Abnormal Mitochondrial Function and Impaired Granulosa Cell Differentiation in Androgen Receptor Knockout Mice
Abstract
:1. Introduction
2. Results
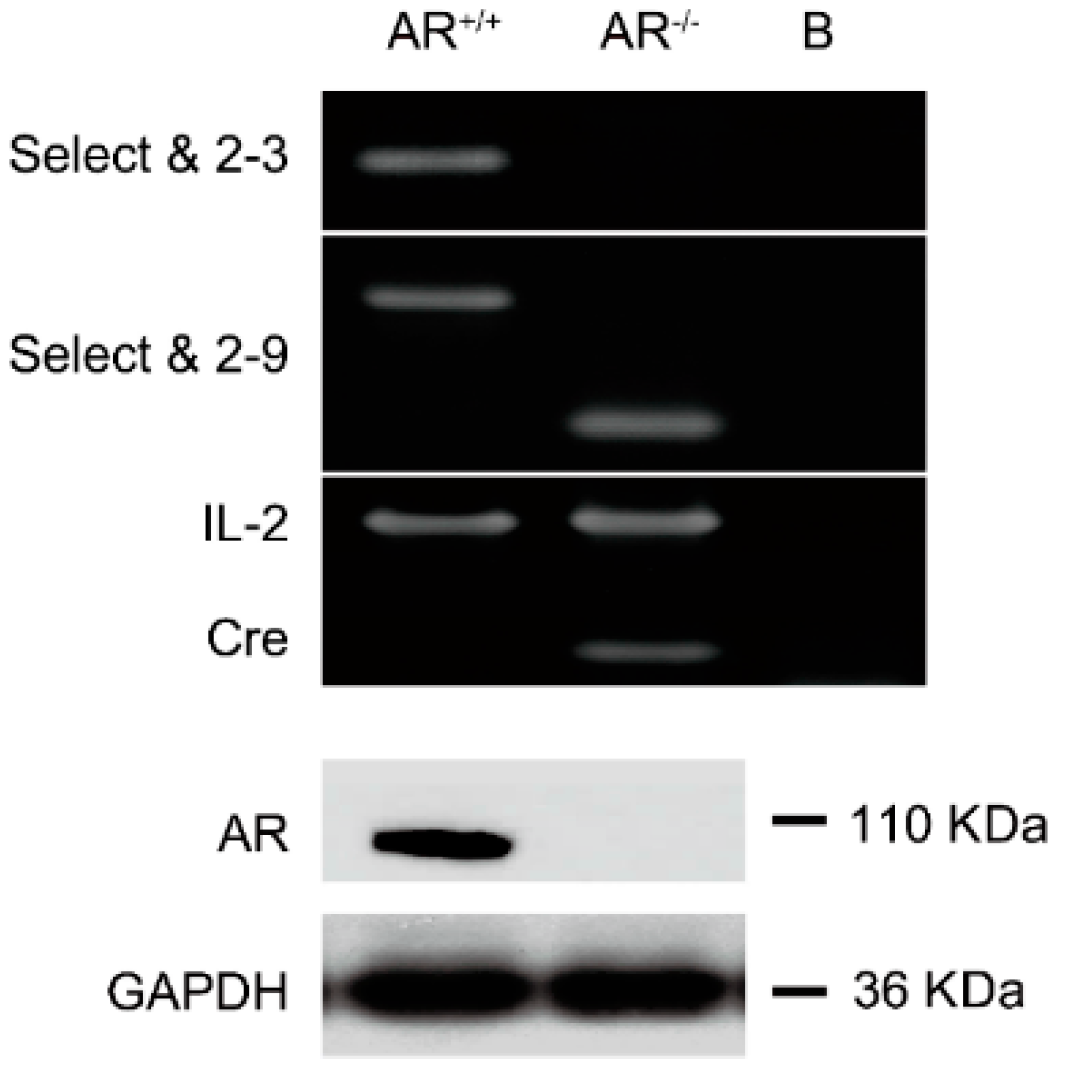
2.1. Confirmation of Knockout Androgen Receptor (AR) in the Ovaries in the AR−/− Mice
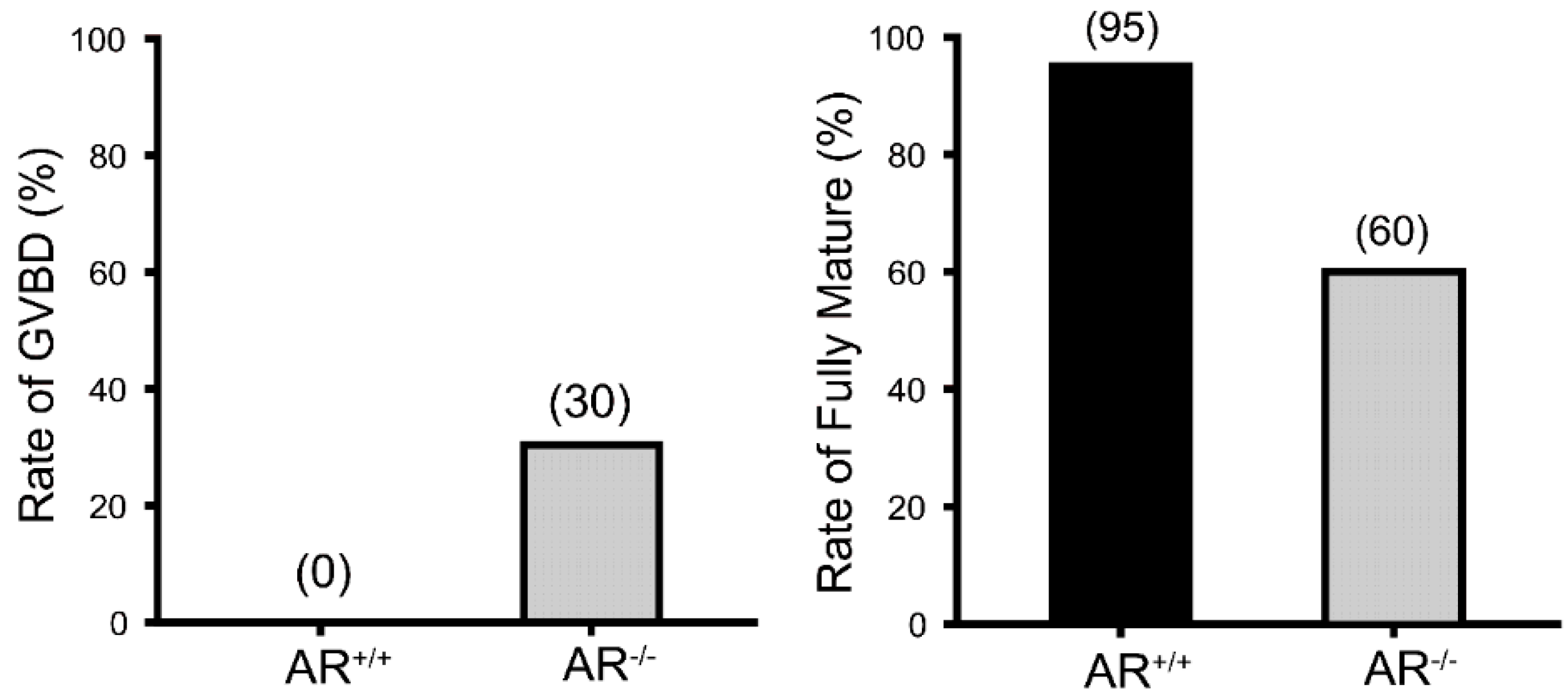
2.2. Reduced Oocyte in Vitro Maturation Rate in AR−/− Mice
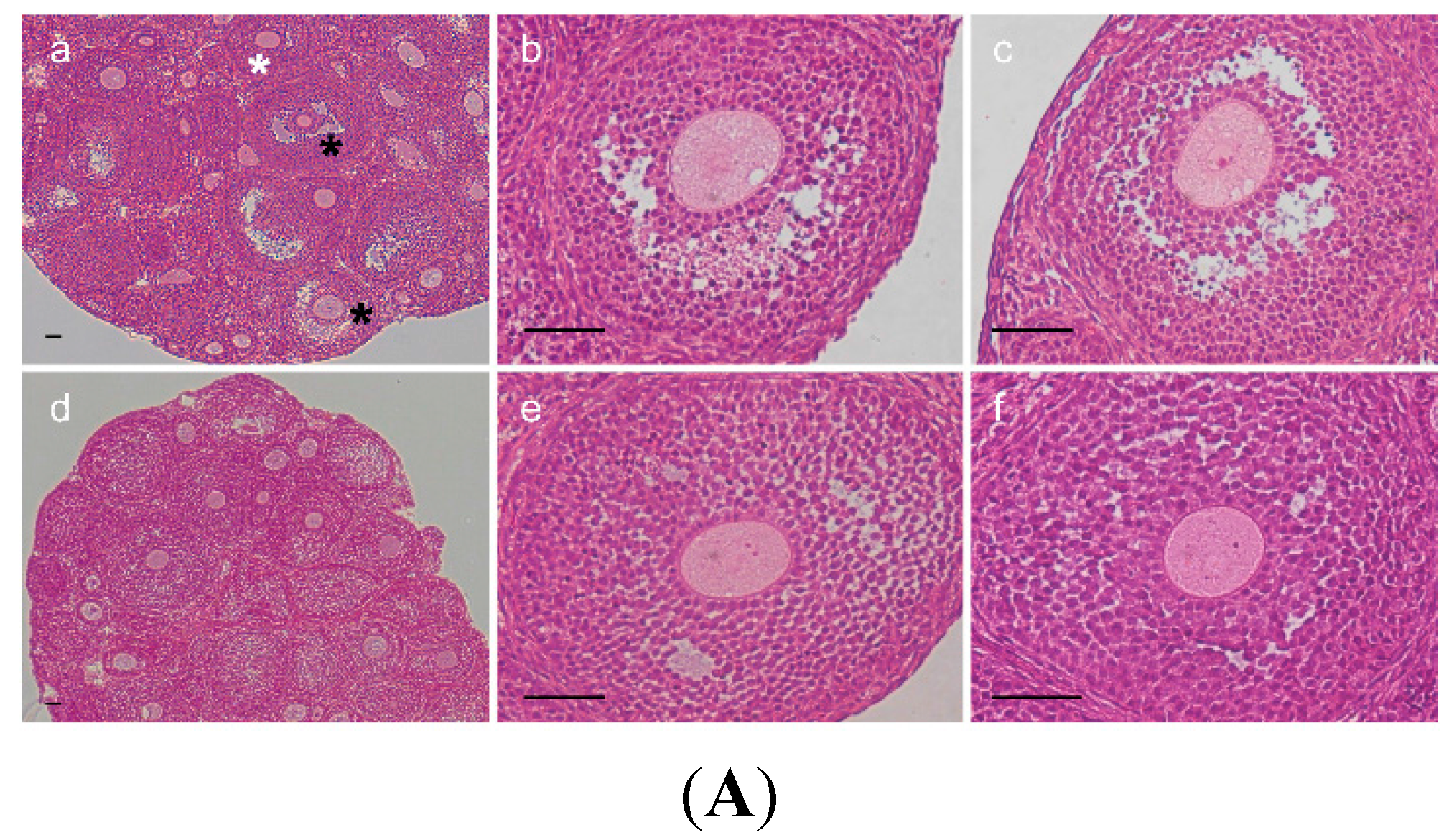
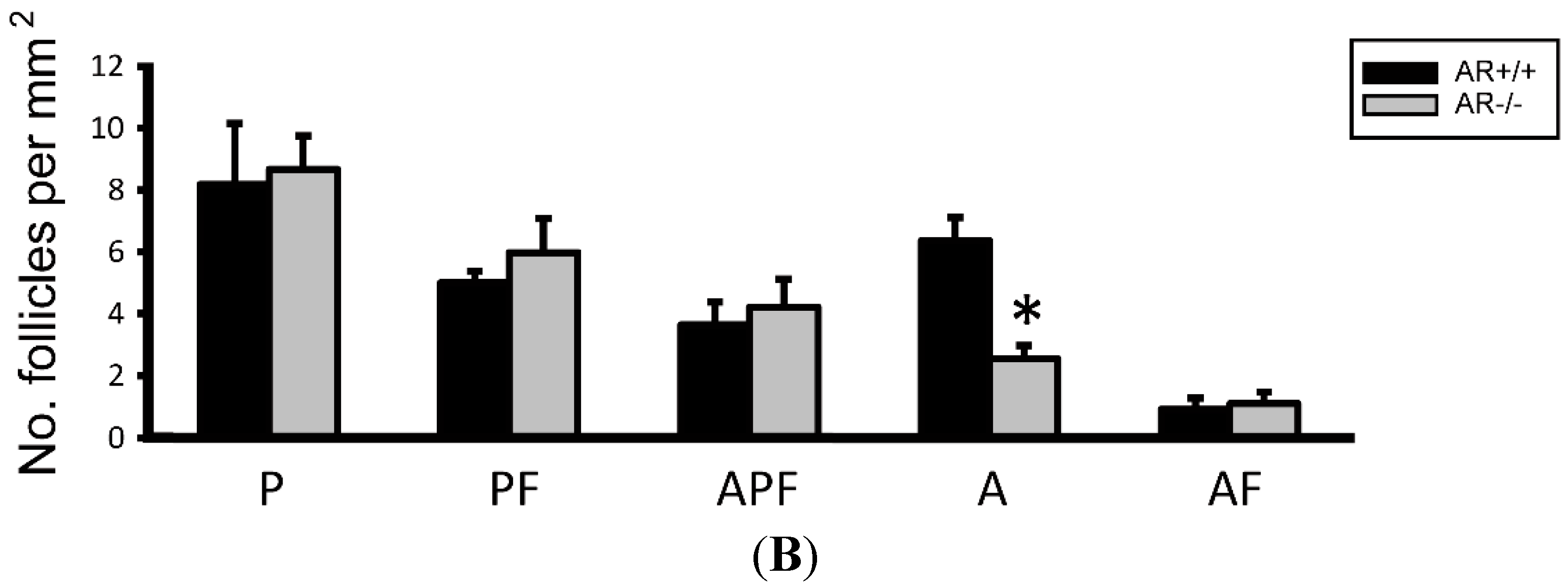
2.3. Change of Ovarian Follicle Morphology in AR−/− Mice
2.4. Alteration of Mitochondrial Morphology and Ultrastructure in Granulosa Cells from AR−/− Mice
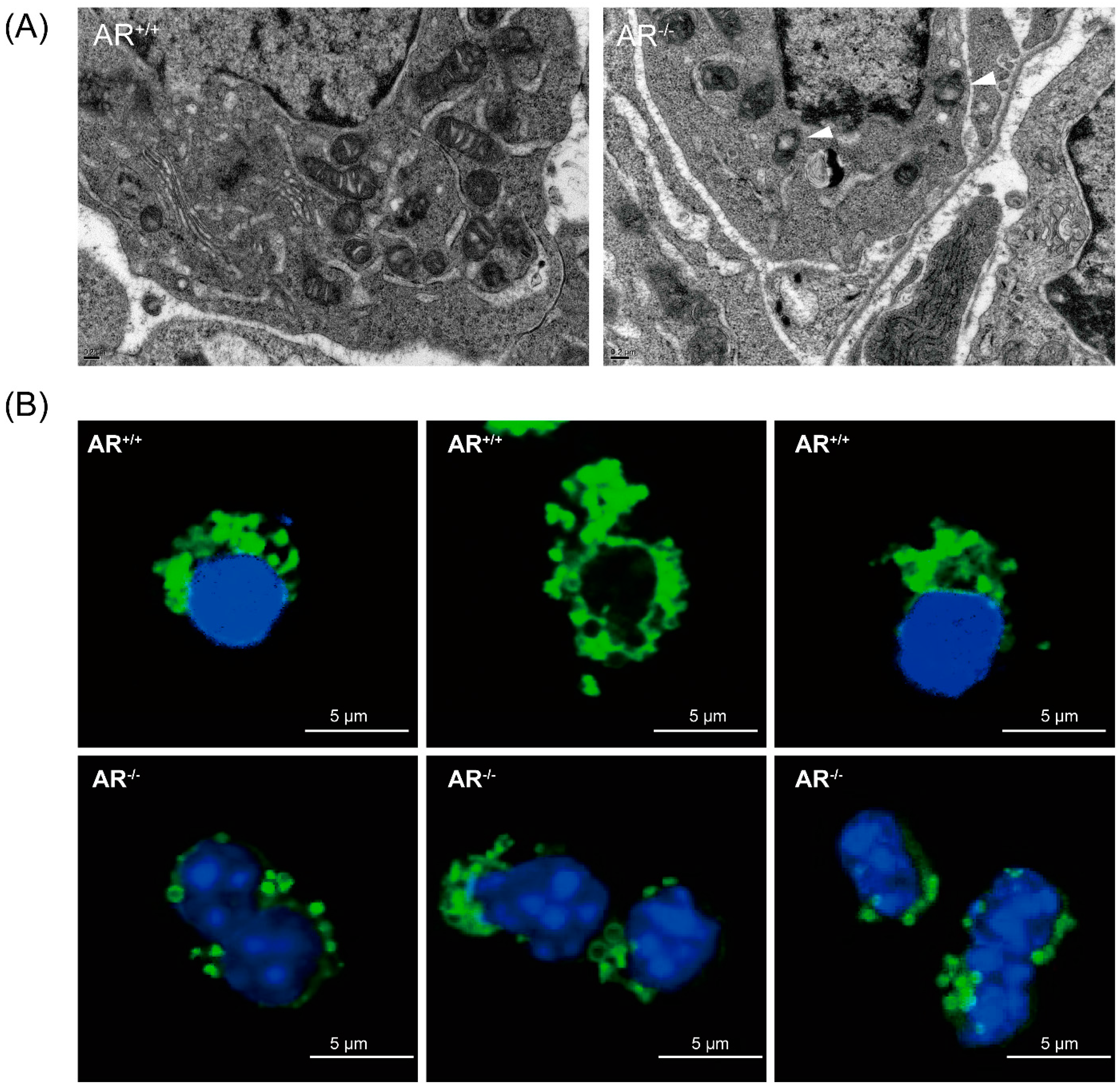
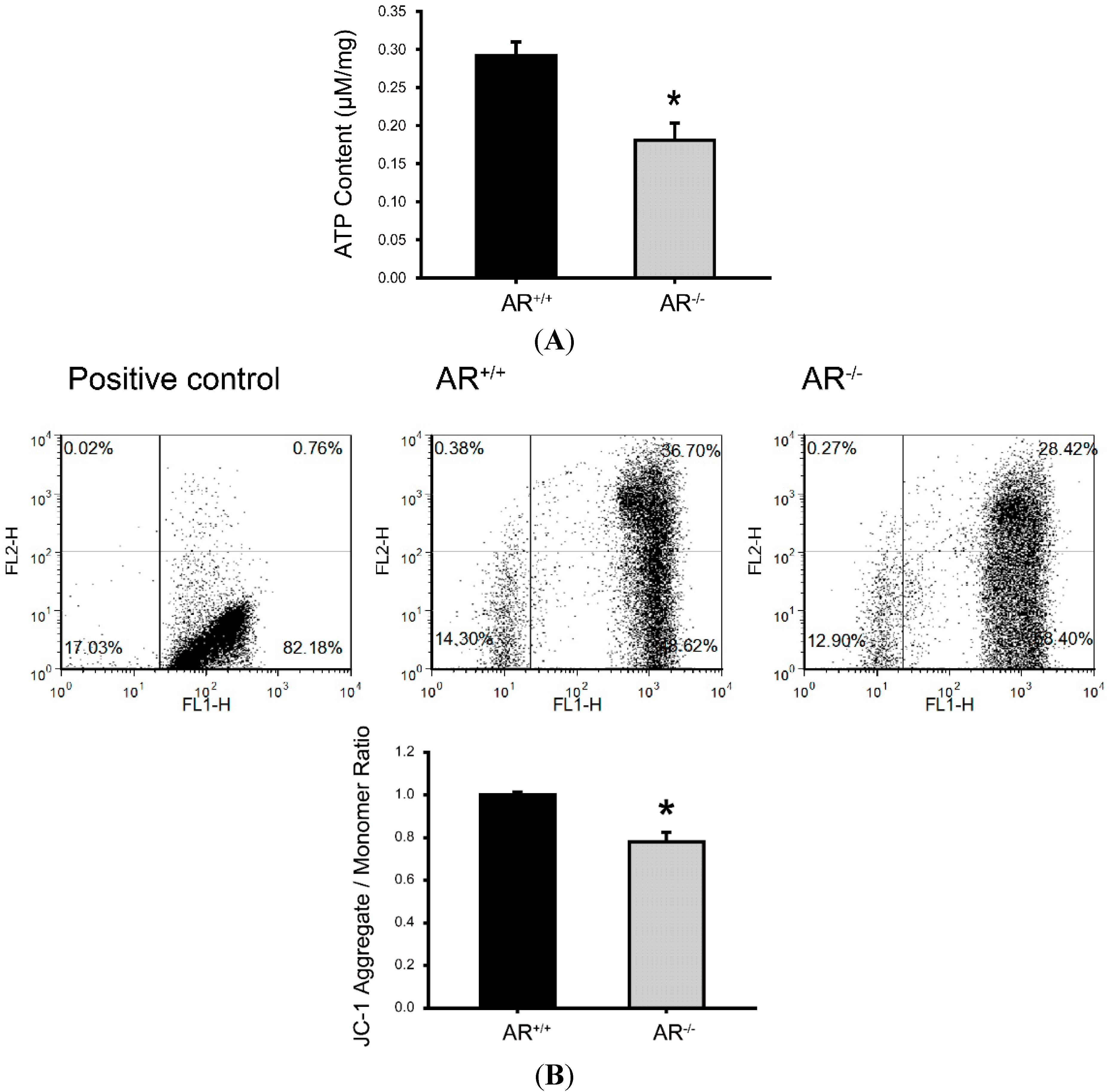
2.5. Metabolic Dysfunction of Mitochondria and Reduced Mitochondrial Membrane Potential in Granulosa Cells of AR−/− Mice
2.6. Decreased Mitochondrial Biogenesis in Granulosa Cells of AR−/− Mice
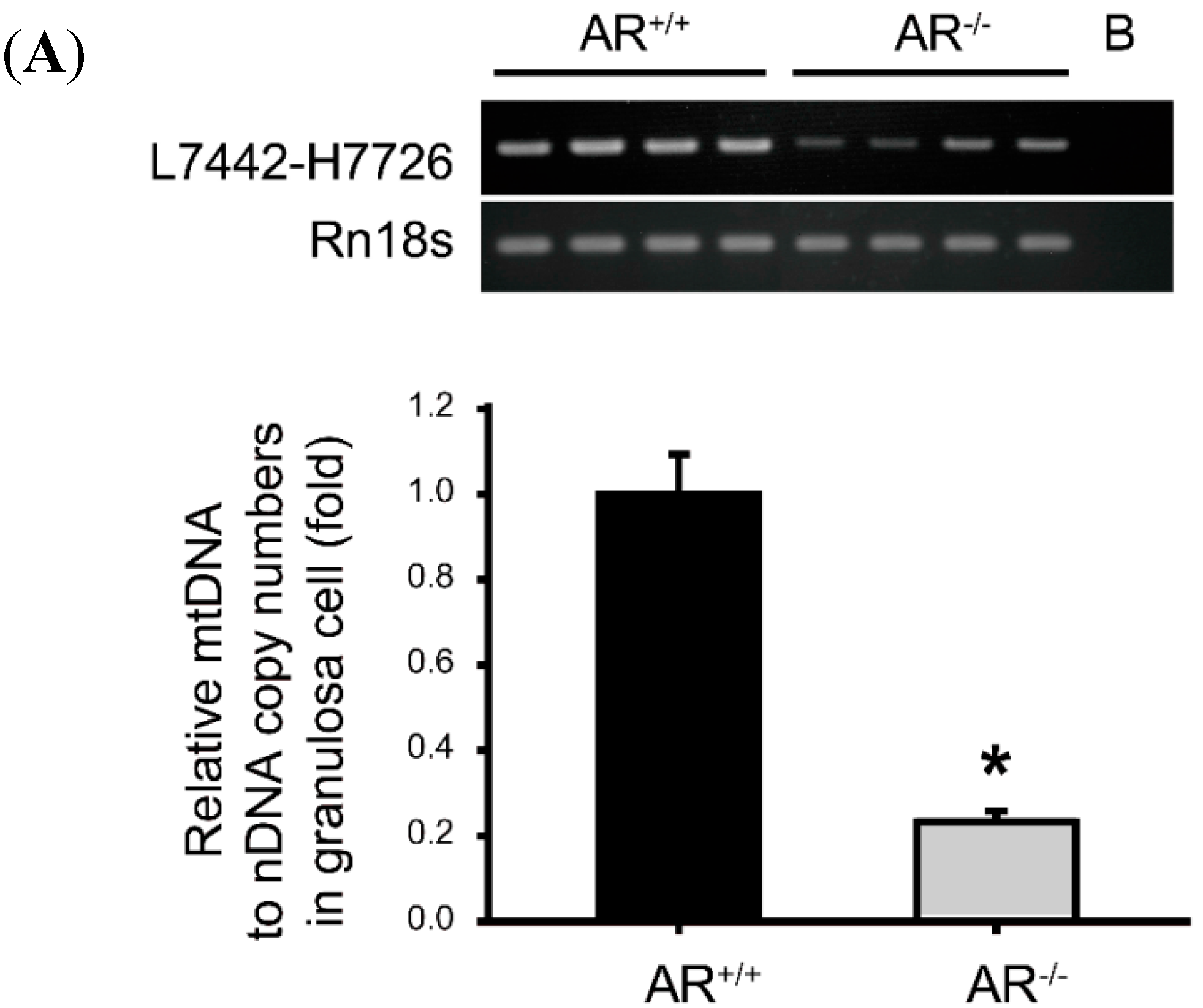
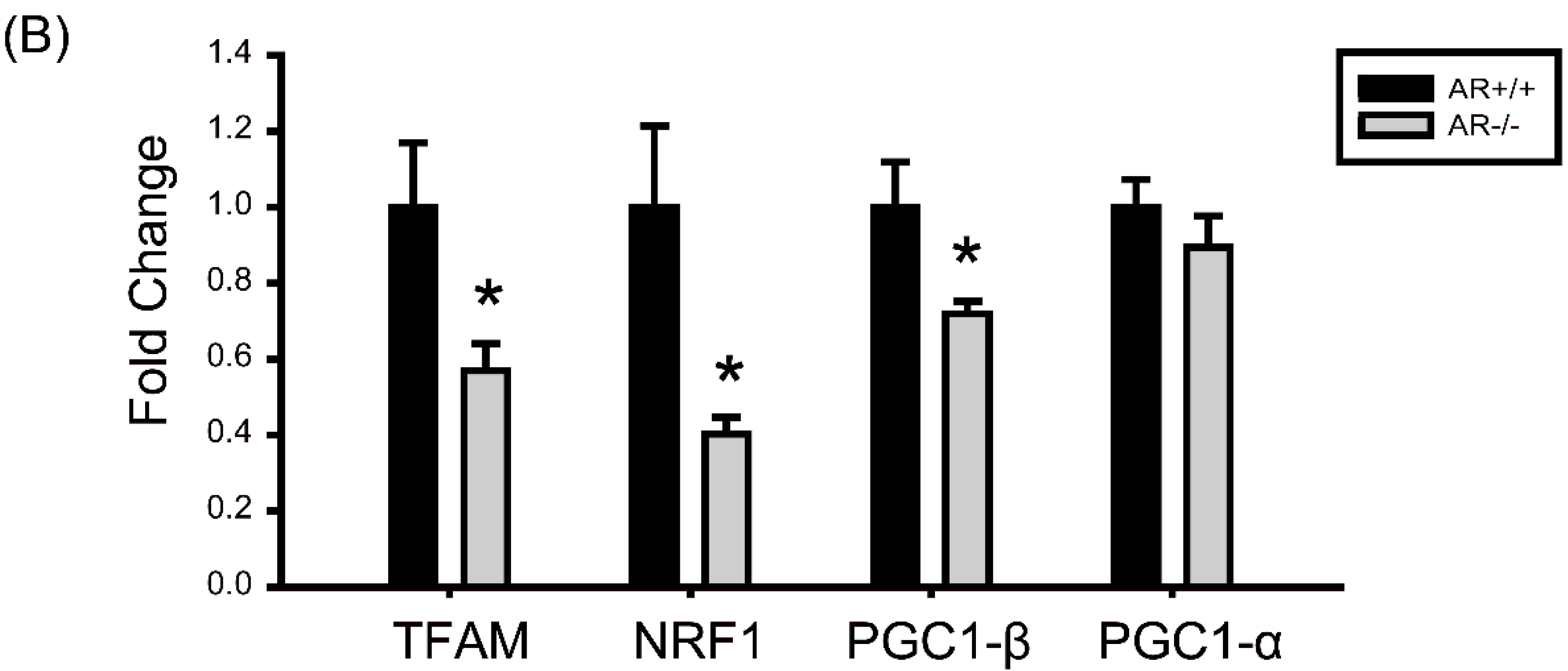
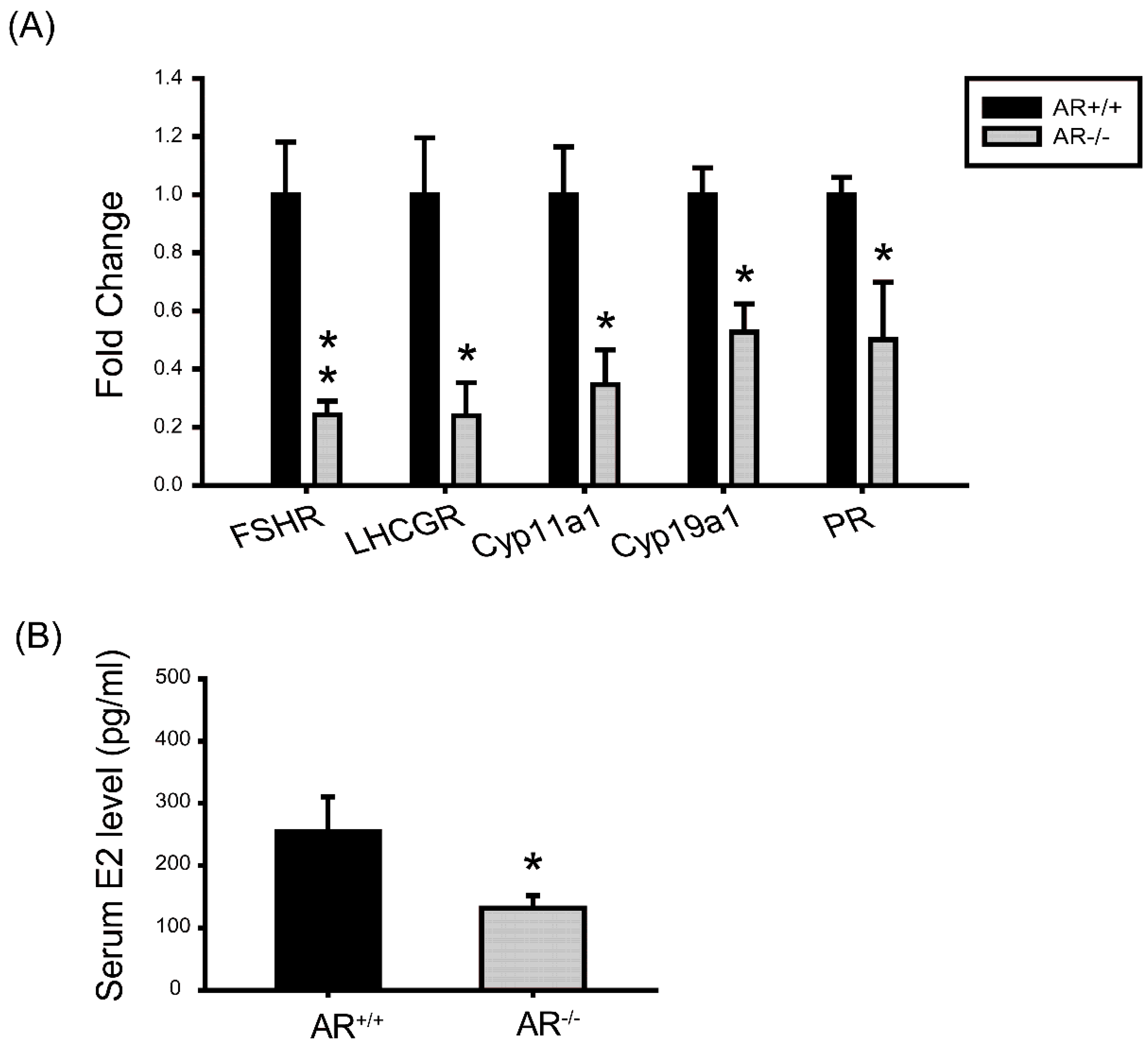
2.7. Molecular Changes in Granulosa Cells of AR−/− Mice and the Effect of AR Deficiency on Serum E2 Levels
3. Discussion
4. Materials and Methods
4.1. Generation of Female AR−/− Mice
4.2. Western Blot Analysis
4.3. Tissue Sampling, Oocyte in Vitro Maturation Assay, RNA Extraction and Analysis
4.4. Histology
4.5. Transmission Electron Microscopy (TEM)
4.6. Immunofluorescence
4.7. Determination of Mitochondrial DNA (mtDNA) Content in Granulosa Cells
4.8. Metabolite Analytic Assays
4.9. Detection of Mitochondrial Membrane Potential by Flow Cytometry
4.10. Assessment of Serum Hormone Levels
4.11. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chang, C.S.; Kokontis, J.; Liao, S.T. Molecular cloning of human and rat complementary DNA encoding androgen receptors. Science 1988, 240, 324–326. [Google Scholar] [CrossRef] [PubMed]
- Heinlein, C.A.; Chang, C. Androgen receptor in prostate cancer. Endocr. Rev. 2004, 25, 276–308. [Google Scholar] [CrossRef] [PubMed]
