In Vitro Selection of a Single-Stranded DNA Molecular Recognition Element against Atrazine
Abstract
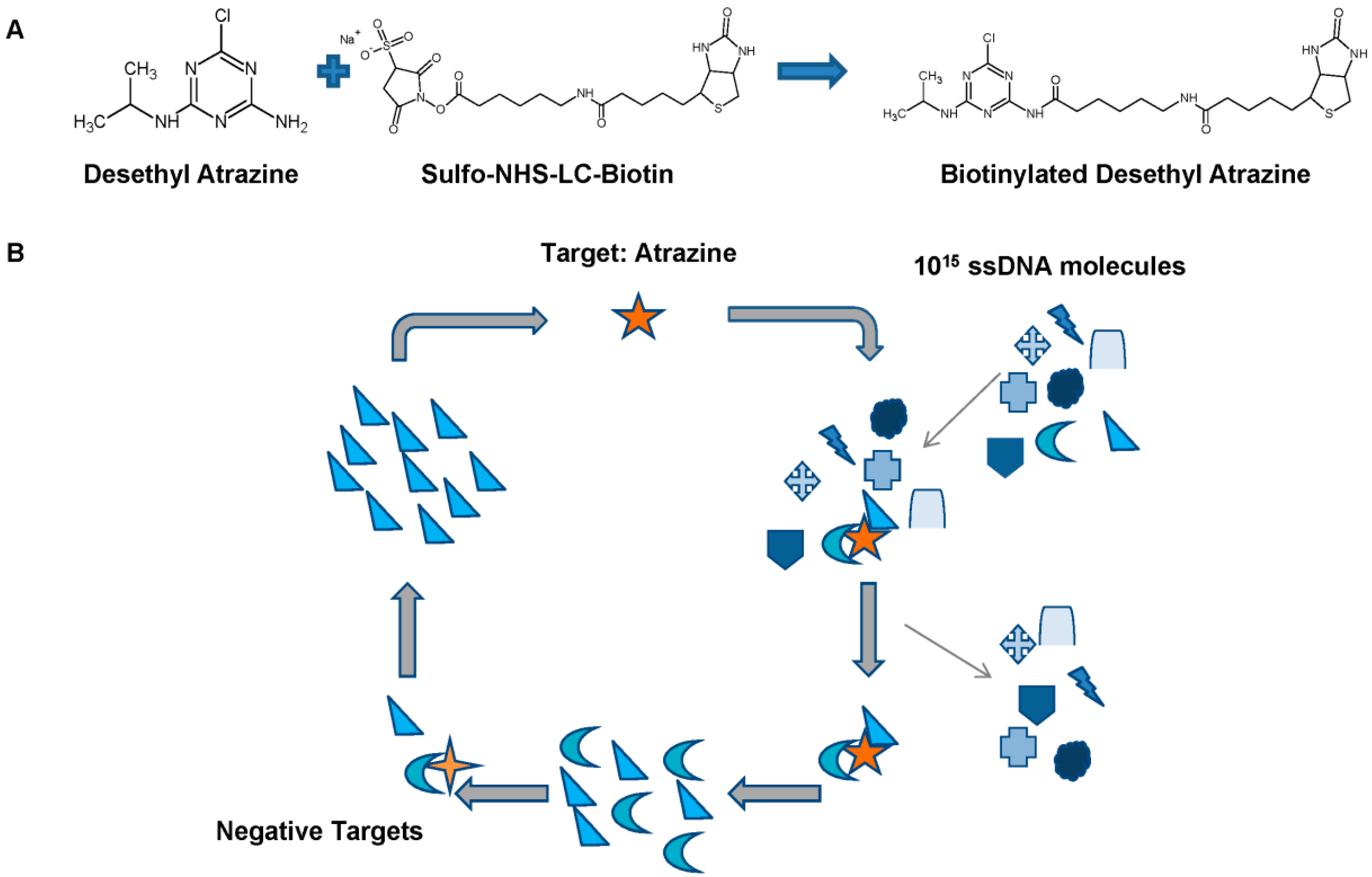
:1. Introduction
2. Results and Discussion
2.1. Selection of Atrazine-Specific MREs
| Round | Positive Selection | Negative Selection |
|---|---|---|
| 1 | Immobilized Target (IT) 25 h | Immobilization Substrate (IS) 23 h |
| 2 | IT 23 h | - |
| 3 | IT 18 h | IS 18 h |
| 4 | IT 12 h | - |
| 5 | IT 10 h | IS 16 h |
| 6 | IT w/methanol buffer 12 h | - |
| 7 | IT 10 h, Competitive Elution w/1 mM Atrazine (CE) 5 min | - |
| 8 | IT 2 h, CE 2 min | IT 2 h, CE w/propanil, 2,4-D acid malathion, 5 min |
| 9 | IT 5 min, CE immediate | IT 5 min, CE w/DACT 5 min |
| 10 | IT 1 min, CE immediate | IT 5 min, CE w/Simazine 5 min |
| 11 | IT immediate, CE immediate | IS 18 h |
