The CENP-T C-Terminus Is Exclusively Proximal to H3.1 and not to H3.2 or H3.3
Abstract
:1. Introduction
2. Results
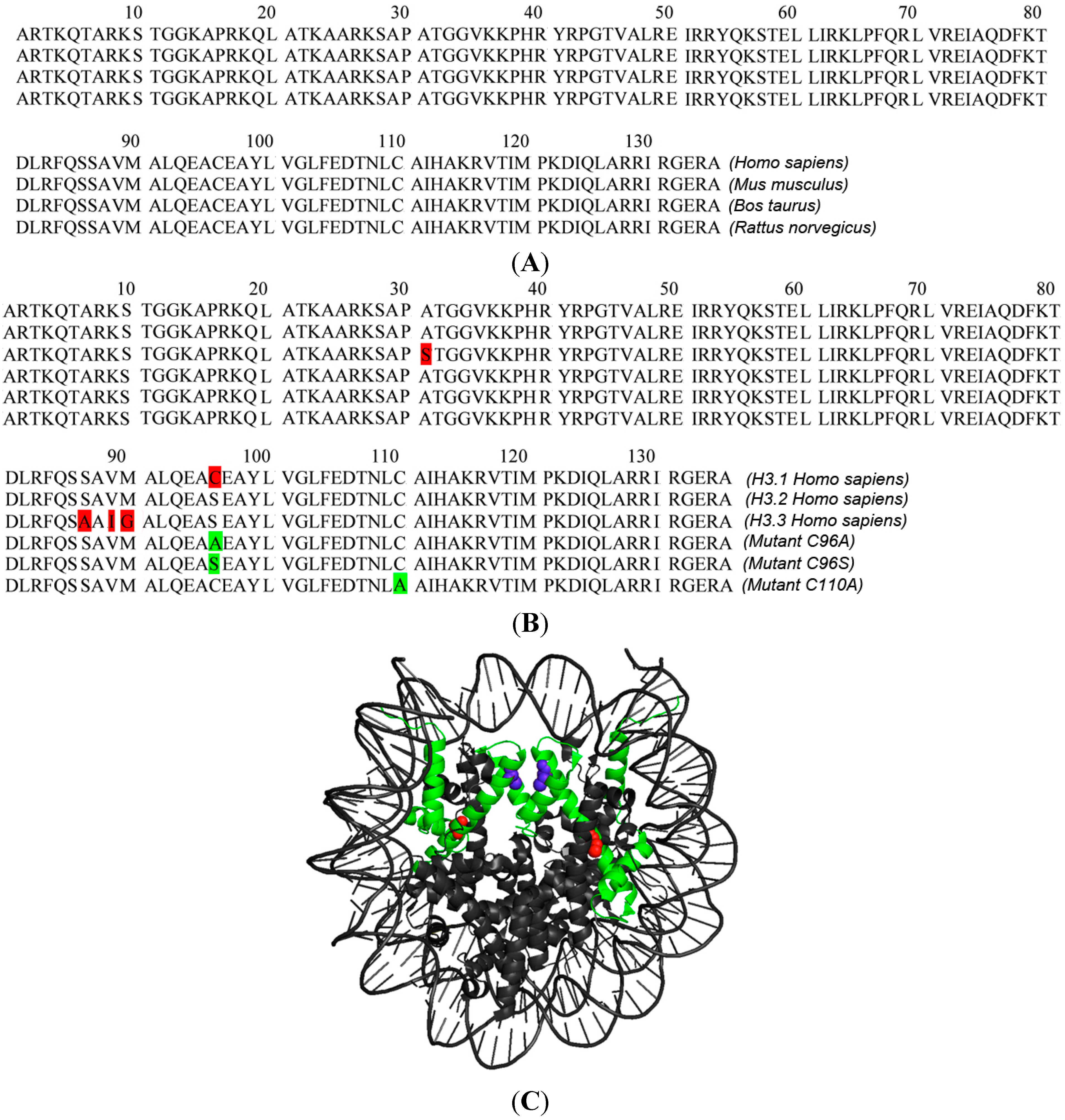
| Donor Fusion | Acceptor Fusion | Mean EFRET % | Mean EVAR% | ΔE % | p-Value | FRET | Number of Kinetochores Bleached (Nkin) |
|---|---|---|---|---|---|---|---|
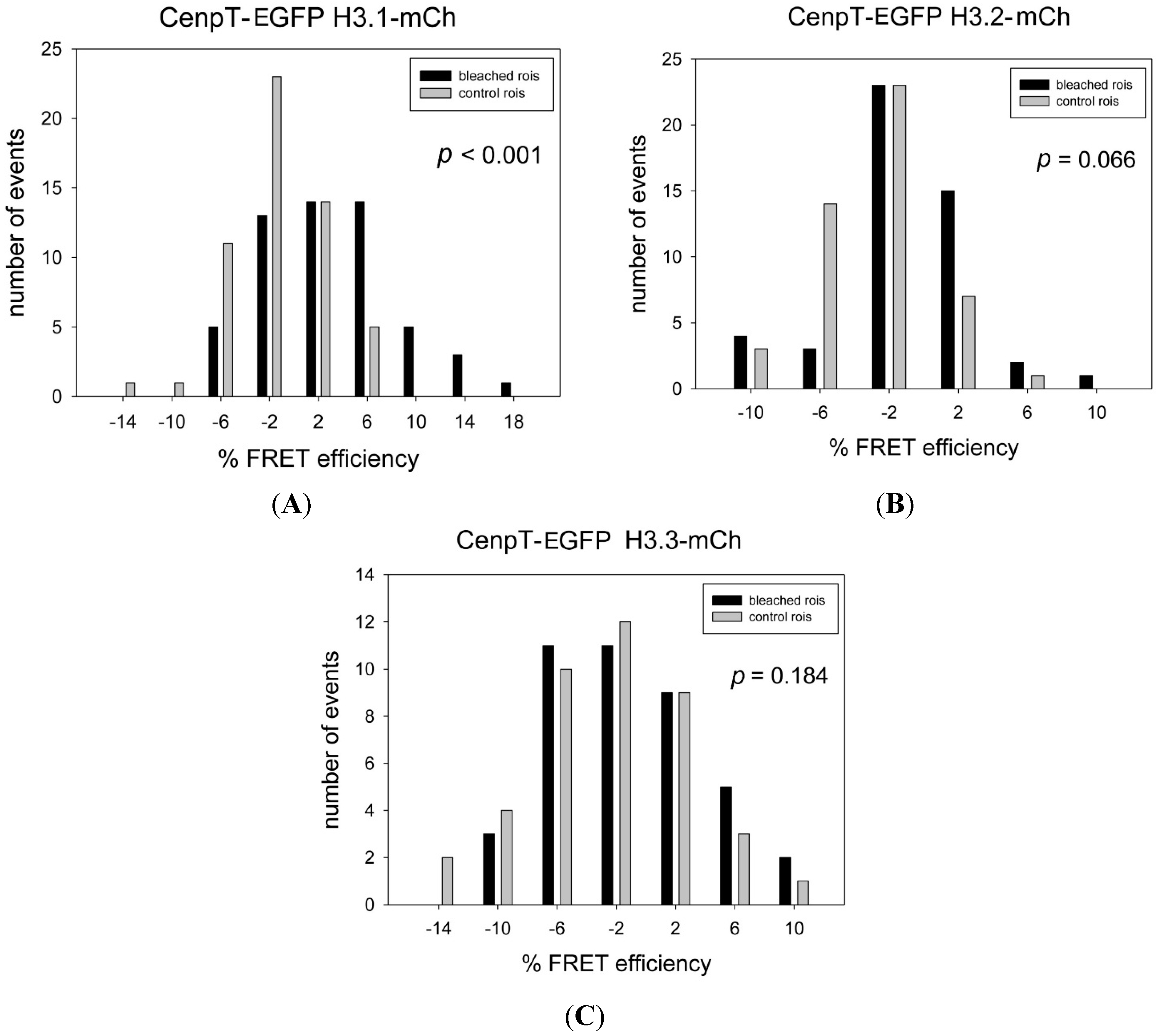
| CENP-T-EGFP * | H3.1-mCh * | 3.03 | −1.41 | 4.45 | <0.001 | ++ | 55 |
| CENP-T-EGFP | H3.2-mCh | −1.54 | −2.89 | 1.35 | 0.066 | − | 48 |
| CENP-T-EGFP | H3.3-mCh | −1.16 | −2.66 | 1.50 | 0,184 | − | 41 |
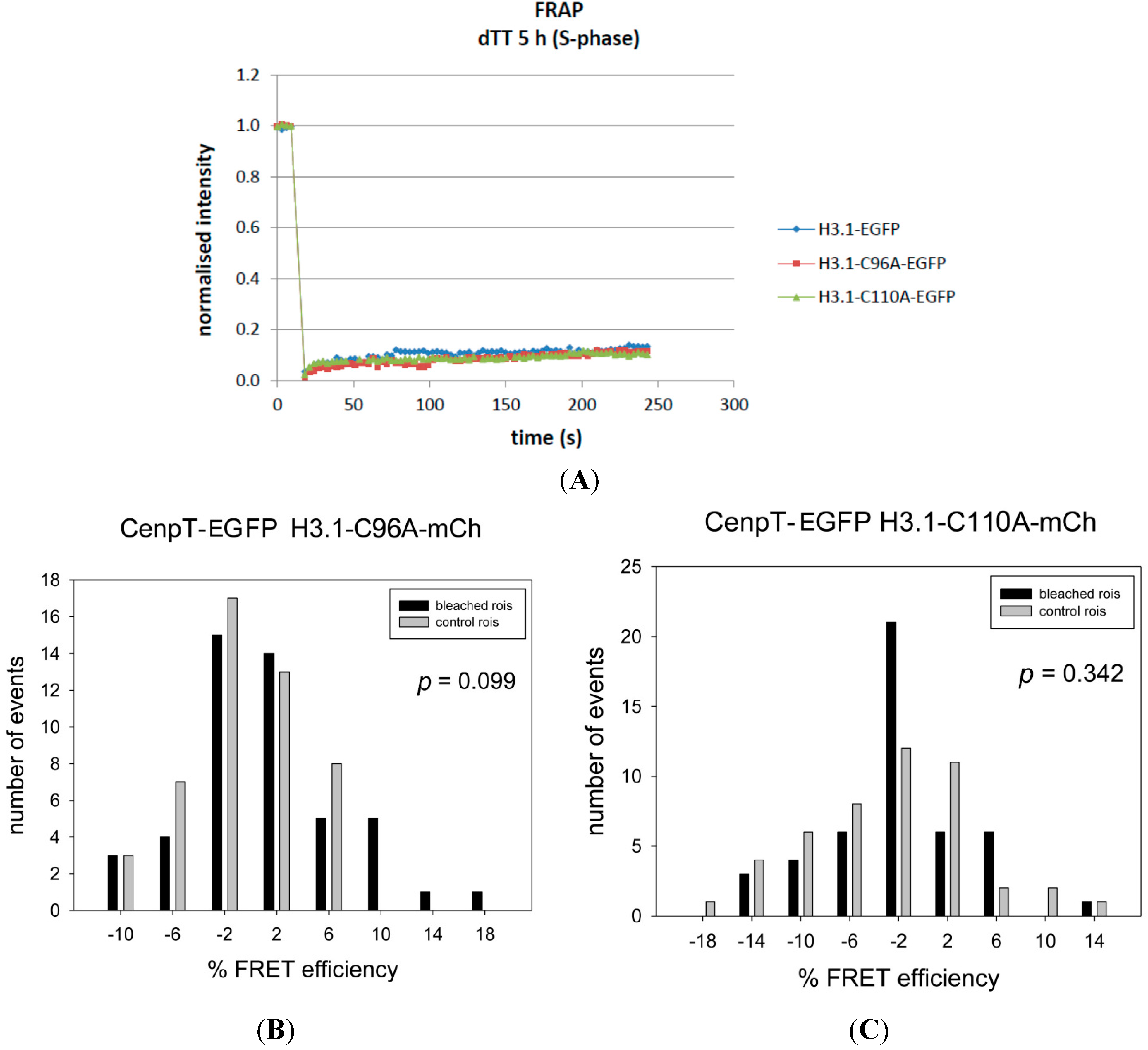
| CENP-T-EGFP | H3.1-C96A-mCh | 0.83 | −0.92 | 1.75 | 0.099 | − | 48 |
| CENP-T-EGFP | H3.1-C110A-mCh | −1.82 | −3.09 | 1.27 | 0.342 | − | 47 |
| CENP-TΔN-EGFP | H3.1-mCh | 3.53 | 1.97 | 1.56 | 0.107 | − | 43 |
| EGFP-CENP-W | H3.1-mCh | 2.00 | 0.27 | 1.73 | 0.251 | − | 36 |
| EGFP-CENP-W | H3.2-mCh | 2.05 | 1.85 | 0.20 | 0.801 | − | 85 |
| EGFP-CENP-W | H3.3-mCh | 0.49 | 1.51 | −1.02 | 0.371 | − | 45 |
| CENP-W-EGFP | mCh-H3.1 | 3.70 | −2.10 | 5.80 | <0.001 | ++ | 37 |
| EGFP-CENP-W | CENP-T-mCh | 7.79 | 0.19 | 7.6 | <0.001 | ++ | 31 |
| EGFP-CENP-W | CENP-B-mCh | 4.37 | 0.75 | 3.60 | <0.001 | ++ | 49 |
| CENP-W-EGFP | mCh-CENP-B | 5.07 | 0.67 | 4.40 | <0.001 | ++ | 30 |
| CENP-W-EGFP | CENP-B-mCh | 1.81 | 0.79 | 1.02 | 0.200 | − | 42 |
| EGFP-CENP-M | mCh-CENP-S | 1.75 | −0.22 | 1.97 | 0.064 | − | 32 |
| EGFP-CENP-M | CENP-S-mCh | 0.87 | 0.05 | 0.82 | 0.427 | − | 39 |
| CENP-M-EGFP | mCh-CENP-S | 4.00 | −0.78 | 4.78 | <0.001 | ++ | 38 |
| CENP-M-EGFP | CENP-S-mCh | 3.49 | −0.42 | 3.91 | <0.001 | ++ | 43 |
| EGFP-CENP-M | H3.1-mCh | 3.58 | −1.42 | 4.99 | <0.001 | ++ | 41 |
| EGFP-CENP-M | H3.1-C96A-mCh | −0.75 | −2.36 | 1,61 | 0.105 | − | 51 |
| EGFP-CENP-M | H3.1-C110A-mCh | 0.54 | −1.49 | 2.03 | 0.011 | − | 53 |
| CENP-M-EGFP | H3.1-mCh | 2.91 | −0.24 | 3.15 | 0.004 | + | 41 |
| CENP-M-mCh | EGFP-CENP-U | 6.08 | −1.78 | 7.86 | <0.001 | ++ | 54 |

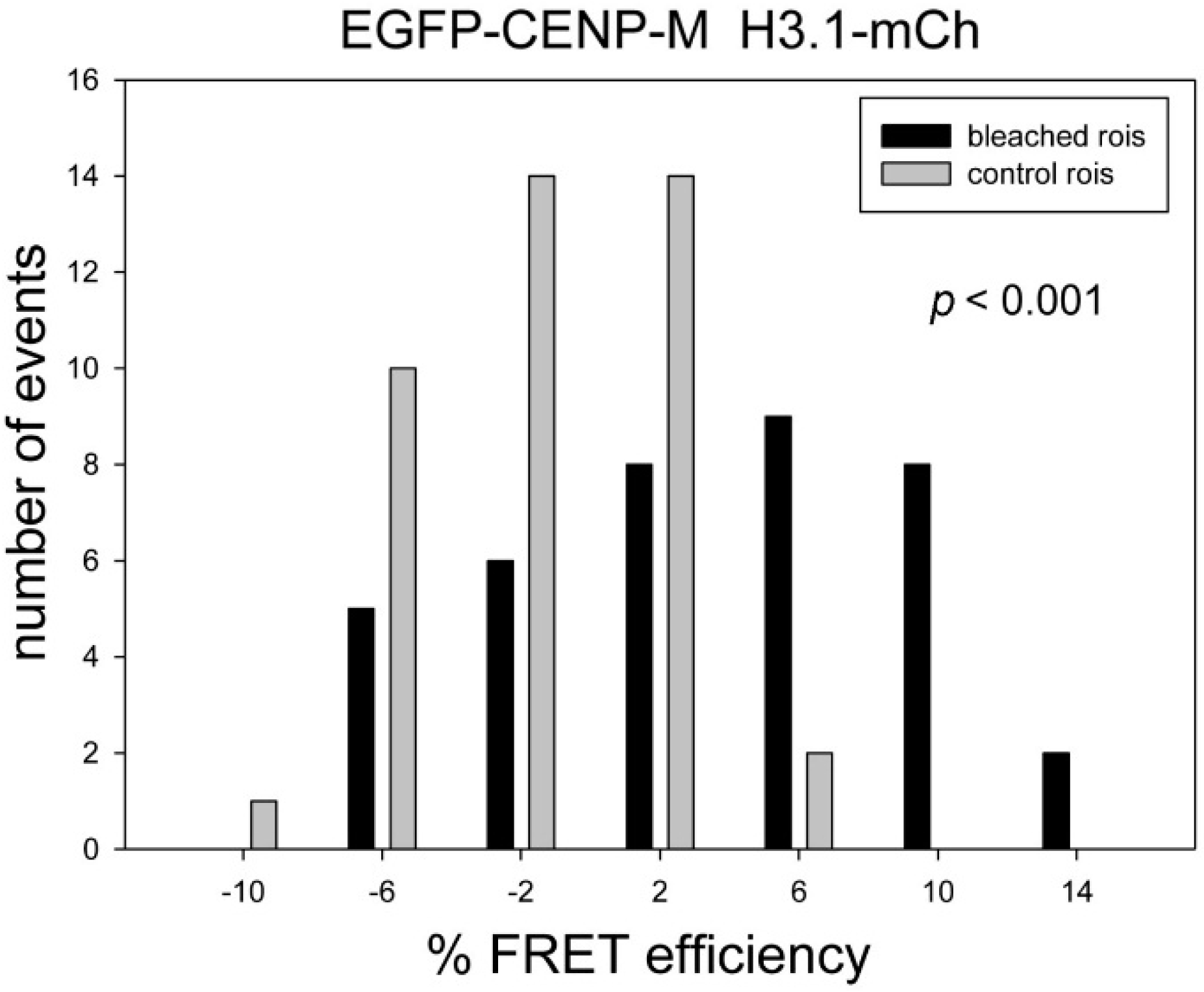
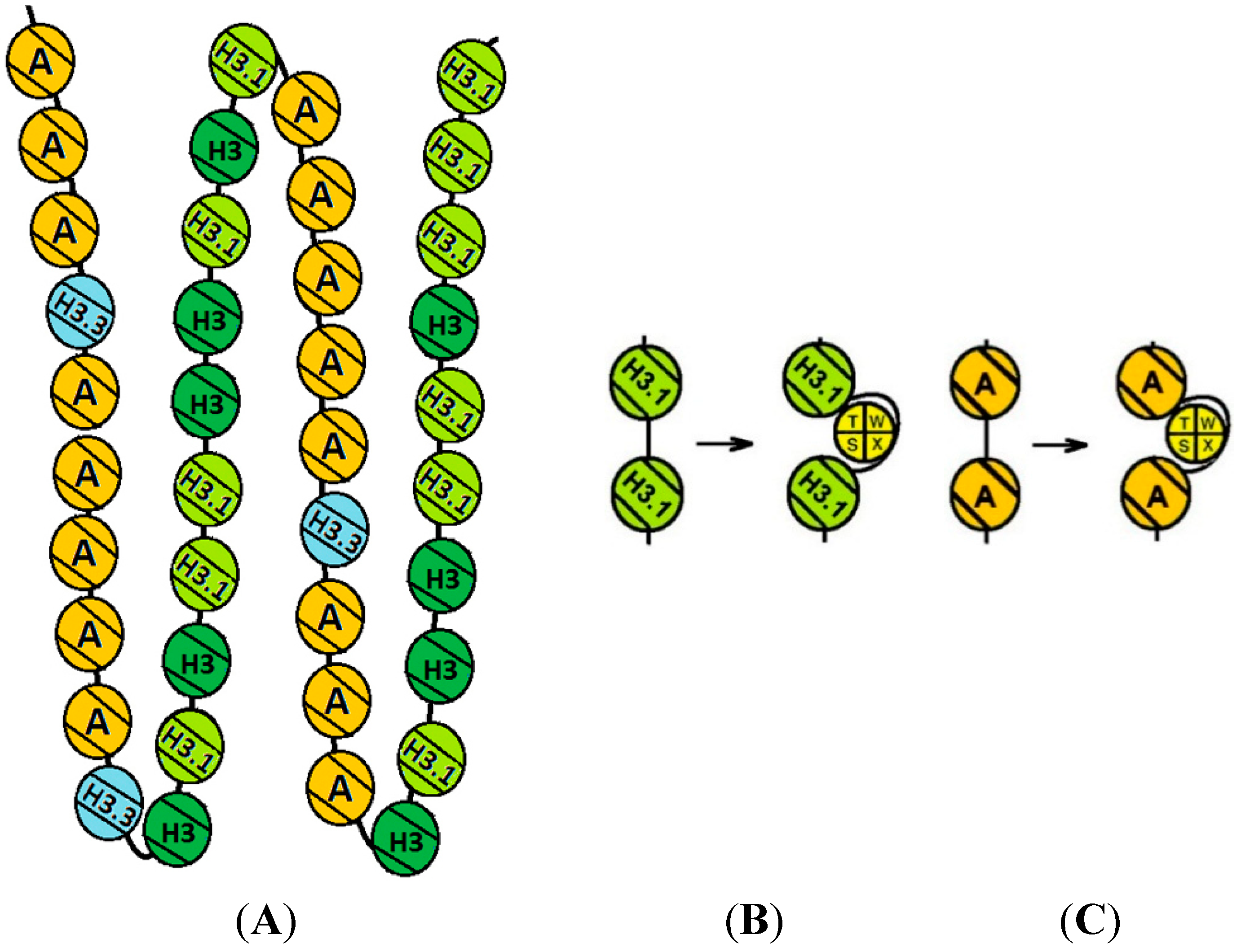
3. Discussion
4. Experimental Section
4.1. Cell Culture
4.2. Plasmids
4.3. Cell Lines and Transfection
4.4. F3H
4.5. Acceptor Photobleaching Based FRET Measurements (AB-FRET)
4.6. Fluorescence Recovery after Photobleaching (FRAP)
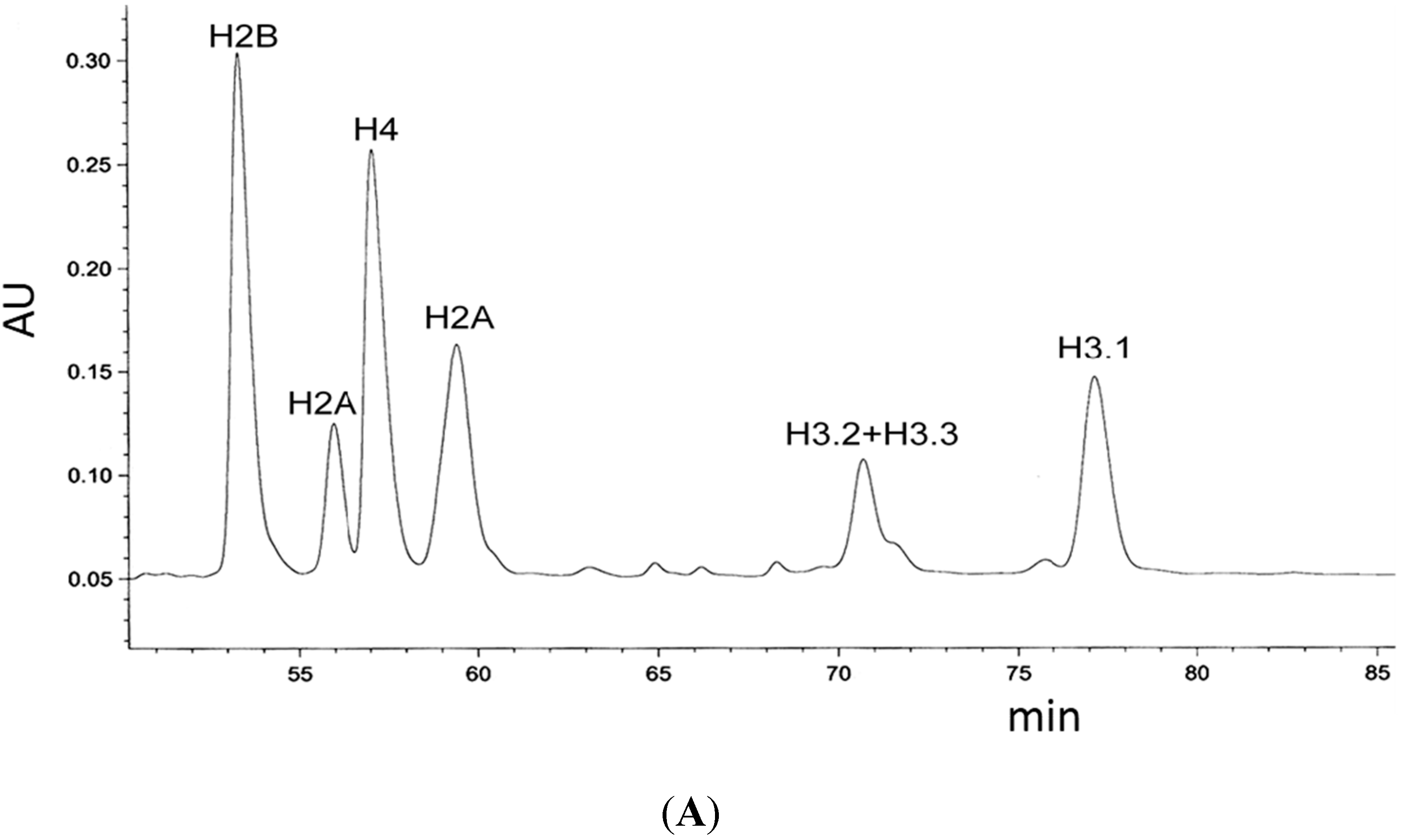
4.7. Analysis of Myc-Tagged H3 Variants by Reverse-Phase HPLC and Fraction Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Santaguida, S.; Musacchio, A. The life and miracles of kinetochores. EMBO J. 2009, 28, 2511–2531. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; O’Quinn, R.P.; Pierce, H.L.; Joglekar, A.P.; Gall, W.E.; DeLuca, J.G.; Carroll, C.W.; Liu, S.T.; Yen, T.J.; McEwen, B.F.; et al. Protein architecture of the human kinetochore microtubule attachment site. Cell 2009, 137, 672–684. [Google Scholar] [CrossRef] [PubMed]
- Przewloka, M.R.; Glover, D.M. The kinetochore and the centromere: A working long distance relationship. Annu. Rev. Genet. 2009, 43, 439–465. [Google Scholar] [CrossRef] [PubMed]
- Perpelescu, M.; Fukagawa, T. The ABCs of CENPs. Chromosoma 2011, 120, 425–446. [Google Scholar] [CrossRef] [PubMed]
- Westermann, S.; Schleiffer, A. Family matters: Structural and functional conservation of centromere-associated proteins from yeast to humans. Trends Cell Biol. 2013, 23, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Cheeseman, I.M.; Chappie, J.S.; Wilson-Kubalek, E.M.; Desai, A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 2006, 127, 983–997. [Google Scholar] [CrossRef] [PubMed]
- Allshire, R.C.; Karpen, G.H. Epigenetic regulation of centromeric chromatin: Old dogs, new tricks? Nat. Rev. Genet. 2008, 9, 923–937. [Google Scholar] [CrossRef] [PubMed]