- Murray, A.A.; Gosden, R.G.; Allison, V.; Spears, N. Effect of androgens on the development of mouse follicles growing in vitro. J. Reprod. Fertil. 1998, 113, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Vendola, K.A.; Zhou, J.; Adesanya, O.O.; Weil, S.J.; Bondy, C.A. Androgens stimulate early stages of follicular growth in the primate ovary. J. Clin. Investig. 1998, 101, 2622–2629. [Google Scholar] [CrossRef] [PubMed]
- Tetsuka, M.; Hillier, S.G. Differential regulation of aromatase and androgen receptor in granulosa cells. J. Steroid Biochem. Mol. Biol. 1997, 61, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Weil, S.; Vendola, K.; Zhou, J.; Bondy, C.A. Androgen and follicle-stimulating hormone interactions in primate ovarian follicle development. J. Clin. Endocrinol. Metab. 1999, 84, 2951–2956. [Google Scholar] [CrossRef] [PubMed]
- Daniel, S.A.; Armstrong, D.T. Enhancement of follicle-stimulating hormone-induced aromatase activity by androgens in cultured rat granulosa cells. Endocrinology 1980, 107, 1027–1033. [Google Scholar] [CrossRef] [PubMed]
- Sen, A.; Prizant, H.; Light, A.; Biswas, A.; Hayes, E.; Lee, H.J.; Barad, D.; Gleicher, N.; Hammes, S.R. Androgens regulate ovarian follicular development by increasing follicle stimulating hormone receptor and microRNA-125b expression. Proc. Natl. Acad. Sci. USA 2014, 111, 3008–3013. [Google Scholar] [CrossRef] [PubMed]
- Hirai, M.; Hirata, S.; Osada, T.; Hagihara, K.; Kato, J. Androgen receptor mRNA in the rat ovary and uterus. J. Steroid Biochem. Mol. Biol. 1994, 49, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Tetsuka, M.; Whitelaw, P.F.; Bremner, W.J.; Millar, M.R.; Smyth, C.D.; Hillier, S.G. Developmental regulation of androgen receptor in rat ovary. J. Endocrinol. 1995, 145, 535–543. [Google Scholar] [CrossRef] [PubMed]
