A Soluble Pyrophosphatase Is Essential to Oogenesis and Is Required for Polyphosphate Metabolism in the Red Flour Beetle (Tribolium castaneum)
Abstract
:1. Introduction
2. Results and Discussion
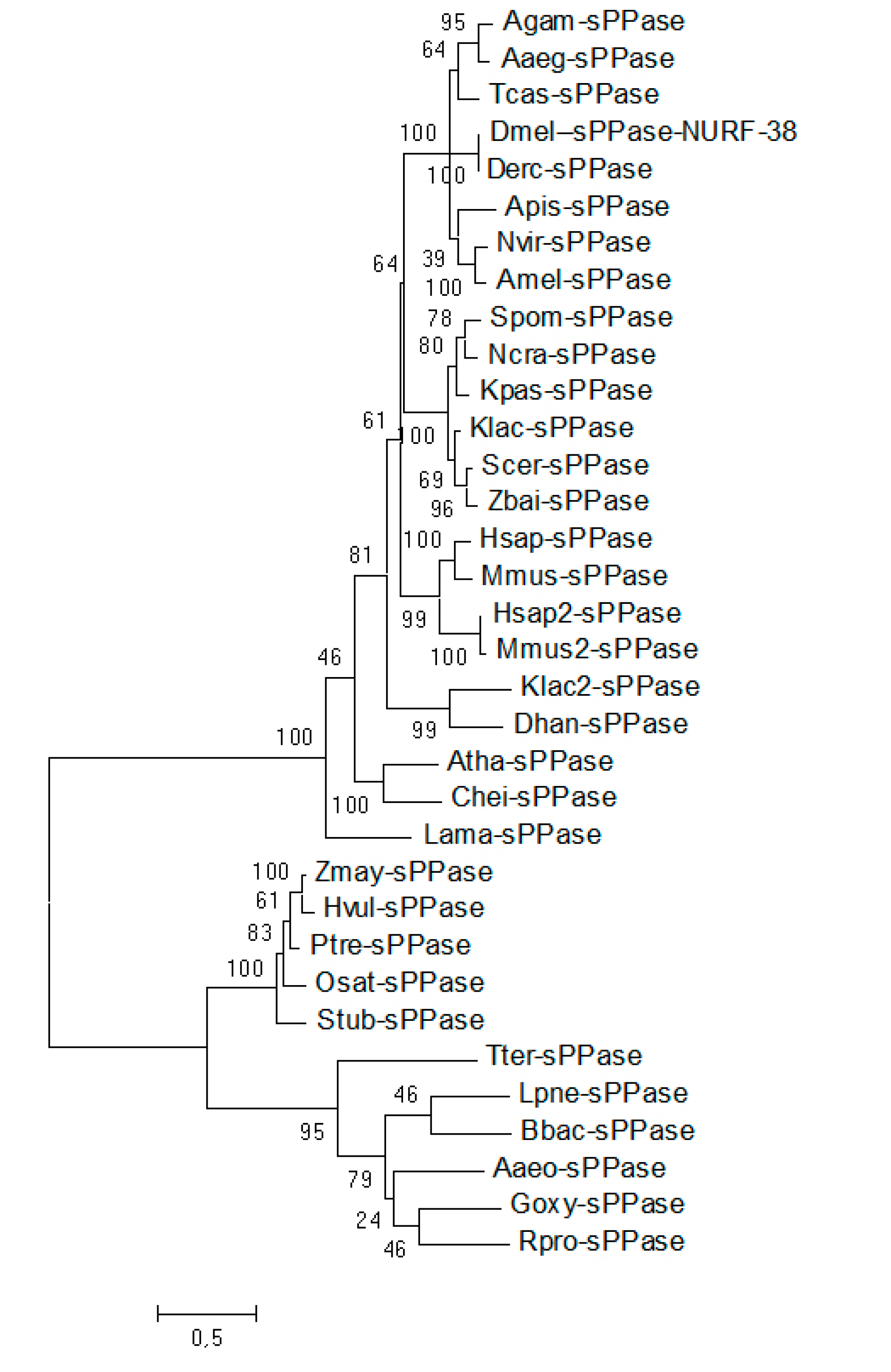
2.1. A Single Soluble Inorganic Pyrophosphatases (sPPases) Can Be Identified in Tribolium castaneum and Other Insect Genomes

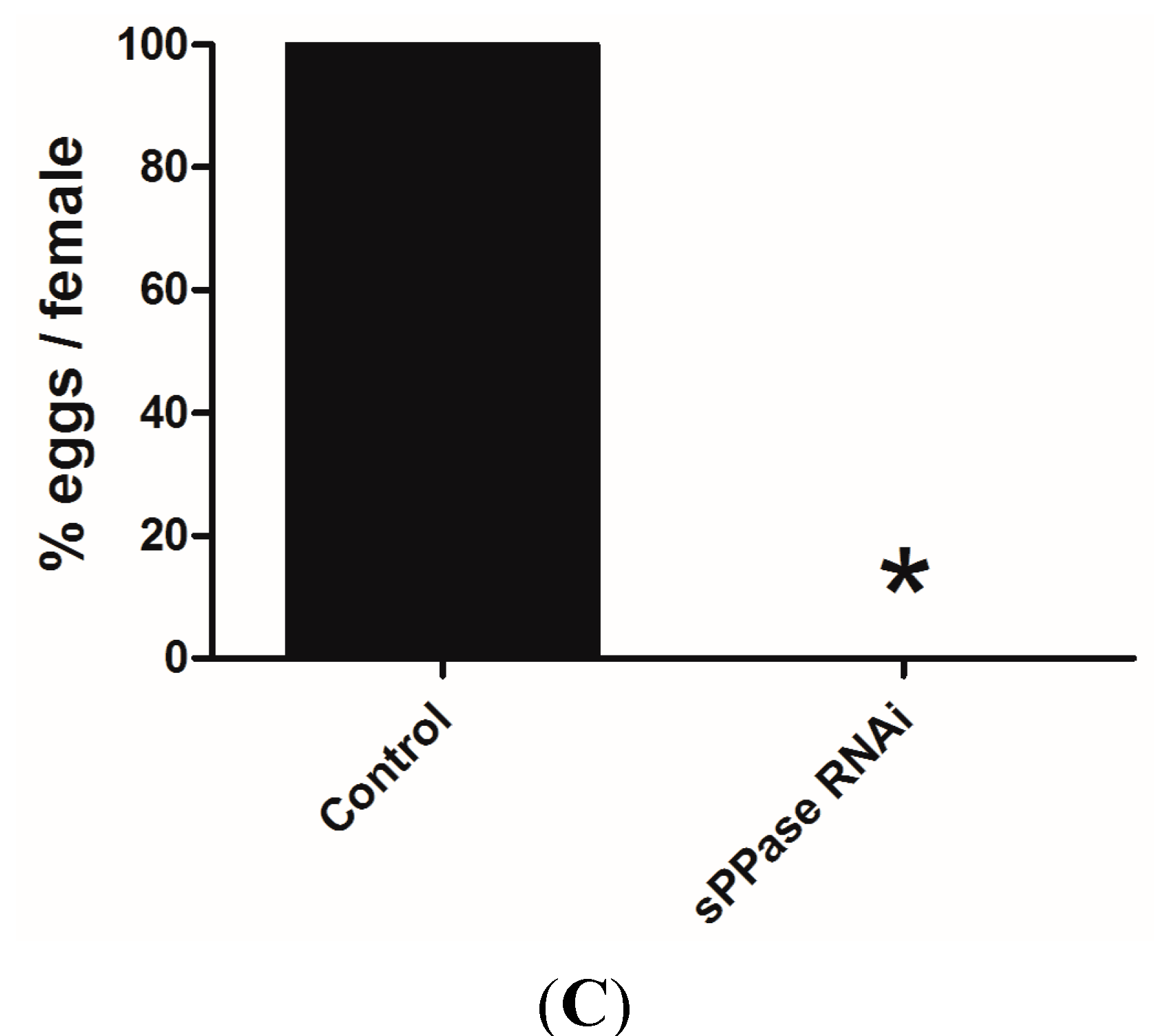
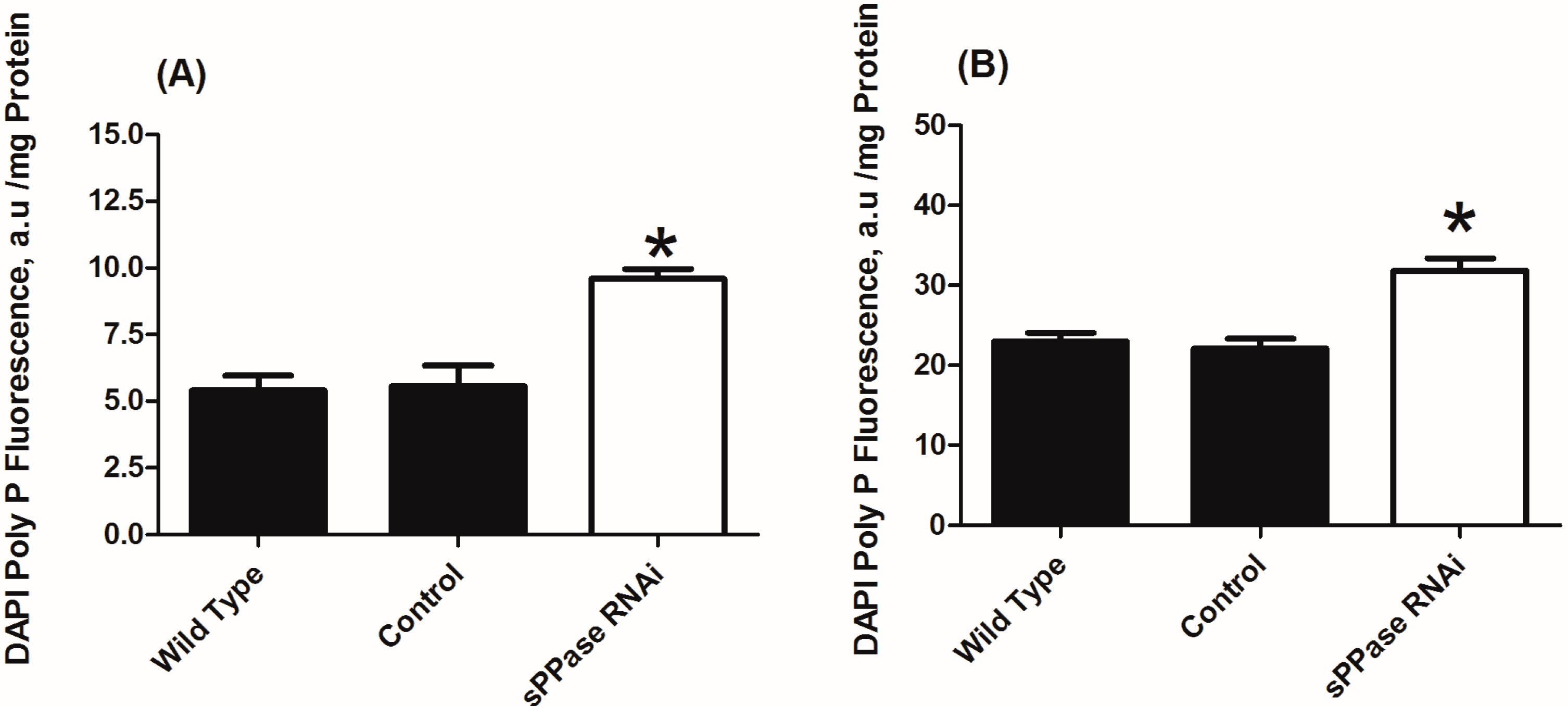
2.2. pRNAi Analysis Shows that Tc-sPPase Is Essential to Oogenesis and Regulates Poly P Metabolism



3. Experimental Section
3.1. Tribolium castaneum Strains
3.2. Sequence Analysis
3.3. Primer Design, in Situ Hybridization and RNAi
3.4. Real-Time PCR
3.5. Cell Fractionation
3.6. Exopolyphosphatase Activity
3.7. Poly P Extraction and Quantification
3.8. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kulaev, I.S.; Vagabov, V.M. Polyphosphate metabolism in microorganisms. Adv. Microb. Physiol. 1983, 24, 83–171. [Google Scholar] [PubMed]
- Kornberg, A.; Rao, N.N.; Ault-Riche, D. Inorganic polyphosphate: A molecule of many functions. Annu. Rev. Biochem. 1999, 68, 89–125. [Google Scholar] [CrossRef] [PubMed]
- Kulaev, I.; Kulakovskaya, T. Polyphosphate and phosphate pump. Annu. Rev. Microbiol. 2000, 54, 709–734. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Fraley, C.D.; Faridi, J.; Kornberg, A.; Roth, R.A. Inorganic polyphosphate stimulates mammalian TOR, a kinase involved in the proliferation of mammary cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 11249–11254. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.A.; Mutch, N.J.; Baskar, D.; Rohloff, P.; Docampo, R.; Morrissey, J.H. Polyphosphate modulates blood coagulation and fibrinolysis. Proc. Natl. Acad. Sci. USA 2006, 103, 903–908. [Google Scholar] [CrossRef]
- Doi, K.; Kubo, T.; Takeshita, R.; Kajihara, S.; Kato, S.; Kawazoe, Y.; Shiba, T.; Akagawa, Y. Inorganic polyphosphate adsorbed onto hydroxyapatite for guided bone regeneration: An animal study. Dent. Mater. J. 2014, 33, 179–186. [Google Scholar] [CrossRef]