- Cheeseman, I.M.; Desai, A. Molecular architecture of the kinetochore–microtubule interface. Nat. Rev. Mol. Cell. Biol. 2008, 9, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.T.; Rattner, J.B.; Jablonski, S.A.; Yen, T.J. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 2006, 175, 41–53. [Google Scholar] [CrossRef] [PubMed]
- McClelland, S.E.; Borusu, S.; Amaro, A.C.; Winter, J.R.; Belwal, M.; McAinsh, A.D.; Meraldi, P. The CENP-A NAC/CAD kinetochore complex controls chromosome congression and spindle bipolarity. EMBO J. 2007, 26, 5033–5047. [Google Scholar] [CrossRef] [PubMed]
- Cheeseman, I.M.; Hori, T.; Fukagawa, T.; Desai, A. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol. Biol. Cell 2008, 19, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Amaro, A.C.; Samora, C.P.; Holtackers, R.; Wang, E.; Kingston, I.J.; Alonso, M.; Lampson, M.; McAinsh, A.D.; Meraldi, P. Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nat. Cell Biol. 2010, 12, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Chen, E.S.; Yanagida, M. Requirement of Mis6 centromere connector for localizing a CENP-A like protein in fission yeast. Science 2000, 288, 2215–2219. [Google Scholar] [CrossRef] [PubMed]
- Okada, M.; Cheeseman, I.M.; Hori, T.; Okawa, K.; McLeod, I.X.; Yates, J.R.; Desai, A.; Fukagawa, T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 2006, 8, 446–457. [Google Scholar] [CrossRef] [PubMed]
- Okada, M.; Okawa, K.; Isobe, T.; Fukagawa, T. CENP-H-containing complex facilitates centromere deposition of CENP-A in cooperation with FACT and CHD1. Mol. Biol. Cell 2009, 20, 3986–3995. [Google Scholar] [CrossRef] [PubMed]
- Hori, T.; Shang, W.H.; Takeuchi, K.; Fukagawa, T. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J. Cell Biol. 2013, 200, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Earnshaw, W.C.; Rothfield, N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 1985, 91, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Palmer, D.K.; O’Day, K.; Wener, M.H.; Andrews, B.S.; Margolis, R.L. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 1987, 104, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Palmer, D.K.; O’Day, K.; Trong, H.L.; Charbonneau, H.; Margolis, R.L. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc. Natl. Acad. Sci. USA 1991, 88, 3734–3738. [Google Scholar] [CrossRef] [PubMed]
- Stoler, S.; Keith, K.C.; Curnick, K.E.; Fitzgerald-Hayes, M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genet. Dev. 1995, 9, 573–586. [Google Scholar] [CrossRef]
- Henikoff, S.; Ahmad, K.; Platero, J.S.; van Steensel, B. Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA 2000, 97, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Yoda, K.; Ando, S.; Morishita, S.; Houmura, K.; Hashimoto, K.; Takeyasu, K.; Okazaki, T. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc. Natl. Acad. Sci. USA 2000, 97, 7266–7271. [Google Scholar] [CrossRef] [PubMed]
- Malik, H.S.; Vermaak, D.; Henikoff, S. Recurrent evolution of DNA-binding motifs in the Drosophila centromeric histone. Proc. Natl. Acad. Sci. USA 2002, 99, 1449–1454. [Google Scholar] [CrossRef] [PubMed]
- Black, B.E.; Foltz, D.R.; Chakravarthy, S.; Luger, K.; Woods, V.L., Jr.; Cleveland, D.W. Structural determinants for generating centromeric chromatin. Nature 2004, 430, 578–582. [Google Scholar] [CrossRef] [PubMed]
- Black, B.E.; Jansen, L.E.; Maddox, P.S.; Foltz, D.R.; Desai, A.B.; Shah, J.V.; Cleveland, D.W. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell 2007, 25, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Black, B.E.; Cleveland, D.W. Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell 2011, 144, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Bui, M.; Dimitriadis, E.K.; Hoischen, C.; An, E.; Quenet, D.; Giebe, S.; Nita-Lazar, A.; Diekmann, S.; Dalal, Y. Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo. Cell 2012, 150, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Fachinetti, D.; Diego-Folco, H.; Nechemia-Arbely, Y.; Valente, L.P.; Nguyen, K.; Wong, A.J.; Zhu, Q.; Holland, A.J.; Desai, A.; Jansen, L.E.T.; et al. A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol. 2013, 15, 1056–1066. [Google Scholar] [CrossRef] [PubMed]
