Proteomics Analysis of Cellular Proteins Co-Immunoprecipitated with Nucleoprotein of Influenza A Virus (H7N9)
Abstract
:1. Introduction
2. Results and Discussion
2.1. Expression and Immunoprecipitation of NP
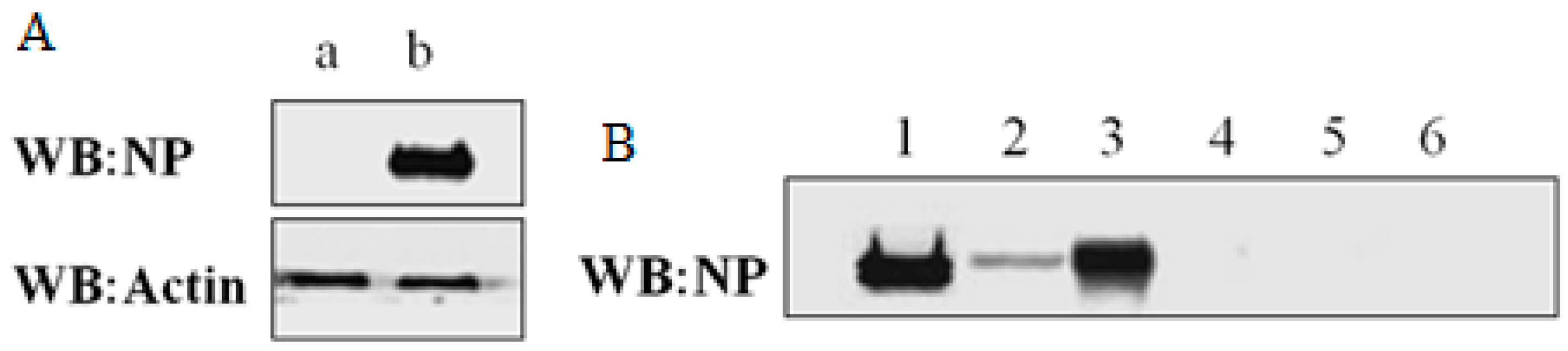
2.2. Identification of Cellular Proteins Co-Immunoprecipitated with NP
| Accession Number | Names | Abbreviation | Average Mass (Da) | Theoretical pI | %Cov(95) | Number of Identified Peptides |
|---|---|---|---|---|---|---|
| P11940 | Polyadenylate-binding protein 1 | PABP1 | 70,670.84 | 9.52 | 39.15 | 29 |
| Q59GN2 | Putative 60S ribosomal protein L39-like 5 | R39L5 | 6322.59 | 12.32 | 19.61 | 4 |
| P02787 | Serotransferrin | TRFE | 77,063.89 | 6.81 | 41.83 | 31 |
| Q12906 | Interleukin enhancer-binding factor 3 | ILF3 | 95,338.37 | 8.86 | 24.61 | 19 |
| P09429 | High mobility group protein B1 | HMGB1 | 24,893.76 | 5.6 | 29.30 | 6 |
| Q08211 | ATP-dependent RNA helicase A | DHX9 | 140,958.5 | 6.41 | 28.90 | 31 |
| P01024 | Complement C3 | CO3 | 187,148.1 | 6.02 | 19.78 | 28 |
| Q00059 | Transcription factor A, mitochondrial | TFAM | 29,096.63 | 9.74 | 34.96 | 10 |
| P26599 | Polypyrimidine tract-binding protein 1 | PTBP1 | 57,221.33 | 9.22 | 21.66 | 10 |
| P25705 | ATP synthase subunit α, mitochondrial | ATPA | 59,750.63 | 9.16 | 21.88 | 9 |
| P38159 | RNA-binding motif protein, X chromosome | RBMX | 42,331.85 | 10.06 | 38.11 | 14 |
| P09874 | Poly [ADP-ribose] polymerase 1 | PARP1 | 113,083.8 | 8.99 | 18.05 | 15 |
| P00738 | Haptoglobin | HPT | 45,205.31 | 6.13 | 34.24 | 12 |
| P43243 | Matrin-3 | MATR3 | 94,623.24 | 5.87 | 20.78 | 18 |
| P68032 | Actin, α cardiac muscle 1 | ACTC | 42,018.97 | 5.23 | 43.24 | 11 |
| P48681 | Nestin | NEST | 177,438.9 | 4.35 | 13.76 | 18 |
| Q96PK6 | RNA-binding protein 14 | RBM14 | 69,491.65 | 9.68 | 16.59 | 10 |
| Q13765 | Nascent polypeptide-associated complex subunit α | NACA | 23,383.9 | 4.52 | 26.76 | 4 |
| Q15459 | Splicing factor 3A subunit 1 | SF3A1 | 88,886.18 | 5.15 | 3.03 | 2 |
| Q14919 | Dr1-associated corepressor | NC2A | 22,349.84 | 5.04 | 31.60 | 6 |
| P17844 | Probable ATP-dependent RNA helicase DDX5 | DDX5 | 69,148.08 | 9.06 | 14.50 | 9 |
| P15927 | Replication protein A 32 kDa subunit | RFA2 | 29,246.85 | 5.74 | 17.78 | 3 |
| O75531 | Barrier-to-autointegration factor | BAF | 10,058.58 | 5.81 | 51.69 | 6 |
| P02790 | Hemopexin | HEMO | 51,676.37 | 6.55 | 16.45 | 5 |
| Q9UQ35 | Serine/arginine repetitive matrix protein 2 | SRRM2 | 299,615.1 | 12.05 | 7.27 | 14 |