- Kawano, M.M. Inorganic polyphosphate induces apoptosis specifically in human plasma cells. Haematologica 2006, 91, 1154A. [Google Scholar] [PubMed]
- Campos, E.; Facanha, A.R.; Costa, E.P.; da Silva Vaz, I., Jr.; Masuda, A.; Logullo, C. Exopolyphosphatases in nuclear and mitochondrial fractions during embryogenesis of the hard tick Rhipicephalus (Boophilus) microplus. Comp. Biochem. Physiol. Part B 2008, 151, 311–316. [Google Scholar] [CrossRef]
- Kulaev, I.S.; Vagabov, V.M.; Kulakovskaya, T.V.; Lichko, L.P.; Andreeva, N.A.; Trilisenko, L.V. The development of A. N. Belozersky’s ideas in polyphosphate biochemistry. Biochemistry (Mosc.) 2000, 65, 271–278. [Google Scholar]
- Kulakovskaya, T.; Kulaev, I. Enzymes of inorganic polyphosphate metabolism. Biomed. Inorg. Polym. 2013, 54, 39–63. [Google Scholar]
- Rao, N.N.; Gomez-Garcia, M.R.; Kornberg, A. Inorganic polyphosphate: Essential for growth and survival. Annu. Rev. Biochem. 2009, 78, 605–647. [Google Scholar] [CrossRef] [PubMed]
- Blum, E.; Py, B.; Carpousis, A.J.; Higgins, C.F. Polyphosphate kinase is a component of the Escherichia coli RNA degradosome. Mol. Microbiol. 1997, 26, 387–398. [Google Scholar] [CrossRef]
- Kuroda, A.; Nomura, K.; Ohtomo, R.; Kato, J.; Ikeda, T.; Takiguchi, N.; Ohtake, H.; Kornberg, A. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease E. coli. Science 2001, 293, 705–708. [Google Scholar]
- Kuroda, A.; Nomura, K.; Takiguchi, N.; Kato, J.; Ohtake, H. Inorganic polyphosphate stimulates lon-mediated proteolysis of nucleoid proteins in Escherichia coli. Cell. Mol. Biol. 2006, 52, 23–29. [Google Scholar]
- McInerney, P.; Mizutani, T.; Shiba, T. Inorganic polyphosphate interacts with ribosomes and promotes translation fidelity in vitro and in vivo. Mol. Microbiol. 2006, 60, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Holbein, S.; Freimoser, F.M.; Werner, T.P.; Wengi, A.; Dichtl, B. Cordycepin-hypersensitive growth links elevated polyphosphate levels to inhibition of poly(A) polymerase in Saccharomyces cerevisiae. Nucleic Acids Res. 2008, 36, 353–363. [Google Scholar] [CrossRef]
