartii. Physiol. Mol. Plant Pathol. 2011, 75, 163–169. [Google Scholar] [CrossRef]
- Gross, M.; Geier, G.; Rudolph, K.; Geider, K. Levan and levansucrase synthesized by the fire blight pathogen Erwinia amylovora. Physiol. Mol. Plant Pathol. 1992, 40, 371–381. [Google Scholar] [CrossRef]
- Geier, G.; Geider, K. Characterization and influence on virulence of the levansucrase gene from the fireblight pathogen Erwinia amylovora. Phys. Mol. Plant Pathol. 1993, 42, 387–404. [Google Scholar] [CrossRef]
- Khokhani, D.; Zhang, C.; Li, Y.; Wang, Q.; Zeng, Q.; Yamazaki, A.; Hutchins, W.; Zhou, S.S.; Chen, X.; Yang, C.H. Discovery of plant phenolic compounds that act as type III secretion system inhibitors orinducers of the fire blight pathogen, Erwinia amylovora. Appl. Environ. Microbiol. 2013, 79, 5424–5436. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Hu, W.; Brown, I.; McGhee, G.; Hart, P.; Jones, A.L.; He, S.Y. Visualization of secreted Hrp and Avr proteins along the Hrp pilus during type III secretion in Erwinia amylovora and Pseudomonas syringae. Mol. Microbiol. 2001, 40, 1129–1139. [Google Scholar] [CrossRef] [PubMed]
- Oh, C.-S.; Beer, S.V. Molecular genetics of Erwinia amylovora involved in the development of fire blight. FEMS Microbiol. Lett. 2005, 253, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Büttner, D.; He, S.Y. Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 2009, 150, 1656–1664. [Google Scholar] [CrossRef] [PubMed]
- Barnhart, M.M.; Chapman, M.R. Curli biogenesis and function. Annu. Rev. Microbiol. 2006, 60, 131–147. [Google Scholar] [CrossRef] [PubMed]
- Epstein, E.A.; Chapman, M.R. Polymerizing the fibre between bacteria and host cells: The biogenesis of functional amyloid fibres. Cell Microbiol. 2008, 10, 1413–1420. [Google Scholar] [CrossRef] [PubMed]
- Sebaihia, M.; Bocsanczy, A.M.; Biehl, B.S.; Quail, M.A.; Perna, N.T.; Glasner, J.D.; DeClerck, G.A.; Cartinhour, S.; Schneider, D.J.; Bentley, S.D.; et al. Complete genome sequence of the plant pathogen Erwinia amylovora strain ATCC 49946. J. Bacteriol. 2010, 192, 2020–2021. [Google Scholar] [CrossRef] [PubMed]
- Smits, T.H.; Rezzonico, F.; Kamber, T.; Blom, J.; Goesmann, A.; Frey, J.E.; Duffy, B. Complete genome sequence of the fire blight pathogen Erwinia amylovora CFBP 1430 and comparison to other Erwinia spp. Mol. Plant Microbe Interact. 2010, 23, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.E.; O’Toole, G.A. Microbial biofilms: From ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 2000, 64, 847–867. [Google Scholar] [CrossRef] [PubMed]
- Sauer, K.; Camper, A.K.; Ehrlich, G.D.; Costerton, J.W.; Davies, D.G. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 2002, 184, 1140–1154. [Google Scholar] [CrossRef] [PubMed]
- Santander, R.D.; Oliver, J.D.; Biosca, E.G. Cellular, physiological, and molecular adaptive responses of Erwinia amylovora to starvation. FEMS Microbiol. Ecol. 2014, 88, 258–271. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, D.; Nakka, S.; Sundin, G.W.; Korban, S.S. Systems level analysis of two-component signal transduction systems in Erwinia amylovora: Role in virulence, regulation of amylovoran biosynthesis and swarming motility. BMC Genomics 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- Rezzonico, F.; Braun-Kiewnick, A.; Mann, R.A.; Goesmann, A.; Rodoni, B.; Duffy, B.; Smits, T.H.M. Lipopolysaccharide biosynthesis genes discriminate between Rubus- and Spiraeoideae-infective genotypes of Erwinia amylovora. Mol. Plant Pathol. 2012, 13, 975–984. [Google Scholar] [CrossRef] [PubMed]
- Fukuoka, S.; Brandenburg, K.; Müller, M.; Lindner, B.; Koch, M.H.; Seydel, U. Physico-chemical analysis of lipid A fractions of lipopolysaccharide from Erwinia carotovora in relation to bioactivity. Biochim. Biophys. Acta 2001, 1510, 185–197. [Google Scholar] [CrossRef]