| P62937 | Peptidyl-prolylcis-trans isomerase A | PPIA | 18,012.49 | 7.68 | 24.24 | 4 |
| P84098 | Ribosomal protein L19 | RL19 | 23,465.97 | 11.48 | 13.47 | 2 |
| Q9NZI8 | Insulin-like growth factor 2 mRNA-binding protein 1 | IF2B1 | 63,480.59 | 9.26 | 22.18 | 10 |
| Q9HCE1 | Putative helicase MOV-10 | MOV10 | 113,671.3 | 9 | 14.86 | 11 |
| Q15717 | ELAV-like protein 1 | ELAV1 | 36,091.88 | 9.23 | 28.22 | 7 |
| Q00325 | Phosphate carrier protein, mitochondrial | MPCP | 40,094.86 | 9.45 | 15.75 | 6 |
| Q07666 | KH domain-containing, RNA-binding, signal transduction-associated protein 1 | KHDR1 | 48,227.34 | 8.73 | 9.71 | 3 |
| Q01658 | Protein Dr1 | NC2B | 19,443.66 | 4.69 | 32.95 | 4 |
| P09661 | U2 small nuclear ribonucleoprotein A' | RU2A | 28,415.57 | 8.71 | 26.27 | 5 |
| P13010 | X-ray repair cross-complementing protein 5 | XRCC5 | 82,704.54 | 5.55 | 5.05 | 3 |
| P36957 | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | ODO2 | 48,755.31 | 9.1 | 12.58 | 5 |
| P22061 | Protein-L-isoaspartate O-methyltransferase | PIMT | 24,636.38 | 6.7 | 19.23 | 6 |
| P35659 | Protein DEK | DEK | 42,674.28 | 8.69 | 12.00 | 4 |
| P02765 | α-2-HS-glycoprotein | FETUA | 39,324.68 | 5.43 | 13.90 | 5 |
| Q9UKM9 | RNA-binding protein Raly | RALY | 32,463.17 | 9.2 | 24.89 | 6 |
| O43809 | Cleavage and polyadenylation-specificity factor subunit 5 | CPSF5 | 26,227.29 | 8.85 | 28.00 | 4 |
| Q08431 | Lactadherin | MFGM | 43,122.99 | 8.47 | 16.80 | 5 |
| P35637 | RNA-binding protein FUS | FUS | 53,425.84 | 9.4 | 11.57 | 4 |
| P22087 | rRNA 2'-O-methyltransferase fibrillarin | FBRL | 33,784.22 | 10.18 | 19.31 | 4 |
| O75475 | PC4 and SFRS1-interacting protein | PSIP1 | 60,103.24 | 9.15 | 11.32 | 5 |
| P40926 | Malate dehydrogenase, mitochondrial | MDHM | 35,503.28 | 8.92 | 20.12 | 6 |
| P62826 | GTP-binding nuclear protein Ran | RAN | 24,423.11 | 7.01 | 15.02 | 4 |
| Q9Y3Y2 | Chromatin target of PRMT1 protein | CHTOP | 26,396.57 | 12.24 | 19.35 | 4 |
| Q9NR30 | Nucleolar RNA helicase 2 | DDX21 | 87,344.4 | 9.32 | 10.09 | 6 |
| P84090 | Enhancer of rudimentary homolog | ERH | 12,258.94 | 5.62 | 37.50 | 3 |
| Q9Y383 | Putative RNA-binding protein Luc7-like 2 | LC7L2 | 46,513.9 | 10.02 | 14.54 | 5 |
| P55769 | NHP2-like protein 1 | NH2L1 | 14,173.55 | 8.72 | 26.52 | 3 |
| P42167 | Lamina-associated polypeptide 2, isoforms β/γ | LAP2B | 50,670.26 | 9.39 | 12.56 | 4 |
