2.1. The hDbr1 Protein Interacts with Xab2
We previously identified the components of the IL complex, which is an intermediate complex for post-splicing intron turnover steps [
17]. Although hDbr1 could debranch lariat introns in the IS complex, it was not able to debranch them in the IL complex [
17]. These results strongly suggest that factor(s) in the IL complex inhibits hDbr1 association with the intron lariat complex and the IS complex factor(s) confers hDbr1 association on it. However, the factors in the IS complex have yet to be identified [
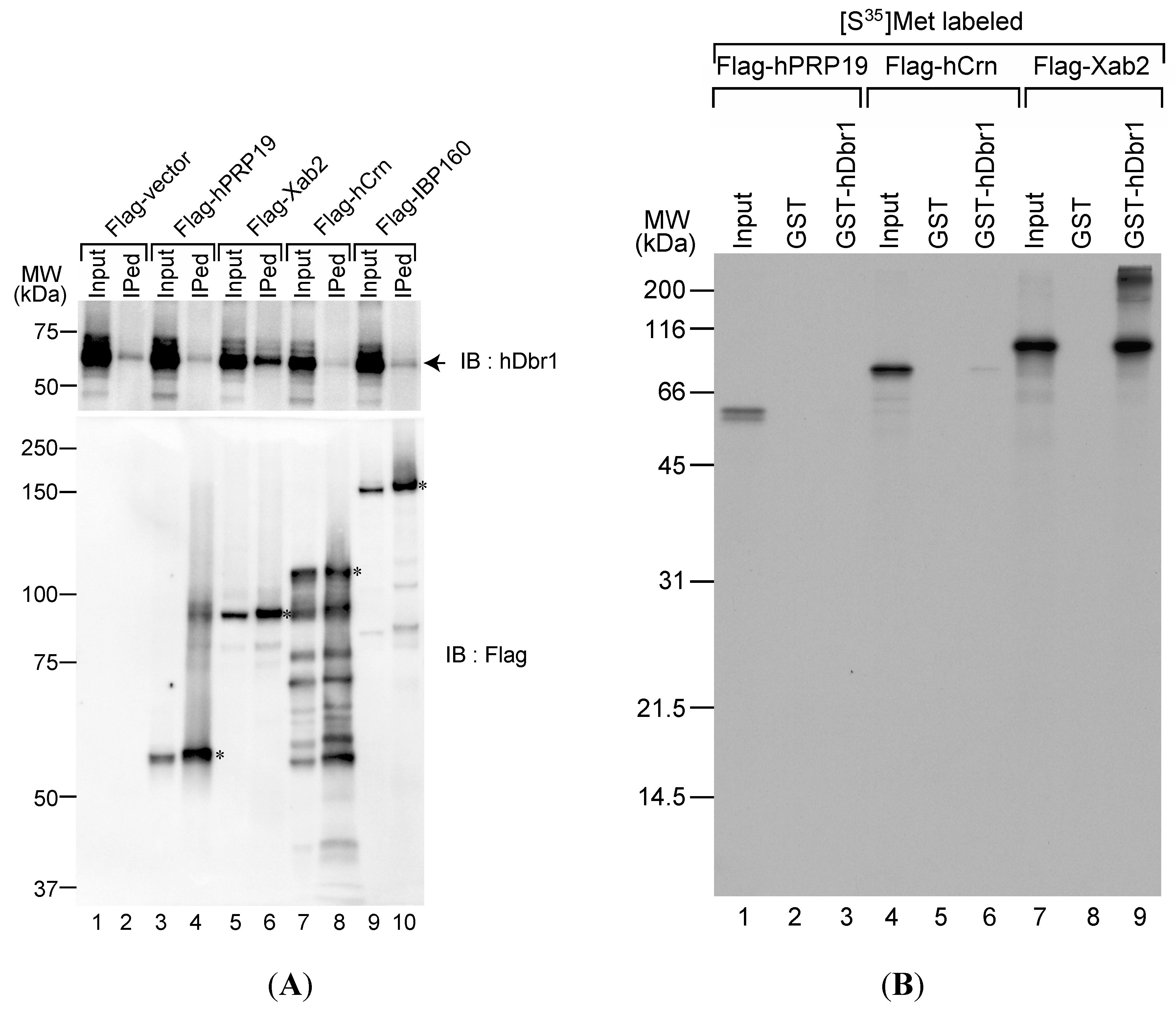
17]. Therefore, we attempted to determine interactions between the IL complex proteins and hDbr1, because the IL complex likely contains the IS complex components. We carried out immunoprecipitation experiments with Flag-tagged IL complex proteins followed by Western blotting using specific antibodies against hDbr1 protein. We transfected cDNAs encoding hPrp19, Xab2, hCrn and IBP160 into HEK293T cells. Flag-vector was used as a control. As shown in
Figure 1A, transfected cDNAs were successfully expressed in HEK293T cells and Flag-tagged proteins were produced (lower panel). We carried out immunoprecipitation from total cell lysates by using anti-Flag M2 antibody to test the immunoprecipitation efficiency. The antibody precipitated full-length Flag-tagged proteins from the lysates (
Figure 1A, lower panel). Under these conditions, anti-hDbr1 antibody was used to test whether or not endogenous hDbr1 protein is co-precipitated with those proteins.