- Zhang, Y.; Bak, D.D.; Heid, H.; Geider, K. Molecular characterization of a protease secreted by Erwinia amylovora. J. Mol. Biol. 1999, 289, 1239–1251. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.W.; Chen, X.L. Extracellular metalloproteases from bacteria. Appl. Microbiol. Biotechnol. 2011, 92, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Vanneste, J.L. Pathogenesis and host specificity in plant diseases: Histopathological, biochemical, genetic and molecular bases. In Procaryotes; Singh, U.S., Singh, R.P., Kohmoto, K., Eds.; Pergamon Press: Oxford, UK, 1995; Volume 1, pp. 21–41. [Google Scholar]
- Feistner, G.J.; Stahl, D.C.; Gabrik, A.H. Proferrioxamine siderophores of Erwinia amylovora. A capillary liquid chromatographic/electrospray tandem mass spectrometry study. Org. Mass Spectrom. 1993, 28, 163–175. [Google Scholar] [CrossRef]
- Kachadourian, R.; Dellagi, A.; Laurent, J.; Bricard, L.; Kunesch, G.; Expert, D. Desferrioxamine-dependent iron transport in Erwinia amylovora CFBP 1430: Cloning of the gene encoding the ferrioxamine receptor FoxR. BioMetals 1996, 9, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Aldridge, P.; Metzger, M.; Geider, K. Genetics of sorbitol metabolism in Erwinia amylovora and its influence on bacterial virulence. Mol. Gen. Genet. 1997, 256, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Grant, C.R.; Rees, T. Sorbitol metabolism by apple seedlings. Phytochemistry 1981, 20, 1505–1511. [Google Scholar] [CrossRef]
- Wallart, R.A. Distribution of sorbitol in Rosaceae. Phytochemistry 1980, 19, 2603–2610. [Google Scholar] [CrossRef]
- Bogs, J.; Geider, K. Molecular analysis of sucrose metabolism of Erwinia amylovora and influence on bacterial virulence. J. Bacteriol. 2000, 182, 5351–5358. [Google Scholar] [CrossRef] [PubMed]
- Dreo, T.; Ravnikar, M.; Frey, J.E.; Smits, T.; Duffy, B. In silico analysis of variable number of tandem repeats in Erwinia amylovora genomes. Acta Hortic. 2011, 896, 115–118. [Google Scholar]
- Rezzonico, F.; Smits, T.H.; Duffy, B. Diversity, evolution, and functionality of clustered regularly interspaced short palindromic repeat (CRISPR) regions in the fire blight pathogen Erwinia amylovora. Appl. Environ. Microbiol. 2011, 77, 3819–3829. [Google Scholar] [CrossRef] [PubMed]
- McManus, P.S.; Jones, A.L. Genetic fingerprinting of Erwinia amylovora strains isolated from tree-fruit crops and Rubus spp. Phytopathology 1995, 85, 1547–1553. [Google Scholar] [CrossRef]
- Smits, T.H.; Rezzonico, F.; Duffy, B. Evolutionary insights from Erwinia amylovora genomics. J. Biotechnol. 2011, 155, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Smits, T.H.; Guerrero-Prieto, V.M.; Hernández-Escarcega, G.; Blom, J.; Goesmann, A.; Rezzonico, F.; Duffy, B.; Stockwell, V.O. Whole-Genome Sequencing of Erwinia amylovora Strains from Mexico Detects Single Nucleotide Polymorphisms in rpsL Conferring Streptomycin Resistance and in the avrRpt2 Effector Altering Host Interactions. Genome Announc. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- De Léon-Door, A.P.; Romo Chacón, A.; Acosta Muñiz, C. Detection of streptomycin resistance in Erwinia amylovora strains isolated from Apple ochards in Chihuahua, Mexico. Eur. J. Plant Pathol. 2013, 137, 223–229. [Google Scholar] [CrossRef]
- Asselin, J.E.; Bonasera, J.M.; Kim, J.F.; Oh, C.S.; Beer, S.V. Eop1 from a Rubus strain of Erwinia amylovora functions as a host-range limiting factor. Phytopathology 2011, 101, 935–944. [Google Scholar] [CrossRef] [PubMed]
- Mann, R.A.; Blom, J.; Bühlmann, A.; Plummer, K.M.; Beer, S.V.; Luck, J.E.; Goesmann, A.; Frey, J.E.; Rodoni, B.C.; Duffy, B.; et al. Comparative analysis of the Hrp pathogenicity island of Rubus- and Spiraeoideae-infecting Erwinia amylovora strains identifies the IT region as a remnant of an integrative conjugativeelement. Gene 2012, 504, 6–12. [Google Scholar] [CrossRef] [PubMed]
- Egan, F.; Reen, F.J.; O’Gara, F. Tle Distribution and Diversity in Metagenomic Datasets Reveals Niche Specialisation. Environ. Microbiol. Rep. 2015, 7, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Nakka, S.; Qi, M.; Zhao, Y. The PmrAB system in Erwinia amylovora renders the pathogen more susceptible to polymyxin B and more resistant to excess iron. Res. Microbiol. 2010, 161, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Molina, L.; Rezzonico, F.; Défago, G.; Duffy, B. Autoinduction in Erwinia amylovora: Evidence of an acyl-homoserine lactone signal in the fire blight pathogen. J. Bacteriol. 2005, 187, 3206–3213. [Google Scholar] [CrossRef] [PubMed]