- Barnhart, M.C.; Kuich, P.H.; Stellfox, M.E.; Ward, J.A.; Bassett, E.A.; Black, B.E.; Foltz, D.R. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol. 2011, 194, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Mendiburo, M.J.; Padeken, J.; Fulop, S.; Schepers, A.; Heun, P. Drosophila CENH3 is sufficient for centromere formation. Science 2011, 334, 686–690. [Google Scholar] [CrossRef] [PubMed]
- Foltz, D.R.; Jansen, L.E.T.; Black, B.E.; Bailey, A.O.; Yates, J.R.; Cleveland, D.W. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 2006, 8, 458–469. [Google Scholar] [CrossRef] [PubMed]
- Carroll, C.W.; Silva, M.C.C.; Godek, K.M.; Jansen, L.E.T.; Straight, A.F. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 2009, 11, 896–902. [Google Scholar] [CrossRef] [PubMed]
- Carroll, C.W.; Milks, K.J.; Straight, A.F. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 2010, 189, 1143–1155. [Google Scholar] [CrossRef] [PubMed]
- Guse, A.; Carroll, C.W.; Moree, B.; Fuller, C.J.; Straight, A.F. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature 2011, 477, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Jansen, L.E.; Black, B.E.; Foltz, D.R.; Cleveland, D.W. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 2007, 176, 795–805. [Google Scholar] [CrossRef] [PubMed]
- Bodor, D.L.; Valente, L.P.; Mata, J.F.; Black, B.E.; Jansen, L.E.T. Assembly in G1 phase and long-term stability are unique intrinsic features of CENP-A nucleosomes. Mol. Biol. Cell 2013, 24, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Prendergast, L.; van Vuuren, C.; Kaczmarczyk, A.; Döring, V.; Hellwig, D.; Quinn, N.; Hoischen, C.; Diekmann, S.; Sullivan, K.F. Premitotic assembly of human CENPs -T and -W switches centromeric chromatin to a mitotic state. PLoS Biol. 2011, 9, e1001082. [Google Scholar] [CrossRef] [PubMed]
- Raychaudhuri, N.; Dubruille, R.; Orsi, G.A.; Bagheri, H.C.; Loppin, B.; Lehner, C.F. Transgenerational propagation and quantitative maintenance of paternal centromeres depends on CID/CENP-A presence in Drosophila sperm. PLoS Biol. 2012, 10, e1001434. [Google Scholar] [CrossRef] [PubMed]
- Schuh, M.; Lehner, C.F.; Heidmann, S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr. Biol. 2007, 17, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Hemmerich, P.; Weidtkamp-Peters, S.; Hoischen, C.; Schmiedeberg, L.; Erliandri, I.; Diekmann, S. Dynamics of inner kinetochore assembly and maintenance in living cells. J. Cell Biol. 2008, 180, 1101–1114. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.C.; Bodor, D.L.; Stellfox, M.E.; Martins, N.M.; Hochegger, H.; Foltz, D.R.; Jansen, L.E. CDK activity couples epigenetic centromere inheritance to cell cycle progression. Dev. Cell 2012, 22, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Camahort, R.; Shivaraju, M.; Mattingly, M.; Li, B.; Nakanishi, S.; Zhu, D.; Shilatifard, A.; Workman, J.L.; Gerton, J.L. Cse4 is part of an octameric nucleosome in budding yeast. Mol. Cell 2009, 35, 794–805. [Google Scholar] [CrossRef] [PubMed]
- Dalal, Y.; Wang, H.; Lindsay, S.; Henikoff, S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007, 5, e218. [Google Scholar] [CrossRef] [PubMed]
- Dimitriadis, E.K.; Weber, C.; Gill, R.K.; Diekmann, S.; Dalal, Y. Tetrameric organization of vertebrate centromeric nucleosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 20317–20322. [Google Scholar] [CrossRef] [PubMed]
- Tachiwana, H.; Kagawa, W.; Shiga, T.; Osakabe, A.; Miya, Y.; Saito, K.; Hayashi-Takanaka, Y.; Oda, T.; Sato, M.; Park, S.-Y.; et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature 2011, 476, 232–235. [Google Scholar] [CrossRef] [PubMed]