Figure 1A demonstrates that only Flag-Xab2 precipitated endogenous hDbr1 protein (upper panel, lane 6). We then tested protein–protein interaction between Xab2 and hDbr1
in vitro.
In vitro translated proteins were incubated with either GST or GST-hDbr1 protein and bound proteins were analyzed by SDS-PAGE. As shown in
Figure 1B,
in vitro translated Xab2 bound to GST-hDbr1, while hPRP19 did not bind and hCrn bound very weakly (lanes 3, 6 and 9). Taken together, these results indicate that hDbr1 can interact with Xab2 among the IL complex components we tested.
Figure 1.
The hDbr1 protein associates with xeroderma pigmentosum, complementeation group A (XPA)-binding protein 2 (Xab2) in vivo and in vitro. (A) The in vivo association of several IL complex proteins with hDbr1 protein was analyzed by immunoprecipitation assay. HEK293T whole-cell extract was prepared after transfection with Flag-vector, Flag-hPrp19, Flag-Xab2, Flag-hCrn or Flag-IBP160 plasmid. Flag-tagged proteins were precipitated using anti-Flag M2 agarose from whole-cell extract, and the precipitates (IPed) were subjected to Western blotting analyses using anti-hDbr1 antibody (upper panel) and an anti-Flag polyclonal antibody (lower panel). A portion (5%) of the precipitated proteins (lanes marked as Input) was also immunoblotted. The position of endogenous hDbr1 protein is indicated by an arrow in the upper panel. The full-length Flag-tagged proteins are marked with asterisks in the lower panel. The positions of the protein size markers are described on the left of the panel; and (B) In vitro binding of hDbr1 to Xab2. Flag-tagged hPrp19, hCrn and Xab2 proteins, produced and labeled with [35S]methionine in vitro, were incubated with 5 μg each of GST or GST-hDbr1. After precipitation and washing, bound proteins were eluted, separated on 12.5% SDS-PAGE gel and visualized by fluorography. The lanes marked Input contain 10% of the total protein used in each binding reaction. The position of molecular mass markers is shown on the left of the panel.
Figure 1.
The hDbr1 protein associates with xeroderma pigmentosum, complementeation group A (XPA)-binding protein 2 (Xab2) in vivo and in vitro. (A) The in vivo association of several IL complex proteins with hDbr1 protein was analyzed by immunoprecipitation assay. HEK293T whole-cell extract was prepared after transfection with Flag-vector, Flag-hPrp19, Flag-Xab2, Flag-hCrn or Flag-IBP160 plasmid. Flag-tagged proteins were precipitated using anti-Flag M2 agarose from whole-cell extract, and the precipitates (IPed) were subjected to Western blotting analyses using anti-hDbr1 antibody (upper panel) and an anti-Flag polyclonal antibody (lower panel). A portion (5%) of the precipitated proteins (lanes marked as Input) was also immunoblotted. The position of endogenous hDbr1 protein is indicated by an arrow in the upper panel. The full-length Flag-tagged proteins are marked with asterisks in the lower panel. The positions of the protein size markers are described on the left of the panel; and (B) In vitro binding of hDbr1 to Xab2. Flag-tagged hPrp19, hCrn and Xab2 proteins, produced and labeled with [35S]methionine in vitro, were incubated with 5 μg each of GST or GST-hDbr1. After precipitation and washing, bound proteins were eluted, separated on 12.5% SDS-PAGE gel and visualized by fluorography. The lanes marked Input contain 10% of the total protein used in each binding reaction. The position of molecular mass markers is shown on the left of the panel.