- Gill, A.; Jamnongjit, M.; Hammes, S.R. Androgens promote maturation and signaling in mouse oocytes independent of transcription: A release of inhibition model for mammalian oocyte meiosis. Mol. Endocrinol. 2004, 18, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.C.; Wang, P.H.; Yeh, S.; Wang, R.S.; Xie, C.; Xu, Q.; Zhou, X.; Chao, H.T.; Tsai, M.Y.; Chang, C. Subfertility and defective folliculogenesis in female mice lacking androgen receptor. Proc. Natl. Acad. Sci. USA 2004, 101, 11209–11214. [Google Scholar] [CrossRef] [PubMed]
- Shiina, H.; Matsumoto, T.; Sato, T.; Igarashi, K.; Miyamoto, J.; Takemasa, S.; Sakari, M.; Takada, I.; Nakamura, T.; Metzger, D.; et al. Premature ovarian failure in androgen receptor-deficient mice. Proc. Natl. Acad. Sci. USA 2006, 103, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Walters, K.A.; Allan, C.M.; Jimenez, M.; Lim, P.R.; Davey, R.A.; Zajac, J.D.; Illingworth, P.; Handelsman, D.J. Female mice haploinsufficient for an inactivated androgen receptor (AR) exhibit age-dependent defects that resemble the AR null phenotype of dysfunctional late follicle development, ovulation, and fertility. Endocrinology 2007, 148, 674–684. [Google Scholar] [CrossRef]
- Sen, A.; Hammes, S.R. Granulosa cell-specific androgen receptors are critical regulators of ovarian development and function. Mol. Endocrinol. 2010, 24, 1393–1403. [Google Scholar] [CrossRef] [PubMed]
- Walters, K.A.; Middleton, L.J.; Joseph, S.R.; Hazra, R.; Jimenez, M.; Simanainen, U.; Allan, C.M.; Handelsman, D.J. Targeted loss of androgen receptor signaling in murine granulosa cells of preantral and antral follicles causes female subfertility. Biol. Reprod. 2012, 87, 151. [Google Scholar] [CrossRef] [PubMed]
- Spears, N.; Murray, A.A.; Allison, V.; Boland, N.I.; Gosden, R.G. Role of gonadotrophins and ovarian steroids in the development of mouse follicles in vitro. J. Reprod. Fertil. 1998, 113, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.Y.; Fortune, J.E. Testosterone stimulates the primary to secondary follicle transition in bovine follicles in vitro. Biol. Reprod. 2006, 75, 924–932. [Google Scholar] [CrossRef] [PubMed]