- Sillero, M.A.; de Diego, A.; Silles, E.; Osorio, H.; Sillero, A. Polyphosphates strongly inhibit the tRNA dependent synthesis of poly(A) catalyzed by poly(A) polymerase from Saccharomyces cerevisiae. FEBS Lett. 2003, 550, 41–45. [Google Scholar] [CrossRef]
- Jimenez-Nunez, M.D.; Moreno-Sanchez, D.; Hernandez-Ruiz, L.; Benitez-Rondan, A.; Ramos-Amaya, A.; Rodriguez-Bayona, B.; Medina, F.; Brieva, J.A.; Ruiz, F.A. Myeloma cells contain high levels of inorganic polyphosphate which is associated with nucleolar transcription. Haematologica 2012, 97, 1264–1271. [Google Scholar] [CrossRef]
- Lemercier, G.; Espiau, B.; Ruiz, F.A.; Vieira, M.; Luo, S.; Baltz, T.; Docampo, R.; Bakalara, N. A pyrophosphatase regulating polyphosphate metabolism in acidocalcisomes is essential for Trypanosoma brucei virulence in mice. J. Biol. Chem. 2004, 279, 3420–3425. [Google Scholar] [CrossRef]
- Baykov, A.A.; Cooperman, B.S.; Goldman, A.; Lahti, R. Cytoplasmic inorganic pyrophosphatase. Inorg. Polyphosphates 1999, 23, 127–150. [Google Scholar]
- Gdula, D.A.; Sandaltzopoulos, R.; Tsukiyama, T.; Ossipow, V.; Wu, C. Inorganic pyrophosphatase is a component of the Drosophila nucleosome remodeling factor complex. Genes Dev. 1998, 12, 3206–3216. [Google Scholar] [CrossRef]
- Espiau, B.; Lemercier, G.; Ambit, A.; Bringaud, F.; Merlin, G.; Baltz, T.; Bakalara, N. A soluble pyrophosphatase, a key enzyme for polyphosphate metabolism in Leishmania. J. Biol. Chem. 2006, 281, 1516–1523. [Google Scholar] [CrossRef] [PubMed]
- Galizzi, M.; Bustamante, J.M.; Fang, J.; Miranda, K.; Soares Medeiros, L.C.; Tarleton, R.L.; Docampo, R. Evidence for the role of vacuolar soluble pyrophosphatase and inorganic polyphosphate in Trypanosoma cruzi persistence. Mol. Microbiol. 2013, 90, 699–715. [Google Scholar] [CrossRef]