![Ijms 16 03705 g001]()
2.2. Identification of a Novel Protein as a Specific Interactor to hDbr1
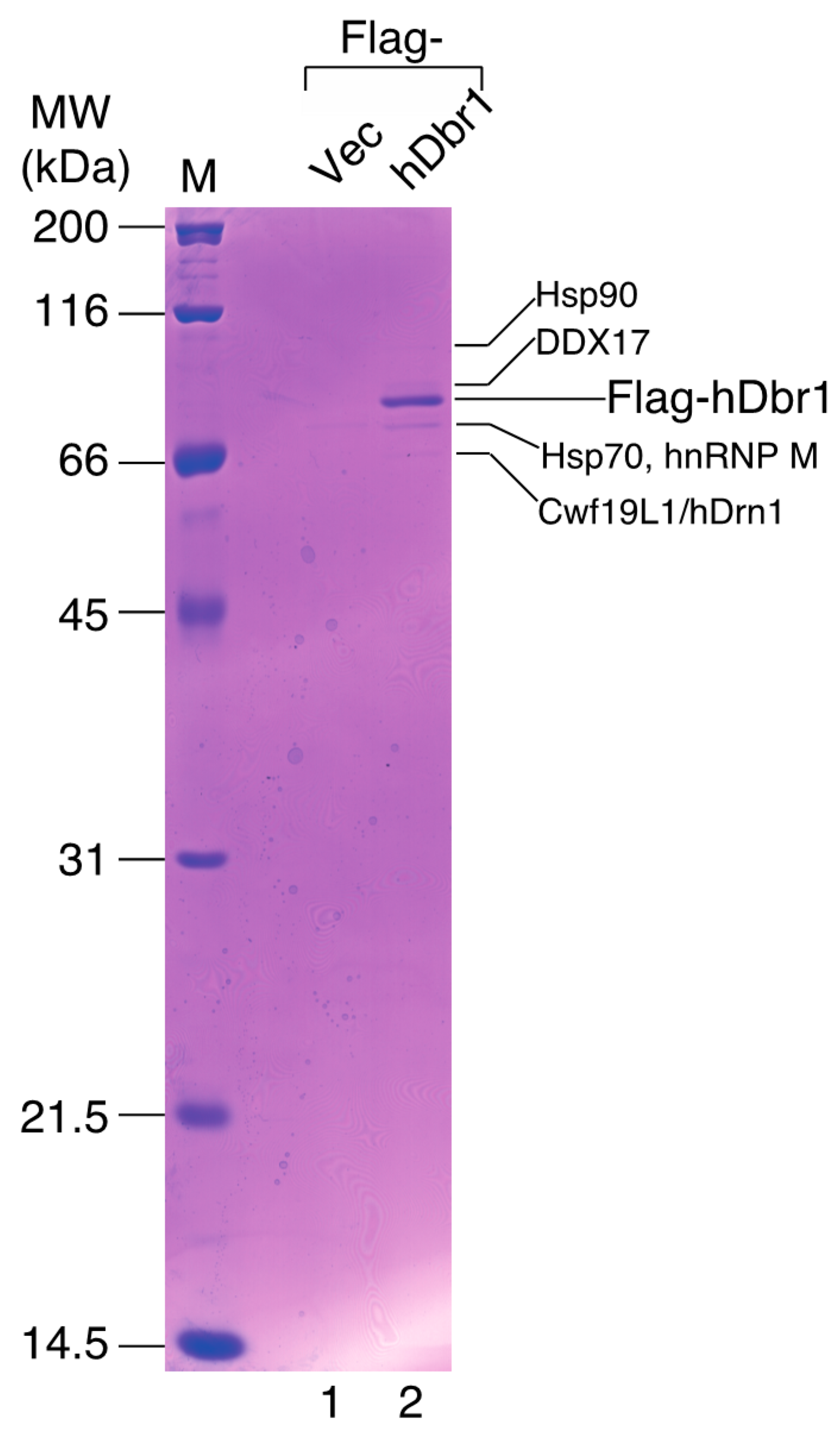
In order to identify more specific binders to hDbr1, we carried out co-immunoprecipitation experiments followed by mass spectrometry analysis. Flag-tagged hDbr1 protein was expressed in HEK293T cells and precipitated by anti-Flag M2 resin. After spectrometry analysis, several proteins were identified, as shown in
Figure 2 (lane 2). Among the interactors, we found an unknown protein, which has recently been designated as Cwf19L1 [
20]. We searched for possible orthologs of Cwf19L1 in other organisms using protein databases and found that this protein is well conserved among many species, from budding yeast to humans, suggesting its important role(s) (
Figure 3), since it shows high homology to the yeast
Ygr093w gene product. This gene is also described as debranching enzyme-associated ribonuclease 1 in the
Saccharomyces gene database (SGD) [
19], so we designated it as human Drn1 (hDrn1) and further characterized the interaction between hDbr1 and hDrn1. HSP70 and HSP90 were also identified as the interactors to hDbr1 in
Figure 2 (lane 2). Although these two proteins were identified in lariat-intron complex [
17], we found that these proteins also bind to other overexpressed proteins and MS2 protein likely non-specifically (data not shown). Thus, we did not analyze these proteins further in this work.
Figure 2.
Identification of the proteins interacting with hDbr1 by MALDI-TOF mass spectrometry. Immunoprecipitation of the hDbr1 complexes with an antibody against Flag tag (M2; Sigma, St. Louis, MO, USA) from HEK293T cell extracts overexpressing Flag-hDbr1 (lane 2). The whole-cell extract prepared from the Flag-pcDNA3-transfected cells was used as a control (Flag-Vec, lane 1). Proteins were separated by 12.5% SDS-PAGE and visualized by Coomassie Brilliant Blue staining. The proteins identified by MALDI-TOF mass spectrometry are indicated by their names. The positions of the size marker proteins are indicated by their sizes (kDa) on the left.
Figure 2.
Identification of the proteins interacting with hDbr1 by MALDI-TOF mass spectrometry. Immunoprecipitation of the hDbr1 complexes with an antibody against Flag tag (M2; Sigma, St. Louis, MO, USA) from HEK293T cell extracts overexpressing Flag-hDbr1 (lane 2). The whole-cell extract prepared from the Flag-pcDNA3-transfected cells was used as a control (Flag-Vec, lane 1). Proteins were separated by 12.5% SDS-PAGE and visualized by Coomassie Brilliant Blue staining. The proteins identified by MALDI-TOF mass spectrometry are indicated by their names. The positions of the size marker proteins are indicated by their sizes (kDa) on the left.
Figure 3.
Amino acid sequence alignment of Drn1 proteins from various organisms. Drn1 proteins from
Saccharomyces cerevisiae (NP_011607.3),
Schizosaccharomyces pombe (NP_593012.1),
Homo sapiens (NP_060764.3),
Mus musculus (NP_001074546.1),
Caenorhabditis elegans (NP_504577.1) and
Danio rerio (NP_001038223.1) were aligned using the ClustalW alignment program (
http://www.genome.jp/tools/clustalw/). Identical and similar residues are indicated by dark shading.
Figure 3.