- Seifer, D.B.; DeJesus, V.; Hubbard, K. Mitochondrial deletions in luteinized granulosa cells as a function of age in women undergoing in vitro fertilization. Fertil. Steril. 2002, 78, 1046–1048. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Frolova, A.I.; Purcell, S.; Adastra, K.; Schoeller, E.; Chi, M.M.; Schedl, T.; Moley, K.H. Mitochondrial dysfunction and apoptosis in cumulus cells of type I diabetic mice. PLoS ONE 2010, 5, e15901. [Google Scholar] [CrossRef] [PubMed]
- Gannon, A.M.; Stampfli, M.R.; Foster, W.G. Cigarette smoke exposure elicits increased autophagy and dysregulation of mitochondrial dynamics in murine granulosa cells. Biol. Reprod. 2013, 88, 63. [Google Scholar] [CrossRef] [PubMed]
- Newmeyer, D.D.; Ferguson-Miller, S. Mitochondria: Releasing power for life and unleashing the machineries of death. Cell 2003, 112, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.L. Steroid hormone synthesis in mitochondria. Mol. Cell. Endocrinol. 2013, 379, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Ramalho-Santos, J.; Varum, S.; Amaral, S.; Mota, P.C.; Sousa, A.P.; Amaral, A. Mitochondrial functionality in reproduction: From gonads and gametes to embryos and embryonic stem cells. Hum. Reprod. Update 2009, 15, 553–572. [Google Scholar] [CrossRef] [PubMed]
- Scarpulla, R.C. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 2008, 88, 611–638. [Google Scholar] [CrossRef] [PubMed]
- Kumar, T.R.; Wang, Y.; Lu, N.; Matzuk, M.M. Follicle stimulating hormone is required for ovarian follicle maturation but not male fertility. Nat. Genet. 1997, 15, 201–204. [Google Scholar] [CrossRef] [PubMed]
- Dierich, A.; Sairam, M.R.; Monaco, L.; Fimia, G.M.; Gansmuller, A.; LeMeur, M.; Sassone-Corsi, P. Impairing follicle-stimulating hormone (FSH) signaling in vivo: Targeted disruption of the FSH receptor leads to aberrant gametogenesis and hormonal imbalance. Proc. Natl. Acad. Sci. USA 1998, 95, 13612–13617. [Google Scholar] [CrossRef] [PubMed]