- Cheeseman, I.M.; Anderson, S.; Jwa, M.; Green, E.M.; Kang, J.S.; Yates, J.R.; Chan, C.S.M.; Drubin, D.G.; Barnes, G. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 2002, 111, 163–172. [Google Scholar] [CrossRef] [PubMed]
- De Wulf, P. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genet. Dev. 2003, 17, 2902–2921. [Google Scholar] [CrossRef]
- Obuse, C.; Yang, H.; Nozaki, N.; Goto, S.; Okazaki, T.; Yoda, K. Proteomics analysis of the centromere compleetx from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells 2004, 9, 105–120. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; McLeod, I.; Anderson, S.; Yates, J.R.; He, X. Molecular analysis of kinetochore architecture in fission yeast. EMBO J. 2005, 24, 2919–2930. [Google Scholar] [CrossRef] [PubMed]
- Izuta, H.; Ikeno, M.; Suzuki, N.; Tomonaga, T.; Nozaki, N.; Obuse, C.; Kisu, Y.; Goshima, N.; Nomura, F.; Nomura, N.; et al. Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes Cells 2006, 11, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Hori, T.; Amano, M.; Suzuki, A.; Backer, C.B.; Welburn, J.P.; Dong, Y.; McEwen, B.F.; Shang, W.-H.; Suzuki, E.; Okawa, K.; et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 2008, 135, 1039–1052. [Google Scholar] [CrossRef] [PubMed]
- Hellwig, D.; Hoischen, C.; Ulbricht, T.; Diekmann, S. Acceptor-photobleaching FRET analysis of core kinetochore and NAC proteins in living human cells. Eur. Biophys. J. 2009, 38, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Moree, B.; Meyer, C.B.; Fuller, C.J.; Straight, A.F. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J. Cell Biol. 2011, 194, 855–871. [Google Scholar] [CrossRef] [PubMed]
- Gascoigne, K.E.; Takeuchi, K.; Suzuki, A.; Hori, T.; Fukagawa, T.; Cheeseman, I.M. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell 2011, 145, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Przewloka, M.R.; Venkei, Z.; Bolanos-Garcia, V.M.; Debski, J.; Dadlez, M.; Glover, D.M. CENP-C is a structural platform for kinetochore assembly. Curr. Biol. 2011, 21, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Screpanti, E.; De Antoni, A.; Alushin, G.M.; Petrovic, A.; Melis, T.; Nogales, E.; Musacchio, A. Direct binding of CENP-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol. 2011, 21, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Dambacher, S.; Deng, W.; Hahn, M.; Sadic, D.; Frohlich, J.; Nuber, A.; Hoischen, C.; Diekmann, S.; Leonhardt, H.; Schotta, G. CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus 2012, 3, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Westhorpe, F.G.; Straight, A.F. Functions of the centromere and kinetochore in chromosome segregation. Curr. Opin. Cell Biol. 2013, 25, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Jiang, J.; Zhou, B.-R.; Rozendaal, M.; Feng, H.; Ghirlando, R.; Xiao, T.S.; Straight, A.F.; Bai, Y. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science 2013, 340, 1110–1113. [Google Scholar] [CrossRef] [PubMed]
- Hinshaw, S.M.; Harrison, S.C. An Iml3-Chl4 heterodimer links the core centromere to factors required for accurate chromosome segregation. Cell Rep. 2013, 5, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.S.; Saitoh, S.; Yanagida, M.; Takahashi, K. A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol. Cell. 2003, 11, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Topp, C.N.; Zhong, C.X.; Dawe, R.K. Centromere-encoded RNAs are integral components of the maize kinetochore. Proc. Natl. Acad. Sci. USA 2004, 101, 15986–15991. [Google Scholar] [CrossRef] [PubMed]
- Bouzinba-Segard, H.; Guais, A.; Francastel, C. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc. Natl. Acad. Sci. USA 2006, 103, 8709–8714. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.H.; Brettingham-Moore, K.H.; Chan, L.; Quach, J.M.; Anderson, M.A.; Northrop, E.L.; Hannan, R.; Saffery, R.; Shaw, M.L.; Williams, E.; et al. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007, 17, 1146–1160. [Google Scholar] [CrossRef] [PubMed]