Amino acid sequence alignment of Drn1 proteins from various organisms. Drn1 proteins from
Saccharomyces cerevisiae (NP_011607.3),
Schizosaccharomyces pombe (NP_593012.1),
Homo sapiens (NP_060764.3),
Mus musculus (NP_001074546.1),
Caenorhabditis elegans (NP_504577.1) and
Danio rerio (NP_001038223.1) were aligned using the ClustalW alignment program (
http://www.genome.jp/tools/clustalw/). Identical and similar residues are indicated by dark shading.
2.3. The hDrn1 Protein Binds Specifically and Directly to hDbr1
We first determined the subcellular localization of hDrn1 in HeLa cells. The hDrn1 protein was expressed as a fusion protein with myc tag peptide in HeLa cells and immunofluorescence analysis was carried out by using anti-myc tag antibody. As shown in
Figure 4A, myc-hDrn1 was localized mainly in the nucleoplasm of HeLa cells and some nucleolus signals could be detected (α-myc panel), which is similar to that of hDbr1 (α-hDbr1 panel). The anti-hDbr1 antibody also exhibited some dot-like signals in the cytoplasm, whose nature is currently unknown (α-hDbr1 panel). Next, we tested whether hDrn1 interacts with hDbr1
in vivo. Flag-tagged hDrn1 protein was expressed in HEK293T cells by transfection and hDrn1 was immunoprecipitated using anti-Flag M2 resin. Flag-pcDNA3 vector was used as a negative control. The precipitated fractions were analyzed by Western blotting. The results in
Figure 4B show that endogenous hDbr1 protein was specifically and efficiently precipitated by Flag-hDrn1, while β-actin was not (
Figure 4B, lane 4). We also confirmed the specific binding of hDrn1 to hDbr1
in vitro. The recombinant Dbr1 protein was expressed in
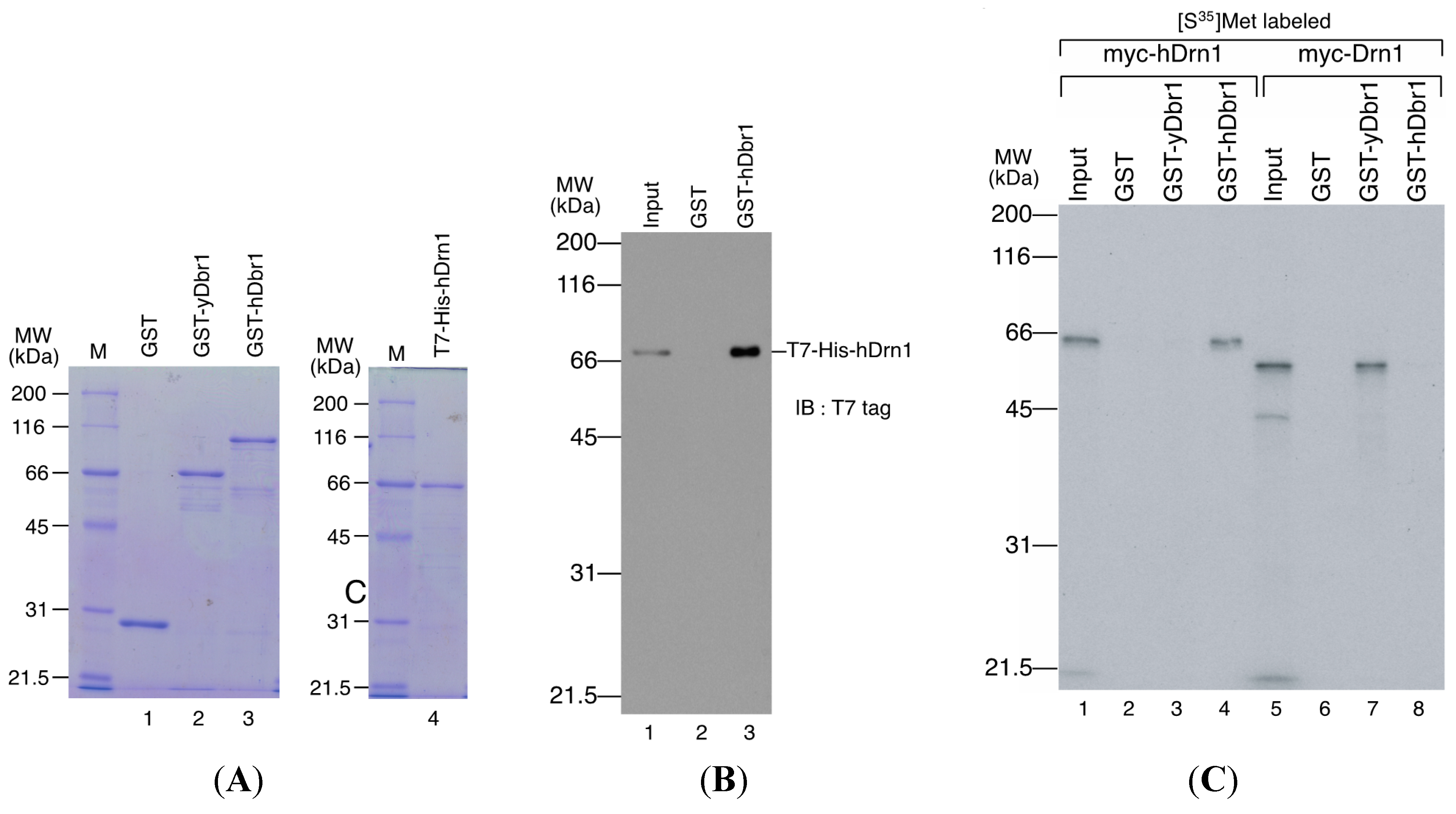
E. coli cells and purified as a fusion protein to GST (
Figure 5A, lane 3). GST-fused yeast Dbr1 protein and GST protein were also prepared for binding assays (
Figure 5A, lanes 1 and 2). For the direct binding assay, we prepared T7- and His-tagged hDrn1 protein (
Figure 5A, lane 4). In order to test whether hDrn1 interacts with hDbr1 directly or not, we carried out
in vitro binding assays using recombinant proteins. T7-His-tagged hDrn1 protein was mixed with GST-hDbr1 protein, and GST was used as a negative control. The results in
Figure 5B indicate that hDrn1 interacts with hDbr1 directly through protein-protein interaction (lane 3). Since hDrn1 showed high similarity to yeast Drn1, we then checked whether the binding between Drn1 and Dbr1 was conserved in these two organisms or not. GST-yDbr1, GST-hDbr1 and GST proteins were incubated with either yDrn1 or hDrn1 prepared by
in vitro translation. As shown in
Figure 5C, human Drn1 bound to GST-hDbr1, but not to yeast Dbr1 and GST (lanes 2–4). In contrast, yeast Drn1 interacted with GST-yDbr1, and did not bind to GST-hDbr1 and GST (