- Brown, S.J.; Shippy, T.D.; Miller, S.; Bolognesi, R.; Beeman, R.W.; Lorenzen, M.D.; Bucher, G.; Wimmer, E.A.; Klingler, M. The red flour beetle, Tribolium castaneum (Coleoptera): A model for studies of development and pest biology. Cold Spring Harb. Protoc. 2009, 2009, pdb–emo126. [Google Scholar] [CrossRef] [PubMed]
- Fraga, A.; Ribeiro, L.; Lobato, M.; Santos, V.; Silva, J.R.; Rezende, G.; Gomes, H.; Moraes, J.; Logullo, C.; Campos, E.; et al. Glycogen and glucose metabolism are essential for early embryonic development of the red flour beetle Tribolium castaneum. PLoS ONE 2013, 8, e65125. [Google Scholar] [CrossRef]
- Shippy, T.D.; Coleman, C.M.; Tomoyasu, Y.; Brown, S.J. Concurrent in situ hybridization and antibody staining in red flour beetle (Tribolium) embryos. Cold Spring Harb. Protoc. 2009, 2009. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.C.; Miyata, K.; Brown, S.J.; Tomoyasu, Y. Dissecting systemic RNA interference in the red flour beetle Tribolium castaneum: Parameters affecting the efficiency of RNAi. PLoS ONE 2012, 7, e47431. [Google Scholar] [CrossRef]
- Posnien, N.; Schinko, J.; Grossmann, D.; Shippy, T.D.; Konopova, B.; Bucher, G. RNAi in the red flour beetle (Tribolium). Cold Spring Harb. Protoc. 2009, 2009. [Google Scholar] [CrossRef] [PubMed]
- Tribolium Genome Sequencing, C.; Richards, S.; Gibbs, R.A.; Weinstock, G.M.; Brown, S.J.; Denell, R.; Beeman, R.W.; Gibbs, R.; Beeman, R.W.; Brown, S.J.; et al. The genome of the model beetle and pest Tribolium castaneum. Nature 2008, 452, 949–955. [Google Scholar] [CrossRef]
- Trauner, J.; Schinko, J.; Lorenzen, M.D.; Shippy, T.D.; Wimmer, E.A.; Beeman, R.W.; Klingler, M.; Bucher, G.; Brown, S.J. Large-scale insertional mutagenesis of a coleopteran stored grain pest, the red flour beetle Tribolium castaneum, identifies embryonic lethal mutations and enhancer traps. BMC Biol. 2009, 7, 73. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Shiba, T.; Doi, K.; Morita, K.; Kubo, T.; Makihara, Y.; Piattelli, A.; Akagawa, Y. Inorganic polyphosphate suppresses lipopolysaccharide-induced inducible nitric oxide synthase (iNOS) expression in macrophages. PLoS ONE 2013, 8, e74650. [Google Scholar] [CrossRef]