- Venturi, V.; Venuti, C.; Devescovi, G.; Lucchese, C.; Friscina, A.; Degrassi, G.; Aguilar, C.; Mazzucchi, U. The plant pathogen Erwinia amylovora produces acyl-homoserine lactone signal molecules in vitro and in planta. FEMS Microbiol. Lett. 2004, 241, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Rezzonico, F.; Duffy, B. The role of luxS in the fire blight pathogen Erwinia amylovora is limited to metabolism and does not involve quorum sensing. Mol. Plant Microbe Interact. 2007, 20, 1284–1297. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Song, J.; Hu, B.; Zhang, L.; Liu, Q.; Liu, F. The luxS gene is involved in AI-2 production, pathogenicity, and some phenotypes in Erwinia amylovora. Curr. Microbiol. 2009, 58, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Castiblanco, L.F.; Edmunds, A.C.; Waters, C.M.; Sundin, G.W. Characterization of quorum sensing and cyclic-di-GMP signaling systems in Erwinia amylovora. Phytopathology 2011, 101, S2. [Google Scholar]
- Edmunds, A.C.; Castiblanco, L.F.; Sundin, G.W.; Waters, C.M. Cyclic Di-GMP modulates the disease progression of Erwinia amylovora. J. Bacteriol. 2013, 195, 2155–2165. [Google Scholar] [CrossRef] [PubMed]
- Ancona, V.; Lee, J.H.; Chatnaparat, T.; Oh, J.; Hong, J.I.; Zhao, Y. The Bacterial Alarmone (p)ppGpp Activates the Type III Secretion System in Erwinia amylovora. J. Bacteriol. 2015, 197, 1433–1443. [Google Scholar] [CrossRef] [PubMed]
- Siamer, S.; Guillas, I.; Shimobayashi, M.; Kunz, C.; Hall, M.N.; Barny, M.A. Expression of the bacterial type III effector DspA/E in Saccharomyces cerevisiae down-regulates the sphingolipid biosynthetic pathway leading to growth arrest. J. Biol. Chem. 2014, 289, 18466–18477. [Google Scholar] [CrossRef] [PubMed]
- Siamer, S.; Patrit, O.; Fagard, M.; Belgareh-Touzé, N.; Barny, M.A. Expressing the Erwinia amylovora type III effector DspA/E in the yeast Saccharomyces cerevisiae strongly alters cellular trafficking. FEBS Open Biol. 2011, 1, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Degrave, A.; Moreau, M.; Launay, A.; Barny, M.A.; Brisset, M.N.; Patrit, O.; Taconnat, L.; Vedel, R.; Fagard, M. The bacterial effector DspA/E is toxic in Arabidopsis thaliana and is required for multiplication and survival of fire blight pathogen. Mol. Plant Pathol. 2013, 14, 506–517. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Kim, J.F.; Beer, S.V. Regulation of hrp genes and type III protein secretion in Erwinia amylovora by HrpX/HrpY, a novel two-component system, and HrpS. Mol. Plant Microbe Interact. 2000, 13, 1251–1262. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.M.; Beer, S.V. hrpL activates Erwinia amylovora hrp gene transcription and is a member of the ECF subfamily of s factors. J. Bacteriol. 1995, 177, 6201–6210. [Google Scholar] [PubMed]
- Pal, K.K.; McSpadden Gardener, B. Biological control of plant pathogens. Plant Health Instr. 2006. [Google Scholar] [CrossRef]
- Choi, S.K.; Jeong, H.; Kloepper, J.W.; Ryu, C.M. Genome Sequence of Bacillus amyloliquefaciens GB03, an Active Ingredient of the First Commercial Biological Control Product. Genome Announc. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piqué, N.; Miñana-Galbis, D.; Merino, S.; Tomás, J.M. Virulence Factors of Erwinia amylovora: A Review. Int. J. Mol. Sci. 2015, 16, 12836-12854. https://doi.org/10.3390/ijms160612836
Piqué N, Miñana-Galbis D, Merino S, Tomás JM. Virulence Factors of Erwinia amylovora: A Review. International Journal of Molecular Sciences. 2015; 16(6):12836-12854. https://doi.org/10.3390/ijms160612836
Chicago/Turabian StylePiqué, Núria, David Miñana-Galbis, Susana Merino, and Juan M. Tomás. 2015. "Virulence Factors of Erwinia amylovora: A Review" International Journal of Molecular Sciences 16, no. 6: 12836-12854. https://doi.org/10.3390/ijms160612836
APA StylePiqué, N., Miñana-Galbis, D., Merino, S., & Tomás, J. M. (2015). Virulence Factors of Erwinia amylovora: A Review. International Journal of Molecular Sciences, 16(6), 12836-12854. https://doi.org/10.3390/ijms160612836