Figure 5C, lanes 6–8). These results indicate that the specific interaction between Dbr1 and Drn1 is conserved in yeast and humans, but human protein does not interact with yeast protein and
vice versa.
We then tried to determine the interaction domains of hDbr1 and hDrn1 by
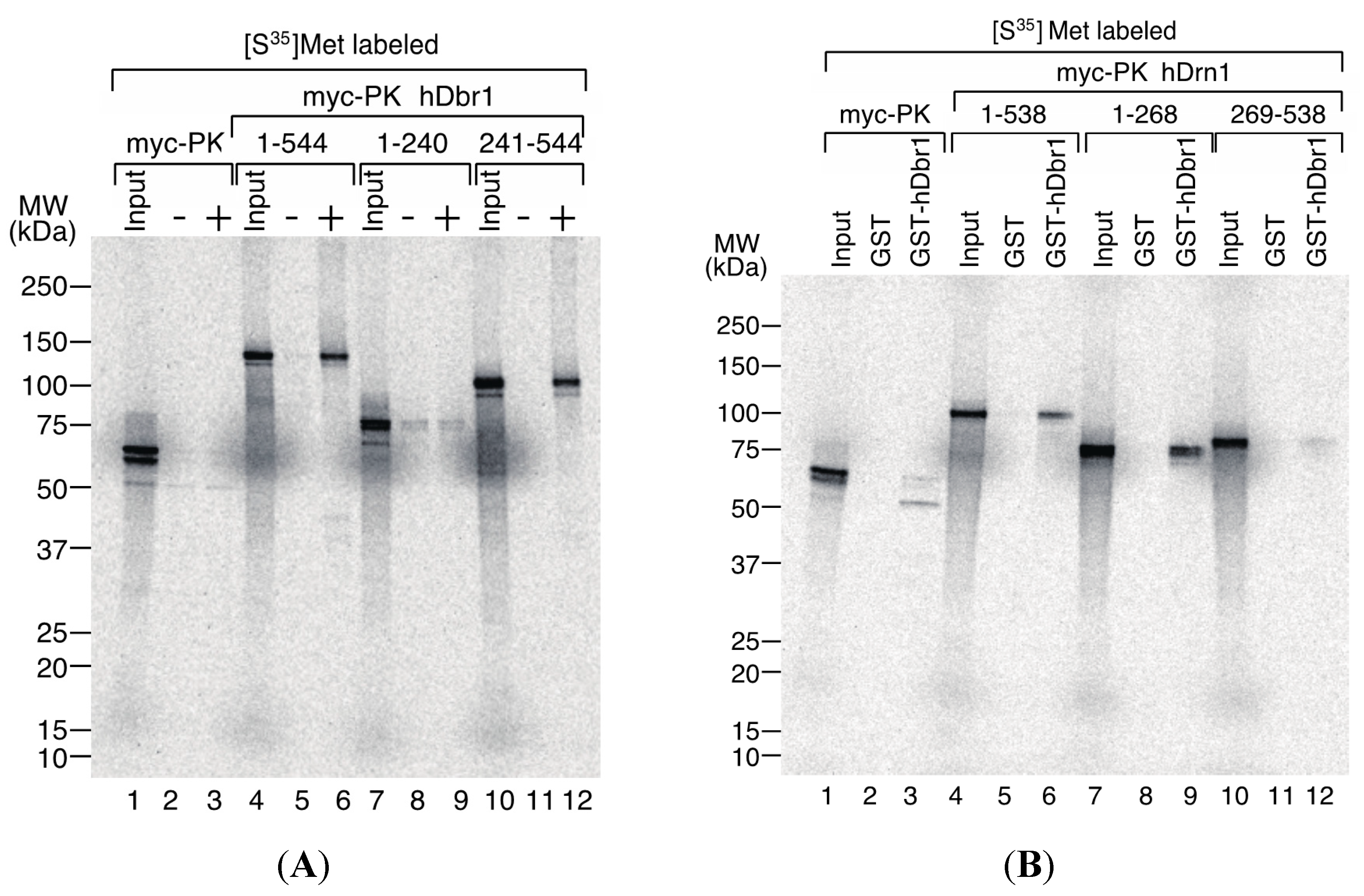
in vitro binding experiments. For the delineation of the binding domain of hDbr1 to hDrn1, T7-His-hDrn1 protein, which was used for the experiments in
Figure 5, was employed. The T7-His-hDrn1 proteins immobilized by anti-T7 tag antibody on protein A beads were incubated with
in vitro translated proteins that have several different portions of hDbr1 protein fused with myc-Pyruvate Kinase (PK) [
16]. The myc-PK protein was used as a negative control (lanes 1–3). As shown in
Figure 6A, myc-PK hDbr1 full length (aa. 1–544) was able to bind to T7-His-hDrn1, while myc-PK alone showed slight binding as the non-specific level (lanes 4–6 and 1–3, respectively). Carboxy-terminal region (aa. 241–544) of hDbr1 bound to T7-His-hDrn1, but amino-terminal portion (aa. 1–240) of hDbr1, which has a catalytic domain for debranching, exhibited weak binding nonspecifically (lanes 10–12 and 7–9, respectively). These results indicate that the binding of hDbr1 to hDrn1 is mostly conferred by carboxy-terminal region. We next investigated which domain of hDrn1 has the binding activity to hDbr1
in vitro. Either full-length (aa. 1–538), amino-terminus (aa. 1–268) or carboxy-terminus (aa. 269–538) of hDrn1 was fused to myc-PK. Those proteins were produced by
in vitro translation and incubated with either GST alone or GST-hDbr1 protein shown in
Figure 5A.
Figure 6B demonstrated that both the full-length and amino-terminus of hDrn1 bound to GST-hDbr1, but not to GST alone (lanes 4–6 and 7–9). In contrast, carboxy-terminus of hDrn1 exhibited very weak binding to GST-hDbr1 as myc-PK alone did (lanes 10–12 and 1–3, respectively). Taken together, amino-terminus region of hDrn1 mostly mediates the binding to hDbr1, which is consistent with the previously reported results with Drn1 by yeast two-hybrid method [
19].
Figure 4.
hDrn1 localizes to the nucleoplasm in HeLa cells by immunofluorescence and co-immunoprecipitates with endogenous hDbr1. (A) The expression vector encoding myc-tagged hDrn1 was transfected into HeLa cells. Twenty-four hours after transfection, the cells were stained with anti-myc (M192-3, MBL, panel FITC, middle), anti-hDbr1 16019-1-AP, Proteintech) and Hoechst 33342 (SIGMA, panel Hoechst, bottom) to label the nuclei. Differential interference contrast (DIC) image of the cells is also shown as Phase panel at the top; and (B) Co-immunoprecipitation of endogenous hDbr1 with Flag-hDrn1 in vivo. Flag-tagged hDrn1 was expressed in HEK293T cells and immunoprecipitated using anti-Flag M2 antibody (SIGMA). Co-precipitated fractions were separated using 12.5% SDS-PAGE gel and analyzed by Western blotting using anti-hDbr1 and anti-Flag polyclonal antibodies (upper panel) and anti-β-actin antibody (MBL) (lower panel). Input lanes contain 5% of the total proteins used for immunoprecipitation assays. The positions of protein mass markers are shown on the left in kDa.