- Ohkuni, K.; Kitagawa, K. Endogenous transcription at the centromere facilitates centromere activity in budding yeast. Curr. Biol. 2011, 21, 1695–1703. [Google Scholar] [CrossRef] [PubMed]
- Quénet, D.; Dalal, Y. A long non-coding RNA is required for targeting centromeric protein A to the human centromere. eLIFE 2014, 3. [Google Scholar] [CrossRef]
- Rošić, S.; Köhler, F.; Erhardt, S. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J. Cell Biol. 2014, 207, 335–349. [Google Scholar] [CrossRef] [PubMed]
- Hellwig, D.; Emmerth, S.; Ulbricht, T.; Döring, V.; Hoischen, C.; Martin, R.; Samora, C.P.; McAinsh, A.D.; Carroll, C.W.; Straight, A.F.; et al. Dynamics of CENP-N kinetochore binding during the cell cycle. J. Cell Sci. 2011, 124, 3871–3883. [Google Scholar] [CrossRef] [PubMed]
- Basilico, F.; Maffini, S.; Weir, J.R.; Prumbaum, D.; Rojas, A.M.; Zimniak, T.; de Antoni, A.; Jeganathan, S.; Voss, B.; van Gerwen, S.; et al. The pseudo GTPase CENP-M drives human kinetochore assembly. eLIFE 2014, 3. [Google Scholar] [CrossRef]
- Liu, S.T.; Hittle, J.C.; Jablonski, S.A.; Campbell, M.S.; Yoda, K.; Yen, T.J. Human CENP-I specifies localization of CENP-F, MAD1 and MAD2 to kinetochores and is essential for mitosis. Nat. Cell Biol. 2003, 5, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Matson, D.R.; Stukenberg, P.T. CENP-I and Aurora B act as a molecular switch that ties RZZ/Mad1 recruitment to kinetochore attachment status. J. Cell Biol. 2014, 205, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Hori, T.; Okada, M.; Maenaka, K.; Fukagawa, T. CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol. Biol. Cell 2008, 19, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Eskat, A.; Deng, W.; Hofmeister, A.; Rudolphi, S.; Emmerth, S.; Hellwig, D.; Ulbricht, T.; Döring, V.; Bancroft, J.M.; McAinsh, A.D.; et al. Step-wise assembly, maturation and dynamic behavior of the human CENP-P/O/R/Q/U kinetochore sub-complex. PLoS One 2012, 7, e44717. [Google Scholar] [CrossRef] [PubMed]
- Schmitzberger, F.; Harrison, S.C. RWD domain: A recurring module in kinetochore architecture shown by a Ctf19-Mcm21 complex structure. Nature 2012, 13, 216–222. [Google Scholar]
- Kagawa, N.; Hori, T.; Hoki, Y.; Hosoya, O.; Tsutsui, K.; Saga, Y.; Sado, T.; Fukagawa, T. The CENP-O complex requirement varies among different cell types. Chromatogr. Res. 2014, 22, 293–303. [Google Scholar]
- Nishino, T.; Takeuchi, K.; Gascoigne, K.E.; Suzuki, A.; Hori, T.; Oyama, T.; Morikawa, K.; Cheeseman, I.M.; Fukagawa, T. CENP-T/W/S/X forms a unique centromeric chromatin structure with a histone-like fold. Cell 2012, 148, 487–501. [Google Scholar] [CrossRef] [PubMed]
- Dornblut, C.; Quinn, N.; Monajambashi, S.; Prendergast, L.; van Vuuren, C.; Münch, S.; Deng, W.; Leonhardt, H.; Cardoso, M.C.; Hoischen, C.; et al. A CENP-S/X complex assembles at the centromere in S and G2 phases of the human cell cycle. Open Biol. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Nishino, T.; Mayanagi, K.; Horikoshi, N.; Osakabe, A.; Tachiwana, H.; Hori, T.; Kurumizaka, H.; Fukagawa, T. The centromeric nucleosome-like CENP-T-W-S-X complex induces positive supercoils into DNA. Nucleic Acids Res. 2014, 42, 1644–1655. [Google Scholar] [CrossRef] [PubMed]
- Nishino, T.; Rago, F.; Hori, T.; Tomii, K.; Cheeseman, I.M.; Fukagawa, T. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J. 2013, 32, 424–436. [Google Scholar] [CrossRef] [PubMed]