- Danilovich, N.; Babu, P.S.; Xing, W.; Gerdes, M.; Krishnamurthy, H.; Sairam, M.R. Estrogen deficiency, obesity, and skeletal abnormalities in follicle-stimulating hormone receptor knockout (FORKO) female mice. Endocrinology 2000, 141, 4295–4308. [Google Scholar] [PubMed]
- Detmer, S.A.; Chan, D.C. Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 2007, 8, 870–879. [Google Scholar] [CrossRef] [PubMed]
- Frazier, A.E.; Kiu, C.; Stojanovski, D.; Hoogenraad, N.J.; Ryan, M.T. Mitochondrial morphology and distribution in mammalian cells. Biol. Chem. 2006, 387, 1551–1558. [Google Scholar] [CrossRef] [PubMed]
- Martinou, J.C.; Youle, R.J. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev. Cell 2011, 21, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.C. Mitochondria: Dynamic organelles in disease, aging, and development. Cell 2006, 125, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- Diaz, F.; Moraes, C.T. Mitochondrial biogenesis and turnover. Cell Calcium 2008, 44, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Kelly, D.P.; Scarpulla, R.C. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004, 18, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Handschin, C.; Spiegelman, B.M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005, 1, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Gittens, J.E.; Barr, K.J.; Vanderhyden, B.C.; Kidder, G.M. Interplay between paracrine signaling and gap junctional communication in ovarian follicles. J. Cell Sci. 2005, 118, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Eppig, J.J. Intercommunication between mammalian oocytes and companion somatic cells. Bioessays 1991, 13, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Matzuk, M.M.; Burns, K.H.; Viveiros, M.M.; Eppig, J.J. Intercellular communication in the mammalian ovary: Oocytes carry the conversation. Science 2002, 296, 2178–2180. [Google Scholar] [CrossRef] [PubMed]