Figure 4.
hDrn1 localizes to the nucleoplasm in HeLa cells by immunofluorescence and co-immunoprecipitates with endogenous hDbr1. (A) The expression vector encoding myc-tagged hDrn1 was transfected into HeLa cells. Twenty-four hours after transfection, the cells were stained with anti-myc (M192-3, MBL, panel FITC, middle), anti-hDbr1 16019-1-AP, Proteintech) and Hoechst 33342 (SIGMA, panel Hoechst, bottom) to label the nuclei. Differential interference contrast (DIC) image of the cells is also shown as Phase panel at the top; and (B) Co-immunoprecipitation of endogenous hDbr1 with Flag-hDrn1 in vivo. Flag-tagged hDrn1 was expressed in HEK293T cells and immunoprecipitated using anti-Flag M2 antibody (SIGMA). Co-precipitated fractions were separated using 12.5% SDS-PAGE gel and analyzed by Western blotting using anti-hDbr1 and anti-Flag polyclonal antibodies (upper panel) and anti-β-actin antibody (MBL) (lower panel). Input lanes contain 5% of the total proteins used for immunoprecipitation assays. The positions of protein mass markers are shown on the left in kDa.
![Ijms 16 03705 g004]()
Figure 5.
hDrn1 binds directly with hDbr1 and the binding of these proteins is conserved among species. (A) Proteins used for in vitro binding assays. GST, GST-yeast Dbr1 (GST-yDbr1), GST-hDbr1 and His-hDrn1 were overexpressed and purified from E. coli cells. Two micrograms of each protein was separated using 12.5% SDS-PAGE gels and stained with Coomassie Brilliant Blue. The molecular mass markers are also shown on the left of the gels with the sizes in kDa; (B) Direct binding of hDrn1 with hDbr1 in vitro. Two micrograms of purified His-tagged hDrn1 was incubated with 5 μg each of GST or GST-hDbr1. Bound proteins were separated by 12.5% SDS-PAGE and detected by Western blotting with anti-T7 tag antibody (Novagen). The molecular mass markers are also shown on the left of the gels; and (C) Specific binding between Dbr1 and Drn1 is conserved between yeast and humans. The myc-tagged yDrn1 and hDrn1 proteins were produced and labeled with [35S]methionine in vitro. Those proteins were incubated with 5 μg each of GST, GST-yDbr1 or GST-hDbr1. After precipitation and washing, bound proteins were eluted, separated on 12.5% SDS-PAGE gel and visualized by fluorography. The lanes marked Input contain 10% of the total protein used in each binding reaction. The position of the molecular mass markers is shown on the left of the panel in kDa.