| 12 | IT immediate, CE w/100 µM atrazine | - |
| R12.28 | TGTACCGTCTGAGCGATTCGTACCATTAGTGGGTGCTCCTTACCTGATGGTCATCTAGCCAGTCAGTGTTAAGGAGTGC |
| R12.57 | TGTACCGTCTGAGCGATTCGTAC  GGGTTTGCACTTTACCTGCGGTGCATCGCAGCCAGTCAGTGTTAAGGAGTGC GGGTTTGCACTTTACCTGCGGTGCATCGCAGCCAGTCAGTGTTAAGGAGTGC |
| R12.59 | TGTACCGTCTGAGCGATTCGTAC  GGGTTTGCACTTTACCTGCGGTGCATCGCAGCCAGTCAGTGTTAAGGAGTGC GGGTTTGCACTTTACCTGCGGTGCATCGCAGCCAGTCAGTGTTAAGGAGTGC |
| R12.23 | TGTACCGTCTGAGCGATTCGTACGAACGGCTTTGTACTGTTTGCACTGGC  AGCCAGTCAGTGTTAAGGAGTGC AGCCAGTCAGTGTTAAGGAGTGC |
| R12.31 | TGTACCGTCTGAGCGATTCGTACTAGGAATCCAGCGGAAAAGGC  TTATCATGAGCCAGTCAGTGTTAAGGAGTGC TTATCATGAGCCAGTCAGTGTTAAGGAGTGC |
| R12.49 | TGTACCGTCTGAGCGATTCGTACAGTTCTAGTAGGCGTTAGCATAAATGTTTG  GCCAGTCAGTGTTAAGGAGTGC GCCAGTCAGTGTTAAGGAGTGC |
| R12.23 | TGTACCGTCTGAGCGATTCGTACGAACGGCTTTGTACTGT  CTGGCGGATTTAGCCAGTCAGTGTTAAGGAGTGC CTGGCGGATTTAGCCAGTCAGTGTTAAGGAGTGC |