- Downs, S.M. The influence of glucose, cumulus cells, and metabolic coupling on ATP levels and meiotic control in the isolated mouse oocyte. Dev. Biol. 1995, 167, 502–512. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, K.; Pendola, F.L.; Eppig, J.J. Oocyte control of metabolic cooperativity between oocytes and companion granulosa cells: Energy metabolism. Dev. Biol. 2005, 279, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Quinn, P.; Wales, R.G. The relationships between the ATP content of preimplantation mouse embryos and their development in vitro during culture. J. Reprod. Fertil. 1973, 35, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Van Blerkom, J.; Davis, P.W.; Lee, J. ATP content of human oocytes and developmental potential and outcome after in vitro fertilization and embryo transfer. Hum. Reprod. 1995, 10, 415–424. [Google Scholar]
- Richards, J.S. Maturation of ovarian follicles: Actions and interactions of pituitary and ovarian hormones on follicular cell differentiation. Physiol. Rev. 1980, 60, 51–89. [Google Scholar] [PubMed]
- Mason, A.J.; Hayflick, J.S.; Zoeller, R.T.; Young, W.S., 3rd.; Phillips, H.S.; Nikolics, K.; Seeburg, P.H. A deletion truncating the gonadotropin-releasing hormone gene is responsible for hypogonadism in the hpg mouse. Science 1986, 234, 1366–1371. [Google Scholar] [CrossRef] [PubMed]
- Hunzicker-Dunn, M.; Maizels, E.T. FSH signaling pathways in immature granulosa cells that regulate target gene expression: Branching out from protein kinase A. Cell Signal. 2006, 18, 1351–1359. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.G.; Bennett, J.; Talla, D.; Stocco, C. Testosterone, not 5α-dihydrotestosterone, stimulates LRH-1 leading to FSH-independent expression of Cyp19 and P450scc in granulosa cells. Mol. Endocrinol. 2011, 25, 656–668. [Google Scholar] [CrossRef] [PubMed]
- McGee, E.A.; Hsueh, A.J. Initial and cyclic recruitment of ovarian follicles. Endocr. Rev. 2000, 21, 200–214. [Google Scholar] [PubMed]
- Segaloff, D.L.; Wang, H.Y.; Richards, J.S. Hormonal regulation of luteinizing hormone/chorionic gonadotropin receptor mRNA in rat ovarian cells during follicular development and luteinization. Mol. Endocrinol. 1990, 4, 1856–1865. [Google Scholar] [CrossRef] [PubMed]