| P63162 | Small nuclear ribonucleoprotein-associated protein N | RSMN | 24,614.04 | 11.2 | 17.16 | 3 |
| P57721 | Poly(rC)-binding protein 3 | PCBP3 | 39,465.25 | 8.22 | 12.47 | 3 |
| P02763 | α-1-acid glycoprotein 1 | A1AG1 | 23,511.56 | 4.93 | 17.41 | 3 |
| P26368 | Splicing factor U2AF 65 kDa subunit | U2AF2 | 53,500.98 | 9.19 | 10.11 | 3 |
| Q6PKG0 | La-related protein 1 | LARP1 | 123,510.3 | 8.91 | 6.57 | 5 |
| P11182 | Lipoamideacyltransferase component of branched-chain α-keto acid dehydrogenase complex, mitochondrial | ODB2 | 53,487.07 | 8.71 | 11.62 | 4 |
| P04637 | Cellular tumor antigen p53 | P53 | 43,653.18 | 6.33 | 5.34 | 2 |
| P14174 | Macrophage migration inhibitory factor | MIF | 12,476.3 | 7.73 | 17.39 | 2 |
| P38919 | Eukaryotic initiation factor 4A-III | IF4A3 | 46,871.03 | 6.3 | 13.14 | 5 |
| Q07021 | Complement component 1 Q subcomponent-binding protein, mitochondrial | C1QBP | 31,362.24 | 4.74 | 12.06 | 2 |
| P12277 | Creatine kinase B-type | KCRB | 42,644.28 | 5.34 | 15.22 | 4 |
| P46013 | Antigen KI-67 | KI67 | 358,693.7 | 9.49 | 2.18 | 3 |
| P00450 | Ceruloplasmin | CERU | 122,205.2 | 5.44 | 3.91 | 3 |
| Q92900 | Regulator of nonsense transcripts 1 | RENT1 | 124,345.3 | 6.18 | 3.81 | 4 |
| O43175 | D-3-phosphoglycerate dehydrogenase | SERA | 56,650.5 | 6.29 | 5.63 | 3 |
| P06454 | Prothymosin α | PTMA | 12,202.96 | 3.66 | 35.51 | 3 |
| Q16576 | Histone-binding protein RBBP7 | RBBP7 | 47,820.08 | 4.89 | 5.53 | 2 |
| P61326 | Protein magonashi homolog | MGN | 17,163.62 | 5.74 | 21.23 | 2 |
| P02774 | Vitamin D-binding protein | VTDB | 52,963.65 | 5.4 | 11.13 | 3 |
| O75955 | Flotillin-1 | FLOT1 | 47,355.28 | 7.08 | 11.48 | 4 |
| Q9Y230 | RuvB-like 2 | RUVB2 | 51,156.57 | 5.49 | 7.34 | 3 |
| P63167 | Dynein light chain 1, cytoplasmic | DYL1 | 10,365.88 | 6.89 | 24.72 | 2 |
| P18754 | Regulator of chromosome condensation | RCC1 | 44,969.02 | 7.18 | 8.07 | 2 |
| O43143 | Pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 | DHX15 | 90,932.83 | 7.12 | 2.77 | 2 |
| P20042 | Eukaryotic translation initiation factor 2 subunit 2 | IF2B | 38,388.41 | 5.6 | 6.61 | 2 |
| Q06787 | Fragile X mental retardation 1, isoform CRA_e | FMR1 | 71,174.48 | 7 | 8.95 | 5 |
| P27824 | Calnexin | CALX | 67,568.3 | 4.46 | 4.56 | 2 |
| P51114 | Fragile X mental retardation syndrome-related protein 1 | FXR1 | 69,720.79 | 5.84 | 3.67 | 1 |
| Q9NY12 | H/ACA ribonucleoprotein complex subunit 1 | GAR1 | 22,347.88 | 10.91 | 12.44 | 2 |