| R12.24 | TGTACCGTCTGAGCGATTCGTACAAGAGACTCGGCTTTTGTAATC  GGTTTTGGAGCCATTCATTGTTAAGGATTGC GGTTTTGGAGCCATTCATTGTTAAGGATTGC |
| R12.26 | TGTACCGTCTGAGCGATTCGTACGGCTAGAGTTGTTATGTTTCGATGGTCATCTGCAAGCCAGTCAGTGTTAAGGAGTGC |
| R12.34 | TGTACCGTCTGAGCGATTCGTACCGGTTCTTGAGCGGCTGAATAGTATTTTTC  GCCAGTCAGTGTTAAGGAGTGC GCCAGTCAGTGTTAAGGAGTGC |
| R12.10 | TGTACCGTCTGAGCGATTCGTACTCATTTGTGGCATTTGAGCGTCAGGGGTAAAGGTAGCCAGTCAGTGTTAAGGAGTGC |
| R12.23 | TGTACCGTCTGAGCGATTCGTACGAACGGCTTTGTACTGTTTGCACTGGCGGATTTAGCCAGTCAGTGTTAAGGAGTGC |
| R12.24 | TGTACCGTCTGAGCGATTCGTACAAGAGACTCGGCTTTTGTAATCTTGCGGTTTTGGAGCCATTCATTGTTAAGGATTGC |
| R12.23 | TGTACCGTCTGAGCGATTCGTACGAACGGCTTTGTACTGTTTGCACTGGCGGATTTAGCCAGTCAGTGTTAAGGAGTGC |
| R12.65 | TACCGTCTGAGCGATTCGTACCATCAGTAGAGTGCGCACTGTAGTAGATGGTCTTAGCCAGTCAGTGTTAAGGAGTGC |