- Amano, M.; Suzuki, A.; Hori, T.; Backer, C.; Okawa, K.; Cheeseman, I.M.; Fukagawa, T. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J. Cell Biol. 2009, 186, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Ohta, S.; Bukowski-Wills, J.C.; Sanchez-Pulido, L.; Alves Fde, L.; Wood, L.; Chen, Z.A.; Platani, M.; Fischer, L.; Hudson, D.F.; Ponting, C.P.; et al. The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell 2010, 142, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Measday, V.; Hailey, D.W.; Pot, I.; Givan, S.A.; Hyland, K.M.; Cagney, G.; Fields, S.; Davis, T.N.; Hieter, P. Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Gen. Dev. 2002, 16, 101–113. [Google Scholar] [CrossRef]
- Pidoux, A.L.; Richardson, W.; Allshire, R.C. Sim4: A novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. J. Cell Biol. 2003, 161, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Pot, I.; Measday, V.; Snydsman, B.; Cagney, G.; Fields, S.; Davis, T.N.; Muller, E.G.D.; Hieter, P. Chl4p and Iml3p are two new members of the budding yeast outer kinetochore. Mol. Biol. Cell 2003, 14, 460–476. [Google Scholar] [CrossRef] [PubMed]
- Westermann, S.; Cheeseman, I.M.; Anderson, S.; Yates, J.R., III; Drubin, D.G.; Barnes, G. Architecture of the buddingyeast kinetochore reveals a conserved molecular core. J. Cell Biol. 2003, 163, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Wieland, G.; Orthaus, S.; Ohndorf, S.; Diekmann, S.; Hemmerich, P. Functional complementation of human centromere protein A (CENP-A) by Cse4p from Saccharomyces cerevisiae. Mol. Cell. Biol. 2004, 24, 6620–6630. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Chang, H.L.; Kagami, A.; Watanabe, Y. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev. Cell 2009, 17, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Schleiffer, A.; Maier, M.; Litos, G.; Lampert, F.; Hornung, P.; Mechtler, K.; Westermann, S. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat. Cell Biol. 2012, 14, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Hornung, P.; Troc, P.; Malvezzi, F.; Maier, M.; Demianova, Z.; Zimniak, T.; Litos, G.; Lampert, F.; Schleiffer, A.; Brunner, M.; et al. A cooperative mechanism drives budding yeast kinetochore assembly downstream of CENP-A. J. Cell Biol. 2014, 206, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Przewloka, M.R.; Zhang, W.; Costa, P.; Archambault, V.; D’Avino, P.P.; Lilley, K.S.; Laue, E.D.; McAinsh, A.D.; Glover, D.M. Molecular analysis of core kinetochore composition and assembly in Drosophila melanogaster. PLoS One 2007, 2, e478. [Google Scholar] [CrossRef] [PubMed]
- Zinkowski, R.P.; Meyne, J.; Brinkley, B.R. The centromere-kinetochore complex: A repeat subunit model. J. Cell Biol. 1991, 113, 1091–1110. [Google Scholar] [CrossRef] [PubMed]
- Blower, M.D.; Sullivan, B.A.; Karpen, G.H. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell 2002, 2, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Dunleavy, E.M.; Almouzni, G.; Karpen, G.H. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G phase. Nucleus 2011, 2, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Hake, S.B.; Garcia, B.A.; Duncan, E.M.; Kauer, M.; Dellaire, G.; Shabanowitz, J.; Bazett-Jones, D.P.; Allis, C.D.; Hunt, D.F. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 2006, 281, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, B.A.; Karpen, G.H. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 2004, 11, 1076–1083. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, S.A.; Vagnarelli, P.; Dong, Y.; Hori, T.; McEwen, B.F.; Fukagawa, T.; Flors, C.; Earnshaw, W.C. A super-resolution map of the vertebrate kinetochore. Proc. Natl. Acad. Sci. USA 2010, 107, 10484–10489. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, J.H.; Rodriguez, M.G.; Martins, N.M.; Kimura, H.; Kelly, D.A.; Masumoto, H.; Larionov, V.; Jansen, L.E.; Earnshaw, W.C. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011, 30, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Boyarchuk, E.; Filipescu, D.; Vassias, I.; Cantaloube, S.; Almouzni, G. The histone variant composition of centromeres is controlled by the pericentric heterochromatin state during the cell cycle. J. Cell Sci. 2014, 127, 3347–3359. [Google Scholar] [CrossRef] [PubMed]