| P59190 | Ras-related protein Rab-15 | RAB15 | 24,375.19 | 5.53 | 10.38 | 2 |
| Q9NZ01 | Very-long-chain enoyl-CoA reductase | TECR | 36,010.78 | 9.50 | 5.84 | 2 |
| P78527 | DNA-dependent protein kinase catalytic subunit | PRKDC | 469,088.8 | 6.75 | 0.56 | 2 |
| P22234 | Multifunctional protein ADE2 | PUR6 | 47,079.22 | 6.94 | 6.30 | 2 |
| O14893 | Gem-associated protein 2 | GEMI2 | 31,585.12 | 5.43 | 12.14 | 2 |
| Q15388 | Mitochondrial import receptor subunit TOM20 homolog | TOM20 | 16,297.88 | 8.81 | 22.76 | 2 |
| P61604 | 10 kDa heat shock protein, mitochondrial | CH10 | 10,931.69 | 8.89 | 46.81 | 2 |
| Q13263 | Transcription intermediary factor 1-β | TIF1B | 88,549.66 | 5.52 | 2.39 | 2 |
| Q04837 | Single-stranded DNA-binding protein, mitochondrial | SSBP | 17,259.67 | 9.59 | 15.54 | 2 |
| Q09161 | Nuclear cap-binding protein subunit 1 | NCBP1 | 91,839.44 | 5.99 | 4.05 | 2 |
| Q9P035 | Very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase 3 | HACD3 | 43,159.55 | 9.04 | 8.84 | 2 |
| P04003 | C4b-binding protein α chain | C4BPA | 67,033.19 | 7.15 | 1.84 | 1 |
| P01042 | Kininogen-1 | KNG1 | 71,957.38 | 6.34 | 2.95 | 1 |
| Q96IX5 | Up-regulated during skeletal muscle growth protein 5 | USMG5 | 6457.57 | 9.78 | 25.86 | 1 |
| P04004 | Vitronectin | VTNC | 54,305.59 | 5.55 | 2.51 | 1 |
| Q9NUD5 | Zinc finger CCHC domain-containing protein 3 | ZCHC3 | 43,618.48 | 8.86 | 3.96 | 1 |
| P85037 | Forkhead box protein K1 | FOXK1 | 75,457.34 | 9.41 | 2.59 | 2 |
| Q96SB3 | Neurabin-2 | NEB2 | 89,192.07 | 4.91 | 6.12 | 3 |
| P35232 | Prohibitin | PHB | 29,804.1 | 5.57 | 8.54 | 2 |
| P02749 | β-2-glycoprotein 1 | APOH | 38,298.16 | 8.34 | 8.70 | 2 |
| Q13838 | Spliceosome RNA helicase DDX39B | DX39B | 48,991.33 | 5.44 | 7.49 | 1 |
| O76021 | Ribosomal L1 domain-containing protein 1 | RL1D1 | 54,972.52 | 10.13 | 2.56 | 1 |
| O43707 | α-actinin-4 | ACTN4 | 104,854 | 5.27 | 1.32 | 1 |
| Q9UN86 | RasGTPase-activating protein-binding protein 2 | G3BP2 | 54,121.13 | 5.41 | 5.39 | 2 |
| Q969G3 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 | SMCE1 | 46,649.42 | 4.84 | 7.34 | 1 |
| Q8WXI9 | Transcriptional repressor p66-β | P66B | 65,260.72 | 9.73 | 1.85 | 1 |
| Q15287 | RNA-binding protein with serine-rich domain 1 | RNPS1 | 34,208.24 | 11.85 | 7.11 | 1 |
| O96019 | Actin-like protein 6A | ACL6A | 47,460.97 | 5.39 | 3.03 | 1 |
| P40425 | Pre-B-cell leukemia transcription factor 1 | PBX2 | 45,881.29 | 7.18 | 10.79 | 1 |