| R12.3 | TGTACCGTCTGAGCGATTCGTACAGTAAGGCACTGGGGCCTTATGCTGTGAGGGATAAGCCAGTCAGTGTTAAGGAGTGC |
| R12.33 | TGTACCGTCTGAGCGATTCGTACTAAGCGACAGAGCACTGTTGCTGTTACAGTATCCAGCCAGTCAGTGTTAAGGAGTGC |
| R12.6 | TGTACCGTCTGAGCGATTCGTACTGGCGTAGGGTCGTATCTCTTTAAGTGCTGTACTAGCCAGTCAGTGTTAAGGAGTGC |
| R12.37 | TGTACCGTCTGAGCGATTCGTACTCGTCTACATTTACTGGGTTGATGATGAGTATTCAGCCAGTCAGTGTTAAGGAGTGC |
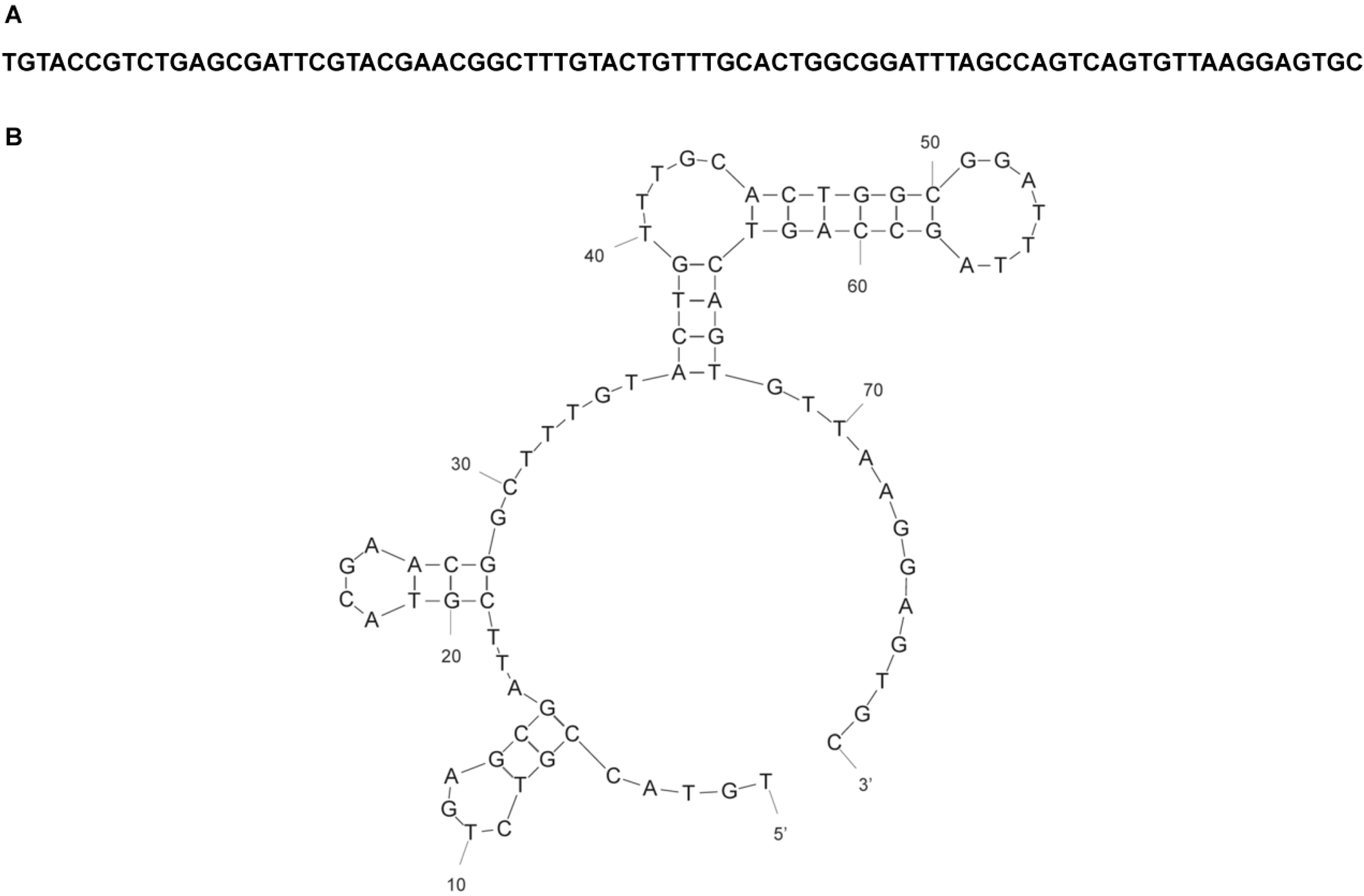
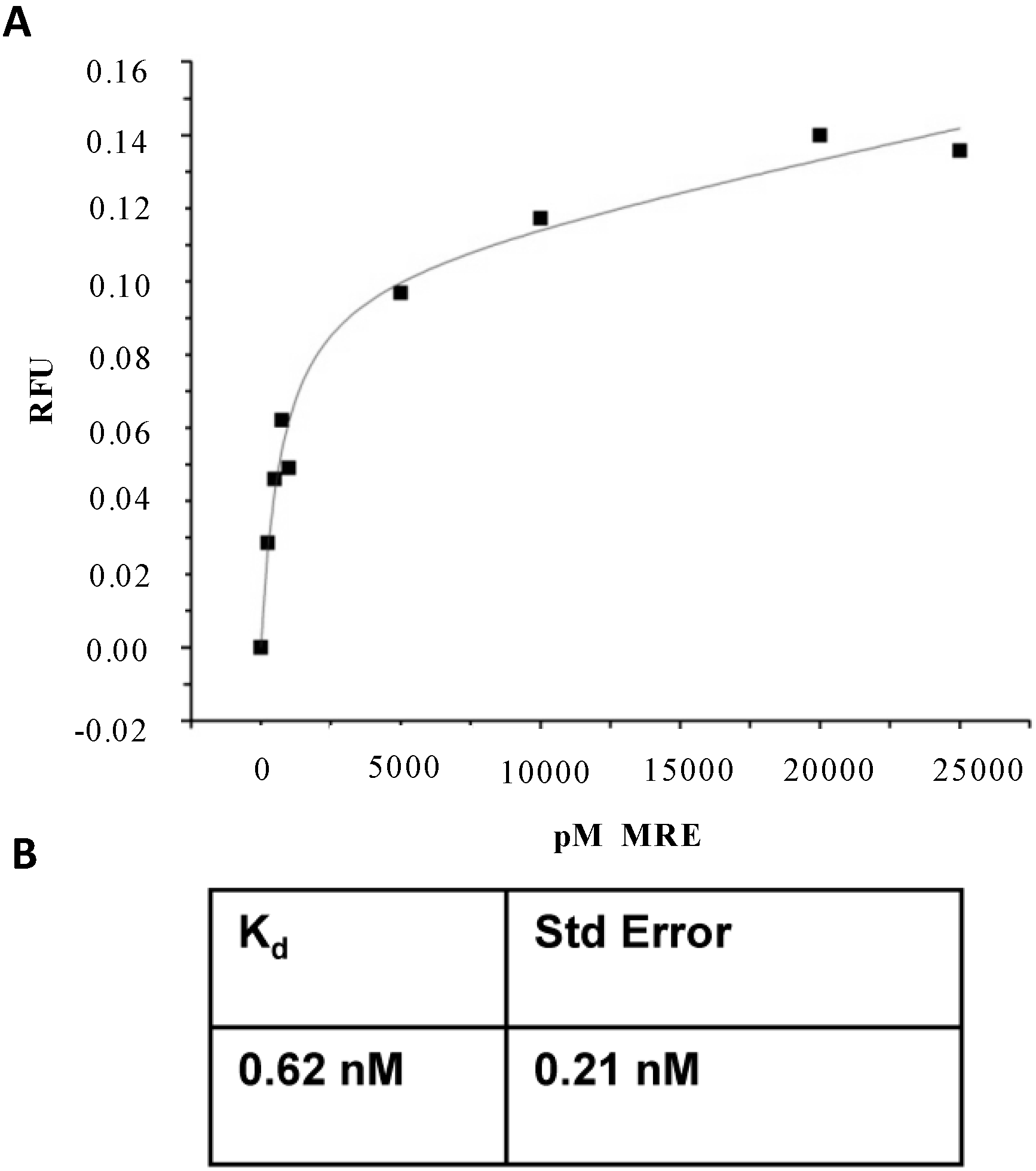
2.2. Affinity and Specificity of Atrazine-Specific MRE
| Eluent | Normalized Average Fluorescence | Standard Deviation | p-Value | Selectivity Ratio |
|---|---|---|---|---|
| Atrazine | 1 | 0.11 | - | - |
| Desethyl Atrazine | −0.054 | 0.44 | 0.01 | Neg. * |
| DACT | 0.47 | 0.10 | 0.002 | 2.1 |
| Simazine | 0.47 | 0.14 | 0.003 | 2.1 |