- Wang, X.; Schroder, H.C.; Diehl-Seifert, B.; Kropf, K.; Schlossmacher, U.; Wiens, M.; Muller, W.E. Dual effect of inorganic polymeric phosphate/polyphosphate on osteoblasts and osteoclasts in vitro. J. Tissue Eng. Regen. Med. 2013, 7, 767–776. [Google Scholar]
- Wang, X.; Schroder, H.C.; Grebenjuk, V.; Diehl-Seifert, B.; Mailander, V.; Steffen, R.; Schlossmacher, U.; Muller, W.E. The marine sponge-derived inorganic polymers, biosilica and polyphosphate, as morphogenetically active matrices/scaffolds for the differentiation of human multipotent stromal cells: Potential application in 3D printing and distraction osteogenesis. Mar. Drugs 2014, 12, 1131–1147. [Google Scholar] [CrossRef]
- Gomes, F.M.; Oliveira, D.M.; Motta, L.S.; Ramos, I.B.; Miranda, K.M.; Machado, E.A. Inorganic polyphosphate inhibits an aspartic protease-like activity in the eggs of Rhodnius prolixus (Stahl) and impairs yolk mobilization in vitro. J. Cell. Physiol. 2010, 222, 606–611. [Google Scholar]
- Gomes, F.M.; Ramos, I.B.; Motta, L.M.; Miranda, K.; Santiago, M.F.; de Souza, W.; Machado, E.A. Polyphosphate polymers during early embryogenesis of Periplaneta americana. J. Insect Physiol. 2008, 54, 1459–1466. [Google Scholar] [CrossRef]
- Ramos, I.; Gomes, F.; Koeller, C.M.; Saito, K.; Heise, N.; Masuda, H.; Docampo, R.; de Souza, W.; Machado, E.A.; Miranda, K. Acidocalcisomes as calcium- and polyphosphate-storage compartments during embryogenesis of the insect Rhodnius prolixus Stahl. PLoS ONE 2011, 6, e27276. [Google Scholar] [CrossRef]
- Van der Zee, M.; Berns, N.; Roth, S. Distinct functions of the Tribolium zerknu llt genes in serosa specification and dorsal closure. Curr. Biol. 2005, 15, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Baumer, D.; Strohlein, N.M.; Schoppmeier, M. Opposing effects of Notch-signaling in maintaining the proliferative state of follicle cells in the telotrophic ovary of the beetle Tribolium. Front. Zool. 2012, 9, 15. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.P.; Posnien, N.; Bolognesi, R.; Fischer, T.D.; Rayl, P.; Oberhofer, G.; Kitzmann, P.; Brown, S.J.; Bucher, G. Asymmetrically expressed axin required for anterior development in Tribolium. Proc. Natl. Acad. Sci. USA 2012, 109, 7782–7786. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.A.; Peel, A.D.; Drechsler, A.; Averof, M.; Roth, S. EGF Signaling and the origin of axial polarity among the insects. Curr. Biol. 2010, 20, 1042–1047. [Google Scholar] [CrossRef] [PubMed]
- Schroder, H.C.; Lorenz, B.; Kurz, L.; Muller, W.E. Inorganic polyphosphate in eukaryotes: Enzymes, metabolism and function. Prog. Mol. Subcell. Biol. 1999, 23, 45–81. [Google Scholar] [PubMed]
- Handel, K.; Grunfelder, C.G.; Roth, S.; Sander, K. Tribolium embryogenesis: A SEM study of cell shapes and movements from blastoderm to serosal closure. Dev. Genes Evol. 2000, 210, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Lethiec, F.; Duroux, P.; Gascuel, O. PHYML Online—A web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005, 33, W557–W559. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Bucher, G.; Scholten, J.; Klingler, M. Parental RNAi in Tribolium (Coleoptera). Curr. Biol. 2002, 12, R85–R86. [Google Scholar] [CrossRef] [PubMed]
- Stylianopoulou, E.; Lykidis, D.; Ypsilantis, P.; Simopoulos, C.; Skavdis, G.; Grigoriou, M. A rapid and highly sensitive method of non radioactive colorimetric in situ hybridization for the detection of mRNA on tissue sections. PLoS ONE 2012, 7, 1. [Google Scholar] [CrossRef]
- Lord, J.C.; Hartzer, K.; Toutges, M.; Oppert, B. Evaluation of quantitative PCR reference genes for gene expression studies in Tribolium castaneum after fungal challenge. J. Microbiol. Methods 2010, 80, 219–221. [Google Scholar] [CrossRef] [PubMed]
- Fiske, C.H.; Subbarow, Y. The nature of the “Inorganic Phosphate” in voluntary muscle. Science 1927, 65, 401–403. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Abramov, A.Y.; Fraley, C.; Diao, C.T.; Winkfein, R.; Colicos, M.A.; Duchen, M.R.; French, R.J.; Pavlov, E. Targeted polyphosphatase expression alters mitochondrial metabolism and inhibits calcium-dependent cell death. Proc. Natl. Acad. Sci. USA 2007, 104, 18091–18096. [Google Scholar] [CrossRef] [PubMed]