| Q5UIP0 | Telomere-associated protein RIF1 | RIF1 | 274,465.6 | 5.39 | 0.57 | 1 |
| P49006 | MARCKS-related protein | MRP | 19,528.8 | 4.65 | 7.69 | 1 |
| P25788 | Proteasome subunit α type-3 | PSA3 | 28,433.23 | 5.19 | 4.71 | 1 |
| Q6PJP8 | DNA cross-link repair 1A protein | DCR1A | 116,399.6 | 8.24 | 1.06 | 1 |
| P62995 | Transformer-2 protein homolog β | TRA2B | 33,665.68 | 11.25 | 11.02 | 1 |
| Q9Y2Q9 | 28S ribosomal protein S28, mitochondrial | RT28 | 20,842.78 | 9.22 | 6.29 | 1 |
| P13797 | Plastin-3 | PLST | 70,811.02 | 5.41 | 1.62 | 1 |
| Q13428 | Treacle protein | TCOF | 152,106 | 9.06 | 0.99 | 1 |
| Q96CT7 | Coiled-coil domain-containing protein 124 | CC124 | 25,835.24 | 9.54 | 9.87 | 2 |
| Q9UHX1 | Poly(U)-binding-splicing factor PUF60 | PUF60 | 59,875.47 | 5.19 | 2.86 | 1 |
| P55072 | Transitional endoplasmic reticulum ATPase | TERA | 89,321.8 | 5.14 | 2.11 | 1 |
| P08579 | U2 small nuclear ribonucleoprotein B'' | RU2B | 25,486.33 | 9.72 | 11.56 | 2 |
| O75533 | Splicing factor 3B subunit 1 | SF3B1 | 145,830.4 | 6.65 | 3.53 | 4 |
| P02647 | Apolipoprotein A-I | APOA1 | 30,777.83 | 5.56 | 62.92 | 20 |
| Q07955 | Serine/arginine-rich-splicing factor 1 | SRSF1 | 27,744.58 | 10.37 | 37.15 | 11 |
| P53999 | Activated RNA polymerase II transcriptional coactivator p15 | TCP4 | 14,395.34 | 9.6 | 60.63 | 8 |
| Q15233 | Non-POU domain-containing octamer-binding protein | NONO | 54,231.54 | 9.01 | 28.87 | 14 |
| P35611 | α-Adducin | ADDA | 80,955.14 | 5.6 | 21.04 | 11 |
| Q16352 | α-Internexin | AINX | 55,390.65 | 5.34 | 47.29 | 20 |
| P52272 | Heterogeneous nuclear ribonucleoprotein M | HNRPM | 77,515.53 | 8.84 | 43.42 | 29 |
| O75165 | DnaJ homolog subfamily C member 13 | DJC13 | 254,414.9 | 6.31 | 6.69 | 18 |
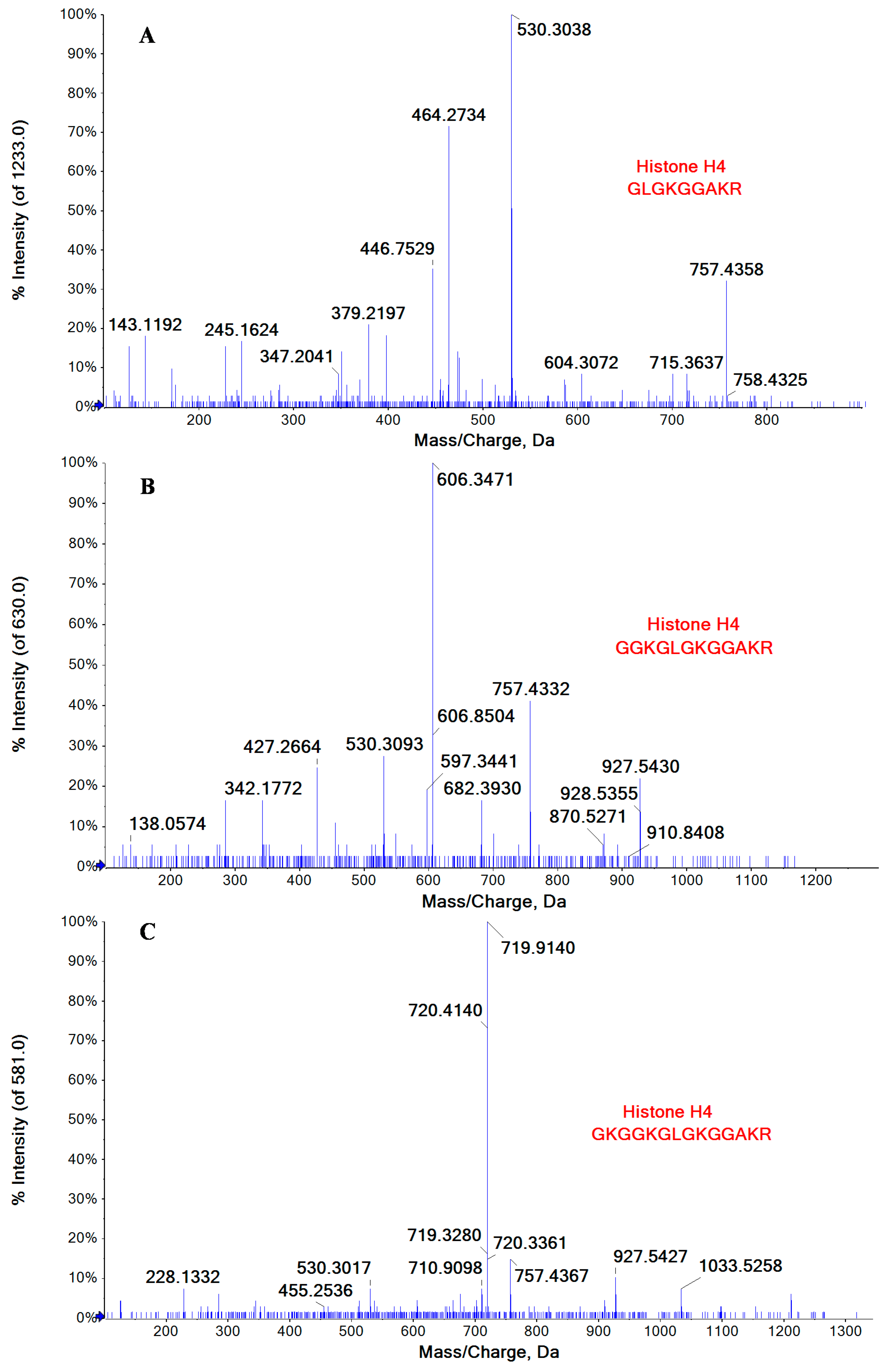
2.3. Lysine Acetylation Modifications Identified in Some of Cellular Proteins Co-Immunoprecipitated with NP
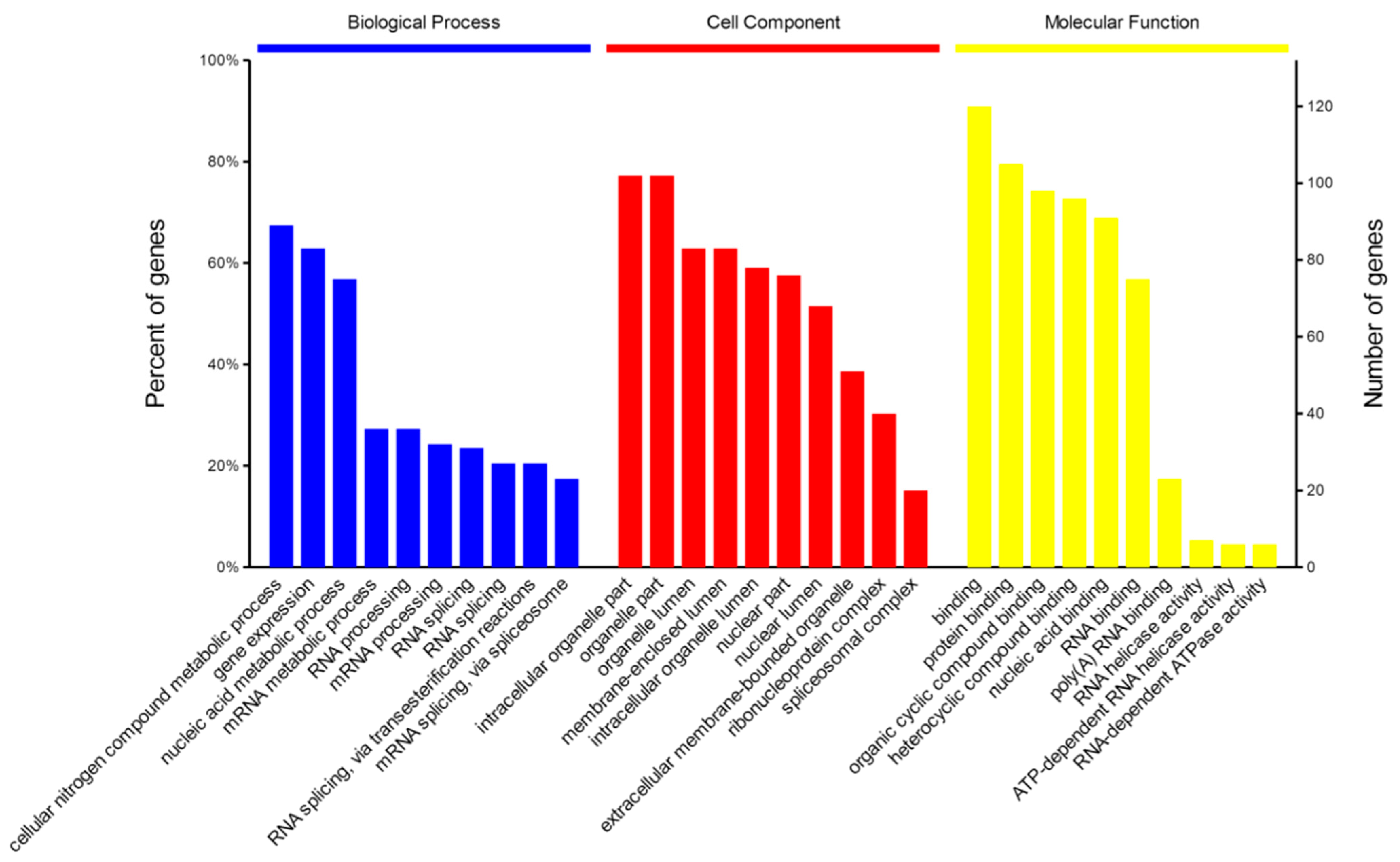
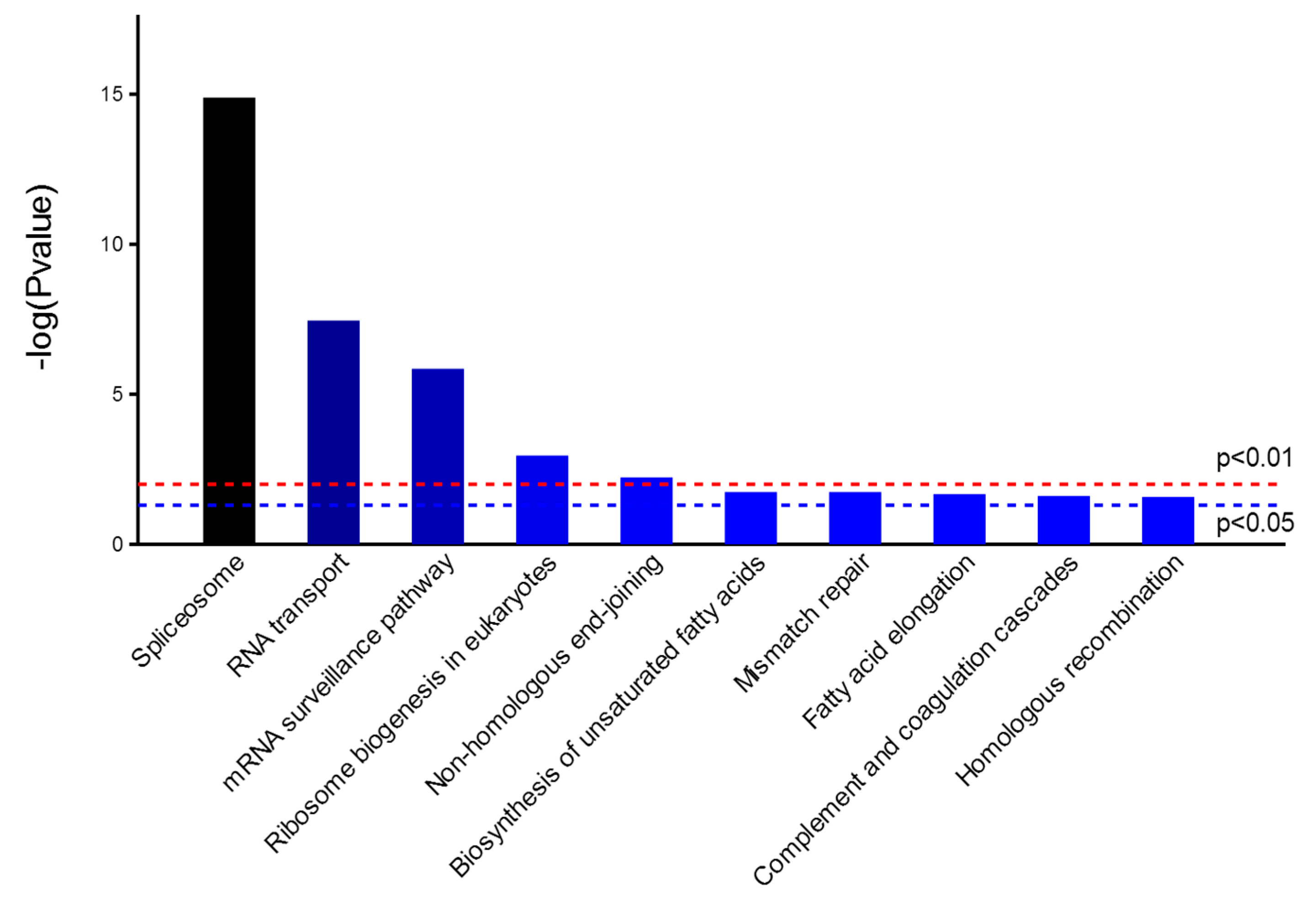
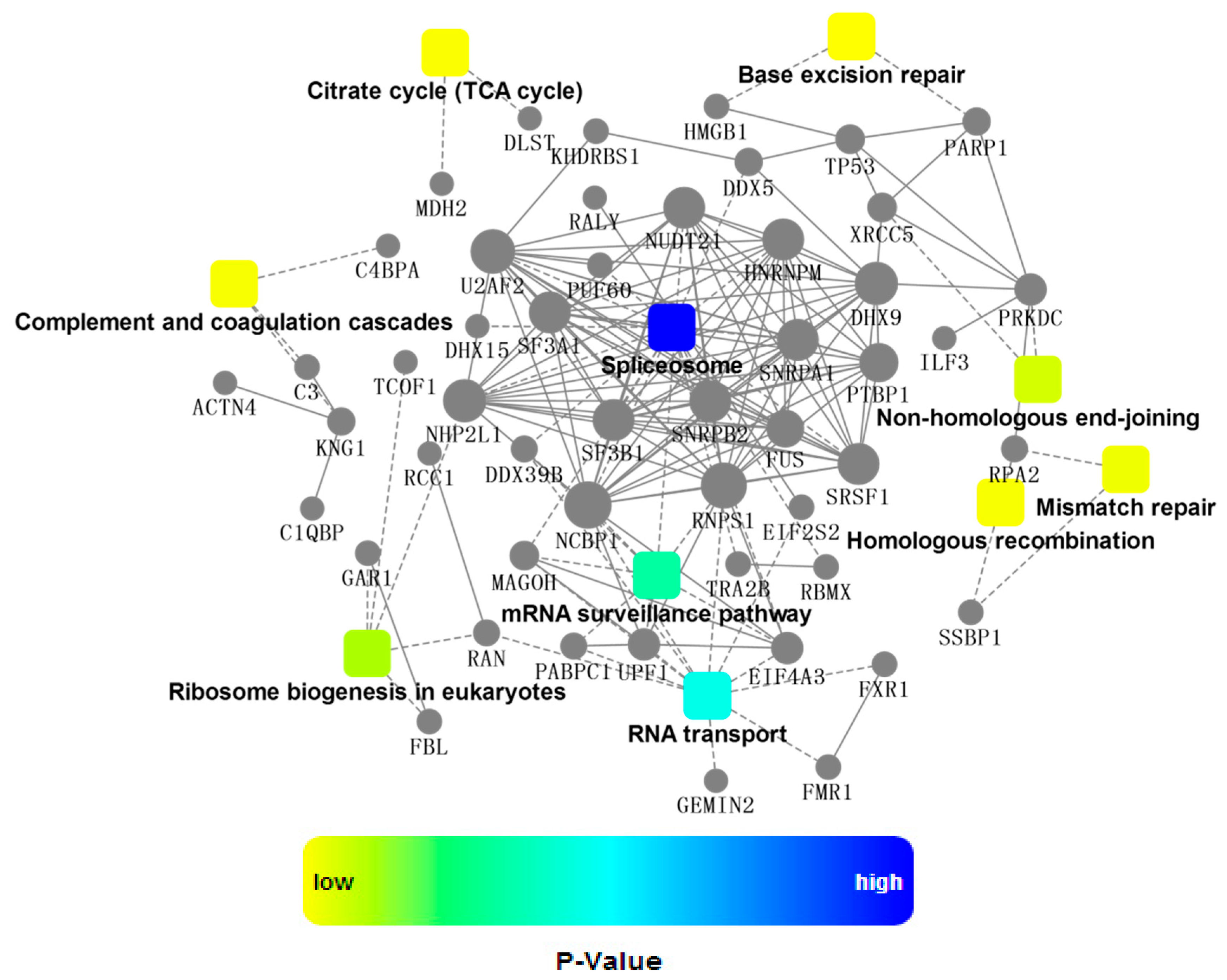
2.4. Bioinformatics Analysis
| Accession | Protein Name | Lysine-Acetylated Peptide | Residues in Protein |
|---|---|---|---|
| P62805 | Histone H4 | GK*GGK*GLGK*GGAK*R | 5–18 |
| GGK*GLGK*GGAK*R | 7–18 | ||
| GLGK*GGAK*R | 10–18 | ||
| Q5TEC6 | Histone H3 | K*STGGK*APR | 10–18 |
| K*QLATK*AAR | 19–27 | ||
| Q15149 | Plectin | IEQEK*AKLEQLFQDEVAK | 2646–2663 |
| P08670 | Vimentin | ASLARLDLERK*VESLQEEIAFLK | 213–235 |