- Kessel, B.; Liu, Y.X.; Jia, X.C.; Hsueh, A.J. Autocrine role of estrogens in the augmentation of luteinizing hormone receptor formation in cultured rat granulosa cells. Biol. Reprod. 1985, 32, 1038–1050. [Google Scholar] [CrossRef] [PubMed]
- Natraj, U.; Richards, J.S. Hormonal regulation, localization, and functional activity of the progesterone receptor in granulosa cells of rat preovulatory follicles. Endocrinology 1993, 133, 761–769. [Google Scholar] [PubMed]
- Park-Sarge, O.K.; Mayo, K.E. Regulation of the progesterone receptor gene by gonadotropins and cyclic adenosine 3',5'-monophosphate in rat granulosa cells. Endocrinology 1994, 134, 709–718. [Google Scholar] [PubMed]
- Yeh, S.; Tsai, M.Y.; Xu, Q.; Mu, X.M.; Lardy, H.; Huang, K.E.; Lin, H.; Yeh, S.D.; Altuwaijri, S.; Zhou, X.; et al. Generation and characterization of androgen receptor knockout (ARKO) mice: An in vivo model for the study of androgen functions in selective tissues. Proc. Natl. Acad. Sci. USA 2002, 99, 13498–13503. [Google Scholar] [CrossRef] [PubMed]
- Yeh, S.; Hu, Y.C.; Wang, P.H.; Xie, C.; Xu, Q.; Tsai, M.Y.; Dong, Z.; Wang, R.S.; Lee, T.H.; Chang, C. Abnormal mammary gland development and growth retardation in female mice and MCF7 breast cancer cells lacking androgen receptor. J. Exp. Med. 2003, 198, 1899–1908. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.Y.; Minahan, K.; Merriman, J.A.; Jones, K.T. Calmodulin-dependent protein kinase γ 3 (CamKIIγ3) mediates the cell cycle resumption of metaphase II eggs in mouse. Development 2009, 136, 4077–4081. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, T.; Peters, H. Proposal for a classification of oocytes and follicles in the mouse ovary. J. Reprod. Fertil. 1968, 17, 555–557. [Google Scholar] [CrossRef] [PubMed]
- Britt, K.L.; Drummond, A.E.; Cox, V.A.; Dyson, M.; Wreford, N.G.; Jones, M.E.; Simpson, E.R.; Findlay, J.K. An age-related ovarian phenotype in mice with targeted disruption of the Cyp 19 (aromatase) gene. Endocrinology 2000, 141, 2614–2623. [Google Scholar] [PubMed]