- Tschernyschkow, S.; Herda, S.; Grünert, G.; Döring, V.; Görlich, D.; Hofmeister, A.; Hoischen, C.; Dittrich, P.; Diekmann, S.; Ibrahim, B. Rule-based modeling and simulations of the inner kinetochore structure. Prog. Biophys. Mol. Biol. 2013, 113, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Diekmann, S.; Hoischen, C. Biomolecular dynamics and binding studies in the living cell. Phys. Life Rev. 2014, 11, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Tramier, M.; Zahid, M.; Mevel, J.C.; Masse, M.J.; Coppey-Moisan, M. Sensitivity of CFP/YFP and GFP/mCherry pairs to donor photobleaching on FRET determination by fluorescence lifetime imaging microscopy in living cells. Microsc. Res. Tech. 2006, 69, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Lam, A.; st-Pierre, F.; Gong, Y.; Marshall, J.D.; Cranfill, P.J.; Baird, M.A.; McKeown, M.R.; Wiedenmann, J.; Davidson, M.W.; Schnitzer, M.J.; et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat. Methods 2012, 9, 1005–1012. [Google Scholar] [CrossRef] [PubMed]
- Orthaus, S.; Biskup, C.; Hoffmann, B.; Hoischen, C.; Ohndorf, S.; Benndorf, K.; Diekmann, S. Assembly of the inner kinetochore proteins CENP-A and CENP-B in living human cells. Chembiochem 2008, 9, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Orthaus, S.; Klement, K.; Happel, N.; Hoischen, C.; Diekmann, S. Linker histone H1 is present in centromeric chromatin of living human cells next to inner kinetochore proteins. Nucleic Acids Res. 2009, 37, 3391–3406. [Google Scholar] [CrossRef] [PubMed]
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Tsunaka, Y.; Kajimura, N.; Tate, S.; Morikawa, K. Alteration of the nucleosomal DNA path in the crystal structure of a human nucleosome core particle. Nucleic Acids Res. 2005, 33, 3424–3434. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Cook, P.R. Kinetics of core histones in living human cells: Little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 2001, 153, 1341–1353. [Google Scholar] [CrossRef] [PubMed]
- Camerini-Otero, R.D.; Felsenfeld, G. Histone H3 disulfide dimers and nucleosome structure. Proc. Natl. Acad. Sci. USA 1977, 74, 5519–5523. [Google Scholar] [CrossRef] [PubMed]
- Hake, S.B.; Allis, C.D. Histone H3 variants and their potential role in indexing mammalian genomes: The “H3 barcode hypothesis”. Proc. Natl. Acad. Sci. USA 2006, 103, 6428–6435. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.; Stillman, B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 1989, 58, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Tagami, H.; Ray-Gallet, D.; Almouzni, G.; Nakatani, A. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent of DNA synthesis. Cell 2004, 116, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Latreille, D.; Bluy, L.; Benkirane, M.; Kiernan, R.E. Identification of histone 3 variant 2 interacting factors. Nucleic Acids Res. 2014, 42, 3542–3550. [Google Scholar] [CrossRef] [PubMed]
- Banks, D.D.; Gloss, L.M. Folding mechanism of the (H3–H4)2 histone tetramer of the core nucleosome. Protein Sci. 2004, 13, 1304–1316. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gimenez, J.L.; Olaso, G.; Hake, S.B.; Bönisch, C.; Wiedemann, S.M.; Markovic, J.; Dasi, F.; Gimeno, A.; Perez-Quilis, C.; Palacios, O.; et al. Histone H3 glutathionylation in proliferating mammalian cells destabilizes nucleosomal structure. Antioxid. Redox Signal. 2013, 19, 1305–1320. [Google Scholar] [CrossRef] [PubMed]