| Q9UHB6 | LIM domain and actin-binding protein 1 | STPAEDDSRDSQVK* | 336–349 |
| P15880 | 40S ribosomal protein S2 | TK*SPYQEFTDHLVK | 262–275 |
| P62158 | Calmodulin | HVMTNLGEK*LTDEEVDEMIR | 108–127 |
| Q86V81 | THO complex subunit 4 | ADK*MDMSLDDIIK | 2–14 |
3. Experimental Section
3.1. Chemicals and Materials
3.2. Plasmid Construction, Amplification and Transfection
3.3. Co-Immunoprecipitation
3.4. SDS-PAGE and Western Blotting
3.5. Filter-Aided Buffer Exchange and Trypsin Digestion
3.6. Nano-LC-MS/MS and Data Processing
3.7. Bioinformatics Analysis
4. Conclusions
Acknowledgments
Supplementary Materials
Author Contributions
Conflicts of Interest
References
- Peiris, J.S.; Yu, W.C.; Leung, C.W.; Cheung, C.Y.; Ng, W.F.; Nicholls, J.M.; Ng, T.K.; Chan, K.H.; Lai, S.T.; Lim, W.L.; et al. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet 2004, 21, 617–619. [Google Scholar] [CrossRef]
- Peiris, M.; Yuen, K.Y.; Leung, C.W.; Chan, K.H.; Ip, P.L.; Lai, R.W.; Orr, W.K.; Shortridge, K.F. Human infection with influenza H9N2. Lancet 1999, 11, 916–917. [Google Scholar] [CrossRef]
- Li, K.S.; Guan, Y.; Wang, J.; Smith, G.J.; Xu, K.M.; Duan, L.; Rahardjo, A.P.; Puthavathana, P.; Buranathai, C.; Nguyen, T.D.; et al. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 2004, 8, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Wu, F.; He, J. Emerging risk of H7N9 influenza in China. Lancet 2013, 4, 1539–1540. [Google Scholar] [CrossRef]
- Gao, R.; Cao, B.; Hu, Y.; Feng, Z.; Wang, D.; Hu, W.; Chen, J.; Jie, Z.; Qiu, H.; Xu, K.; et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 2013, 368, 1888–1897. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Zhang, X.; Hu, J.; Chen, J.; Pan, Q.; Teng, Z.; Zheng, Y.; Mao, S.; Zhang, H.; King, C.C.; et al. A case report of avian influenza H7N9 killing a young doctor in Shanghai, China. BMC Infect. Dis. 2015, 23, 237. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Palese, P.; Shaw, M.L. Fields virology. In Orthomyxoviridae: The Viruses and Their Replication, 5th ed.; Lippincott Williams & Wilkins, Wolters Kluwer Business: Philadelphia, PA, USA, 2007; pp. 1647–1689. [Google Scholar]
- Chenavas, S.; Crépin, T.; Delmas, B.; Ruigrok, R.W.; Slama-Schwok, A. Influenza virus nucleoprotein: structure, RNA binding, oligomerization and antiviral drug target. Future Microbiol. 2013, 8, 1537–1545. [Google Scholar] [CrossRef] [PubMed]