- Weng, S.W.; Lin, T.K.; Liou, C.W.; Chen, S.D.; Wei, Y.H.; Lee, H.C.; Chen, I.Y.; Hsieh, C.J.; Wang, P.W. Peripheral blood mitochondrial DNA content and dysregulation of glucose metabolism. Diabetes Res. Clin. Pract. 2009, 83, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Bonnard, C.; Durand, A.; Peyrol, S.; Chanseaume, E.; Chauvin, M.A.; Morio, B.; Vidal, H.; Rieusset, J. Mitochondrial dysfunction results from oxidative stress in the skeletal muscle of diet-induced insulin-resistant mice. J. Clin. Investig. 2008, 118, 789–800. [Google Scholar] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, R.-S.; Chang, H.-Y.; Kao, S.-H.; Kao, C.-H.; Wu, Y.-C.; Yeh, S.; Tzeng, C.-R.; Chang, C. Abnormal Mitochondrial Function and Impaired Granulosa Cell Differentiation in Androgen Receptor Knockout Mice. Int. J. Mol. Sci. 2015, 16, 9831-9849. https://doi.org/10.3390/ijms16059831
Wang R-S, Chang H-Y, Kao S-H, Kao C-H, Wu Y-C, Yeh S, Tzeng C-R, Chang C. Abnormal Mitochondrial Function and Impaired Granulosa Cell Differentiation in Androgen Receptor Knockout Mice. International Journal of Molecular Sciences. 2015; 16(5):9831-9849. https://doi.org/10.3390/ijms16059831
Chicago/Turabian StyleWang, Ruey-Sheng, Heng-Yu Chang, Shu-Huei Kao, Cheng-Heng Kao, Yi-Chen Wu, Shuyuan Yeh, Chii-Reuy Tzeng, and Chawnshang Chang. 2015. "Abnormal Mitochondrial Function and Impaired Granulosa Cell Differentiation in Androgen Receptor Knockout Mice" International Journal of Molecular Sciences 16, no. 5: 9831-9849. https://doi.org/10.3390/ijms16059831
APA StyleWang, R.-S., Chang, H.-Y., Kao, S.-H., Kao, C.-H., Wu, Y.-C., Yeh, S., Tzeng, C.-R., & Chang, C. (2015). Abnormal Mitochondrial Function and Impaired Granulosa Cell Differentiation in Androgen Receptor Knockout Mice. International Journal of Molecular Sciences, 16(5), 9831-9849. https://doi.org/10.3390/ijms16059831