- Earnshaw, W.C.; Sullivan, K.F.; Machlin, P.S.; Cooke, C.A.; Kaiser, D.A.; Pollard, T.D.; Rothfield, N.F.; Cleveland, D.W. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J. Cell Biol. 1987, 104, 817–829. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, K.; Masumoto, H.; Ikeda, M.; Okazaki, T. Analysis of protein–DNA and protein–protein interactions of centromere protein B (CENP-B) and properties of the DNA–CENP-B complex in the cell cycle. Mol. Cell. Biol. 1995, 15, 1602–1612. [Google Scholar] [PubMed]
- Tanaka, Y.; Nureki, O.; Kurumizaka, H.; Fukai, S.; Kawaguchi, S.; Ikuta, M.; Iwahara, J.; Okazaki, T.; Yokoyama, S. Crystal structure of the CENP-B protein–DNA complex: The DNA-binding domains of CENP-B induce kinks in the CENP-B box DNA. EMBO J. 2001, 20, 6612–6618. [Google Scholar] [CrossRef] [PubMed]
- Monier, K.; Mouradian, S.; Sullivan, K.F. DNA methylation promotes Aurora-B-driven phosphorylation of histone H3 in chromosomal subdomains. J. Cell Sci. 2007, 120, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, S.M.; Mildner, S.N.; Bönisch, C.; Israel, L.; Maiser, A.; Matheisl, S.; Straub, T.; Merkl, R.; Leonhardt, H.; Kremmer, E.; et al. Identification and characterization of two novel primate-specific histone H3 variants, H3.X and H3.Y. J. Cell Biol. 2010, 190, 777–791. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, T.; Hashiguchi, N.; Janicki, S.M.; Tumbar, T.; Belmont, A.S.; Spector, D.L. Visualization of gene activity in living cells. Nat. Cell Biol. 2000, 2, 871–878. [Google Scholar] [CrossRef] [PubMed]
- Rothbauer, U.; Zolghadr, K.; Tillib, S.; Nowak, D.; Schermelleh, L.; Gahl, A.; Backmann, N.; Conrath, K.; Muyldermans, S.; Cardoso, M.C.; et al. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods 2006, 3, 887–889. [Google Scholar] [CrossRef] [PubMed]
- Zolghadr, K.; Mortusewicz, O.; Rothbauer, U.; Kleinhans, R.; Goehler, H.; Wanker, E.E.; Cardoso, M.C.; Leonhardt, H. A fluorescent two-hybrid assay for direct visualization of protein interactions in living cells. Mol. Cell. Proteomics 2008, 7, 2279–2287. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Huang, S. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 2001, 153, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Schmiedeberg, L.; Weisshart, K.; Diekmann, S.; Meyer zu Hoerste, G.; Hemmerich, P. High- and Low-mobility Populations of HP1 in Heterochromatin of Mammalian Cells. Mol. Biol. Cell 2004, 15, 2819–2833. [Google Scholar] [CrossRef] [PubMed]
- Shechter, D.; Dormann, H.L.; Allis, C.D.; Hake, S.B. Extraction, purification and analysis of histones. Nat. Protoc. 2007, 2, 1445–1457. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abendroth, C.; Hofmeister, A.; Hake, S.B.; Kamweru, P.K.; Miess, E.; Dornblut, C.; Küffner, I.; Deng, W.; Leonhardt, H.; Orthaus, S.; et al. The CENP-T C-Terminus Is Exclusively Proximal to H3.1 and not to H3.2 or H3.3. Int. J. Mol. Sci. 2015, 16, 5839-5863. https://doi.org/10.3390/ijms16035839
Abendroth C, Hofmeister A, Hake SB, Kamweru PK, Miess E, Dornblut C, Küffner I, Deng W, Leonhardt H, Orthaus S, et al. The CENP-T C-Terminus Is Exclusively Proximal to H3.1 and not to H3.2 or H3.3. International Journal of Molecular Sciences. 2015; 16(3):5839-5863. https://doi.org/10.3390/ijms16035839
Chicago/Turabian StyleAbendroth, Christian, Antje Hofmeister, Sandra B. Hake, Paul K. Kamweru, Elke Miess, Carsten Dornblut, Isabell Küffner, Wen Deng, Heinrich Leonhardt, Sandra Orthaus, and et al. 2015. "The CENP-T C-Terminus Is Exclusively Proximal to H3.1 and not to H3.2 or H3.3" International Journal of Molecular Sciences 16, no. 3: 5839-5863. https://doi.org/10.3390/ijms16035839
APA StyleAbendroth, C., Hofmeister, A., Hake, S. B., Kamweru, P. K., Miess, E., Dornblut, C., Küffner, I., Deng, W., Leonhardt, H., Orthaus, S., Hoischen, C., & Diekmann, S. (2015). The CENP-T C-Terminus Is Exclusively Proximal to H3.1 and not to H3.2 or H3.3. International Journal of Molecular Sciences, 16(3), 5839-5863. https://doi.org/10.3390/ijms16035839