- Vreede, F.T.; Brownlee, G.G. Influenza virion-derived viral ribonucleoproteins synthesize both mRNA and cRNA in vitro. J. Virol. 2007, 81, 2196–2204. [Google Scholar] [CrossRef] [PubMed]
- Newcomb, L.L.; Kuo, R.L.; Ye, Q.; Jiang, Y.; Tao, Y.J.; Krug, R.M. Interaction of the influenza a virus nucleocapsid protein with the viral RNA polymerase potentiates unprimed viral RNA replication. J. Virol. 2009, 83, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Mayank, A.K.; Nailwal, H.; Tripathi, S.; Patel, J.R.; Bowzard, J.B.; Gaur, P.; Donis, R.O.; Katz, J.M.; Cox, N.J.; et al. Influenza A viral nucleoprotein interacts with cytoskeleton scaffolding protein α-actinin-4 for viral replication. FEBS J. 2014, 281, 2899–2914. [Google Scholar] [CrossRef] [PubMed]
- Generous, A.; Thorson, M.; Barcus, J.; Jacher, J.; Busch, M.; Sleister, H. Identification of putative interactions between swine and human influenza A virus nucleoprotein and human host proteins. Virol. J. 2014, 11, 2509. [Google Scholar] [CrossRef] [PubMed]
- Mayer, D.; Molawi, K.; Martínez-Sobrido, L.; Ghanem, A.; Thomas, S.; Baginsky, S.; Grossmann, J.; García-Sastre, A.; Schwemmle, M. Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. J. Proteome Res. 2007, 6, 672–682. [Google Scholar] [CrossRef] [PubMed]
- Kao, R.Y.; Yang, D.; Lau, L.S.; Tsui, W.H.; Hu, L.; Dai, J.; Chan, M.P.; Chan, C.M.; Wang, P.; Zheng, B.J.; et al. Identification of influenza A nucleoprotein as an antiviral target. Nat. Biotechnol. 2010, 28, 600–605. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.W.; Chang, S.C.; Mok, C.K.; Lo, Y.L.; Kung, Y.N.; Huang, J.H.; Shih, Y.H.; Wang, J.Y.; Chiang, C.; Chen, C.J.; et al. Genomic signatures of human versus avian influenza A viruses. Emerg. Infect. Dis. 2006, 12, 1353–1360. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Cheung, B.; Tan, S.; Li, C.; Li, L.; Liu, S.; Jiang, S. Genomic signature and mutation trend analysis of pandemic (H1N1) 2009 influenza A virus. PLoS ONE 2010, 5, e9549. [Google Scholar] [CrossRef] [PubMed]
- Mänz, B.; Schwemmle, M.; Brunotte, L. Adaptation of avian influenza A virus polymerase in mammals to overcome the host species barrier. J. Virol. 2013, 87, 7200–7209. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Robles, I.; Akarsu, H.; Müller, C.W.; Ruigrok, R.W.; Baudin, F. Interaction of influenza virus proteins with nucleosomes. Virology 2005, 332, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Portela, A.; Digard, P. The influenza virus nucleoprotein: A multifunctional RNA-binding protein pivotal to virus replication. J. Gen. Virol. 2002, 83, 723–734. [Google Scholar] [CrossRef] [PubMed]