- Aschar-Sobbi, R.; Abramov, A.Y.; Diao, C.; Kargacin, M.E.; Kargacin, G.J.; French, R.J.; Pavlov, E. High sensitivity, quantitative measurements of polyphosphate using a new DAPI-based approach. J. Fluoresc. 2008, 18, 859–866. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carvalho, K.; Ribeiro, L.; Moraes, J.; Da Silva, J.R.; Costa, E.P.; Souza-Menezes, J.; Logullo, C.; Da Fonseca, R.N.; Campos, E. A Soluble Pyrophosphatase Is Essential to Oogenesis and Is Required for Polyphosphate Metabolism in the Red Flour Beetle (Tribolium castaneum). Int. J. Mol. Sci. 2015, 16, 6631-6644. https://doi.org/10.3390/ijms16046631
Carvalho K, Ribeiro L, Moraes J, Da Silva JR, Costa EP, Souza-Menezes J, Logullo C, Da Fonseca RN, Campos E. A Soluble Pyrophosphatase Is Essential to Oogenesis and Is Required for Polyphosphate Metabolism in the Red Flour Beetle (Tribolium castaneum). International Journal of Molecular Sciences. 2015; 16(4):6631-6644. https://doi.org/10.3390/ijms16046631
Chicago/Turabian StyleCarvalho, Klébea, Lupis Ribeiro, Jorge Moraes, José Roberto Da Silva, Evenilton P. Costa, Jackson Souza-Menezes, Carlos Logullo, Rodrigo Nunes Da Fonseca, and Eldo Campos. 2015. "A Soluble Pyrophosphatase Is Essential to Oogenesis and Is Required for Polyphosphate Metabolism in the Red Flour Beetle (Tribolium castaneum)" International Journal of Molecular Sciences 16, no. 4: 6631-6644. https://doi.org/10.3390/ijms16046631
APA StyleCarvalho, K., Ribeiro, L., Moraes, J., Da Silva, J. R., Costa, E. P., Souza-Menezes, J., Logullo, C., Da Fonseca, R. N., & Campos, E. (2015). A Soluble Pyrophosphatase Is Essential to Oogenesis and Is Required for Polyphosphate Metabolism in the Red Flour Beetle (Tribolium castaneum). International Journal of Molecular Sciences, 16(4), 6631-6644. https://doi.org/10.3390/ijms16046631