Figure 5.
hDrn1 binds directly with hDbr1 and the binding of these proteins is conserved among species. (A) Proteins used for in vitro binding assays. GST, GST-yeast Dbr1 (GST-yDbr1), GST-hDbr1 and His-hDrn1 were overexpressed and purified from E. coli cells. Two micrograms of each protein was separated using 12.5% SDS-PAGE gels and stained with Coomassie Brilliant Blue. The molecular mass markers are also shown on the left of the gels with the sizes in kDa; (B) Direct binding of hDrn1 with hDbr1 in vitro. Two micrograms of purified His-tagged hDrn1 was incubated with 5 μg each of GST or GST-hDbr1. Bound proteins were separated by 12.5% SDS-PAGE and detected by Western blotting with anti-T7 tag antibody (Novagen). The molecular mass markers are also shown on the left of the gels; and (C) Specific binding between Dbr1 and Drn1 is conserved between yeast and humans. The myc-tagged yDrn1 and hDrn1 proteins were produced and labeled with [35S]methionine in vitro. Those proteins were incubated with 5 μg each of GST, GST-yDbr1 or GST-hDbr1. After precipitation and washing, bound proteins were eluted, separated on 12.5% SDS-PAGE gel and visualized by fluorography. The lanes marked Input contain 10% of the total protein used in each binding reaction. The position of the molecular mass markers is shown on the left of the panel in kDa.
![Ijms 16 03705 g005]()
Figure 6.
Determination of the interaction domains of hDbr1 and hDrn1. (A) 2 μg of T7-His-hDrn1 proteins were incubated with in vitro translated myc-PK, myc-PK hDbr1 (aa. 1–544), myc-PK hDbr1 (aa. 1–240) and myc-PK hDbr1 (aa. 241–544). After washing, bound proteins were eluted, separated on 5%–20% gradient SDS-PAGE and visualized by fluorography. Input lanes contain 5% of the total protein used in each binding experiment. The positions of protein molecular mass markers are shown on the left of the panel; and (B) In vitro translated myc-PK, myc-PK hDrn1 (aa. 1–538), myc-PK hDrn1 (aa. 1–268) and myc-PK hDrn1 (aa. 269–538) were incubated with 5 μg of either GST alone or GST-hDbr1. The bound proteins were analyzed and visualized as in (A). Input lanes contain 5% of the total protein used in each binding experiment. The positions of protein molecular mass markers are shown on the left of the panel.
Figure 6.
Determination of the interaction domains of hDbr1 and hDrn1. (A) 2 μg of T7-His-hDrn1 proteins were incubated with in vitro translated myc-PK, myc-PK hDbr1 (aa. 1–544), myc-PK hDbr1 (aa. 1–240) and myc-PK hDbr1 (aa. 241–544). After washing, bound proteins were eluted, separated on 5%–20% gradient SDS-PAGE and visualized by fluorography. Input lanes contain 5% of the total protein used in each binding experiment. The positions of protein molecular mass markers are shown on the left of the panel; and (B) In vitro translated myc-PK, myc-PK hDrn1 (aa. 1–538), myc-PK hDrn1 (aa. 1–268) and myc-PK hDrn1 (aa. 269–538) were incubated with 5 μg of either GST alone or GST-hDbr1. The bound proteins were analyzed and visualized as in (A). Input lanes contain 5% of the total protein used in each binding experiment. The positions of protein molecular mass markers are shown on the left of the panel.