- Nailwal, H.; Sharma, S.; Mayank, A.K.; Lal, S.K. The nucleoprotein of influenza A virus induces p53 signaling and apoptosis via attenuation of host ubiquitin ligase RNF43. Cell. Death Dis. 2015, 21, e1768. [Google Scholar] [CrossRef] [PubMed]
- Elton, D.; Simpson-Holley, M.; Archer, K.; Medcalf, L.; Hallam, R.; McCauley, J.; Digard, P. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J. Virol. 2001, 75, 408–419. [Google Scholar] [CrossRef] [PubMed]
- Momose, F.; Basler, C.F.; O’Neill, R.E.; Iwamatsu, A.; Palese, P.; Nagata, K. Cellular splicing factor RAF-2p48/NPI-5/BAT1/UAP56 interacts with the influenza virus nucleoprotein and enhances viral RNA synthesis. J. Virol. 2001, 75, 1899–1908. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Song, W.; Wang, P.; Lee, K.; Chan, W.; Chen, H.; Cai, Z. Proteomics analysis of differential expression of cellular proteins in response to avian H9N2 virus infection in human cells. Proteomics 2008, 8, 1851–1858. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, N.; Sun, W.; Li, S.; Yang, J.; Yang, L.; Quan, G.; Gao, X.; Wang, Z.; Cheng, X.; Li, Z.; et al. Proteomics Analysis of Cellular Proteins Co-Immunoprecipitated with Nucleoprotein of Influenza A Virus (H7N9). Int. J. Mol. Sci. 2015, 16, 25982-25998. https://doi.org/10.3390/ijms161125934
Sun N, Sun W, Li S, Yang J, Yang L, Quan G, Gao X, Wang Z, Cheng X, Li Z, et al. Proteomics Analysis of Cellular Proteins Co-Immunoprecipitated with Nucleoprotein of Influenza A Virus (H7N9). International Journal of Molecular Sciences. 2015; 16(11):25982-25998. https://doi.org/10.3390/ijms161125934
Chicago/Turabian StyleSun, Ningning, Wanchun Sun, Shuiming Li, Jingbo Yang, Longfei Yang, Guihua Quan, Xiang Gao, Zijian Wang, Xin Cheng, Zehui Li, and et al. 2015. "Proteomics Analysis of Cellular Proteins Co-Immunoprecipitated with Nucleoprotein of Influenza A Virus (H7N9)" International Journal of Molecular Sciences 16, no. 11: 25982-25998. https://doi.org/10.3390/ijms161125934
APA StyleSun, N., Sun, W., Li, S., Yang, J., Yang, L., Quan, G., Gao, X., Wang, Z., Cheng, X., Li, Z., Peng, Q., & Liu, N. (2015). Proteomics Analysis of Cellular Proteins Co-Immunoprecipitated with Nucleoprotein of Influenza A Virus (H7N9). International Journal of Molecular Sciences, 16(11), 25982-25998. https://doi.org/10.3390/ijms161125934