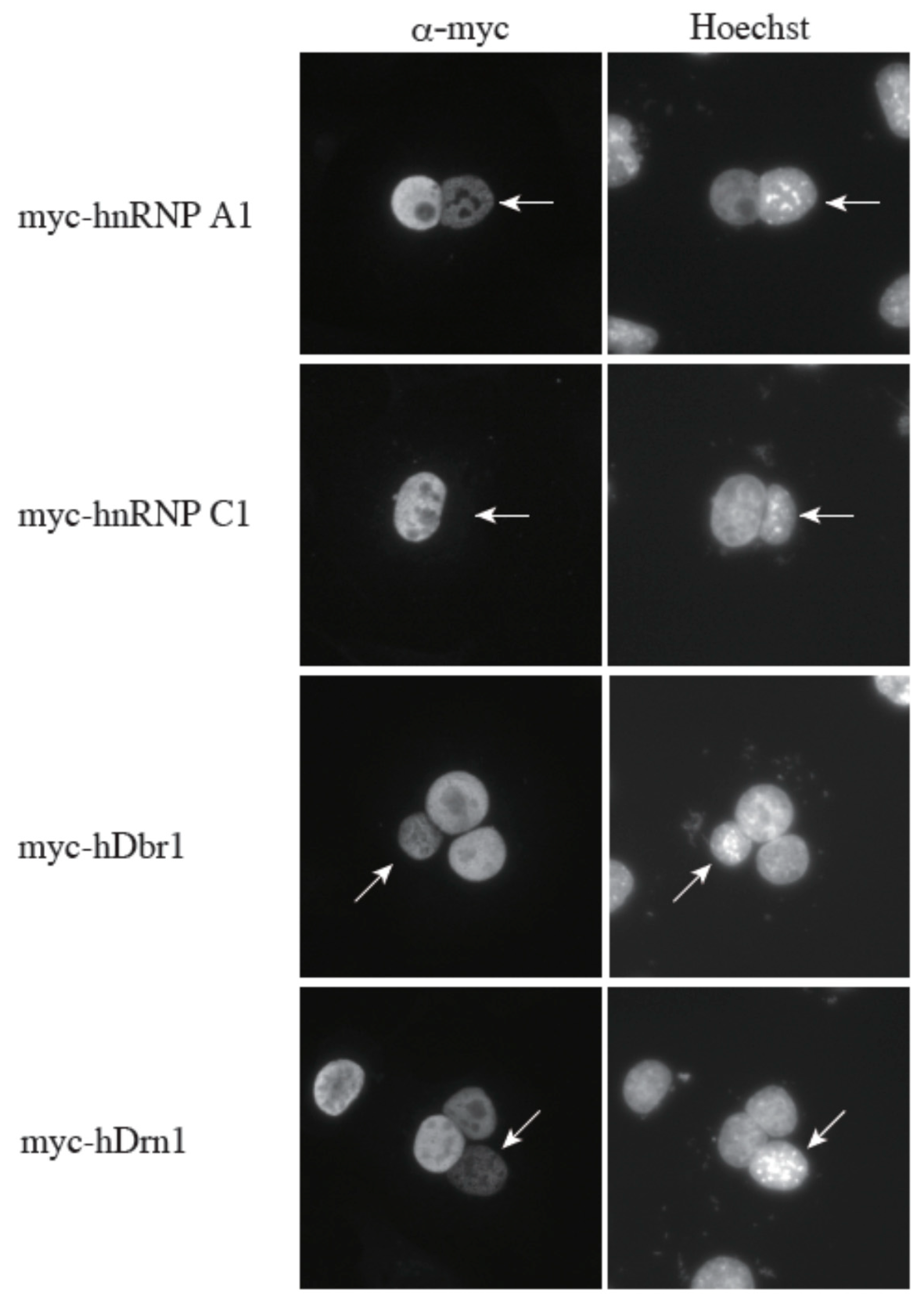
2.4. The hDrn1 Protein Has Shuttling Activity between the Nucleus and the Cytoplasm
We previously demonstrated that hDbr1 shuttles between the nucleus and the cytoplasm [
16]. Since hDrn1 binds to hDbr1, we hypothesized that hDrn1 also has nucleo-cytoplasmic shuttling activity. In order to test this hypothesis, we performed a heterokaryon assay (
Figure 7). Heterokaryons were formed by polyethylene glycol (PEG)-mediated fusions of HeLa cells that had been transfected with the indicated constructs in advance and mouse NIH3T3 cells. Staining of the cells with a DNA dye, Hoechst 33342, allowed us to distinguish between the HeLa and the NIH3T3 nuclei. After cell fusion, subsequent incubations were carried out in the presence of cycloheximide to ensure that the observed signals result from the proteins in the HeLa nuclei and not from the newly synthesized proteins in the cytoplasm. A construct containing the rapid shuttling protein hnRNP A1 was a positive control, while the non-shuttling hnRNP C1 protein served as a negative control. The results shown in
Figure 7 demonstrate that hDrn1 shuttles between the nucleus and the cytoplasm, since the myc-hDrn1 signal was clearly detected in both HeLa and NIH3T3 nuclei, as was that of hDbr1. These results strongly suggest that hDrn1 has cytoplasmic function(s), likely involving hDbr1.
Figure 7.
The hDrn1 protein shuttles between the nucleus and the cytoplasm as hDbr1. Expression vectors encoding hnRNP A1, C1, hDbr1 and hDrn1 with myc-tag were transfected into HeLa cells. After expression of the transfected cDNAs, the cells were fused using PEG with mouse NIH3T3 cells to form heterokaryons and incubated in culture medium containing 100 μg/mL cycloheximide for two hours. The heterokaryon cells were then fixed and stained for immunofluorescence microscopy with anti-myc tag antibody (panel FITC) to localize the proteins, and Hoechst 33342 (panel Hoechst), which distinguishes the human and mouse nuclei within the heterokaryon. The arrows indicate the mouse nuclei.
Figure 7.
The hDrn1 protein shuttles between the nucleus and the cytoplasm as hDbr1. Expression vectors encoding hnRNP A1, C1, hDbr1 and hDrn1 with myc-tag were transfected into HeLa cells. After expression of the transfected cDNAs, the cells were fused using PEG with mouse NIH3T3 cells to form heterokaryons and incubated in culture medium containing 100 μg/mL cycloheximide for two hours. The heterokaryon cells were then fixed and stained for immunofluorescence microscopy with anti-myc tag antibody (panel FITC) to localize the proteins, and Hoechst 33342 (panel Hoechst), which distinguishes the human and mouse nuclei within the heterokaryon. The arrows indicate the mouse nuclei.