| Propanil | 0.65 | 0.13 | 0.01 | 1.5 |
| 2-4,D Acid | 0.64 | 0.11 | 0.009 | 1.5 |
| Malathion | 0.92 | 0.46 | 0.39 | 1.1 |
| MSB | 0.15 | 0.19 | 0.001 | 6.6 |
2.3. Atrazine Detection in Environmental Conditions
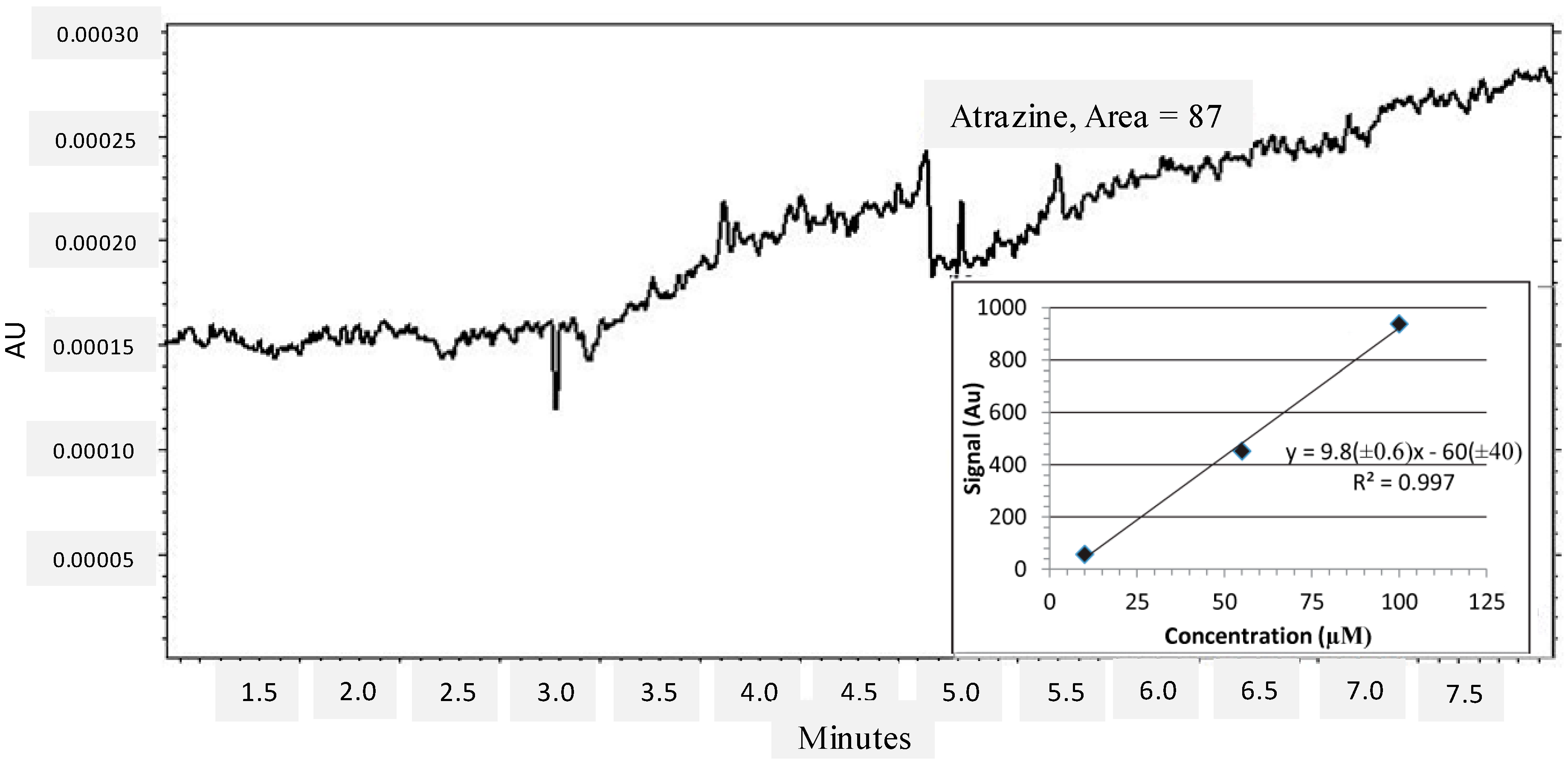
3. Experimental Section
3.1. SELEX for Selection of Atrazine-Specific MREs
3.2. Sequencing of Atrazine-Specific MREs
3.3. Atrazine MRE Binding Assays
3.4. Atrazine Detection in River Water
3.4.1. MRE Bead Immobilization
3.4.2. Sample Processing
3.4.3. Atrazine Quantification
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Brassard, M.; Gill, L.; Stavola, A.; Lin, J.; Turner, L. Atrazine: Analysis of Risks; United States Environmental Protection Agency: Washington, DC, USA, 2003. [Google Scholar]
- Ackerman, F. The economics of atrazine. Int. J. Occup. Environ. Health 2007, 13, 441–449. [Google Scholar]
- Graymore, M.S.F.; Allinson, G. Impacts of atrazine on aquatic ecosystems. Environ. Int. 2001, 26, 483–495. [Google Scholar] [CrossRef]
- Jiang, H.A.C.; Graziano, N.; Roberson, A.; McGuire, M.; Khiari, D. Occurrence and removal of chloro-s-triazines in water treatment plants. Environ. Sci Technol. 2006, 40, 3602–3616. [Google Scholar]
- Atrazine Chemical Summary. In United States Environmental Protection Agency, Toxicity and Exposure Assessment for Children’s Health. Available online: http://www.epa.gov/teach/chem_summ/Atrazine_summary.pdf (accessed on 14 August 2014).
- Sass, J.B.; Colangelo, A. European union bans atrazine, while the united states negotiates continued use. Int. J. Occup. Environ. Health 2006, 12, 260–267. [Google Scholar] [CrossRef]
- Hayes, T.; Haston, K.; Tsui, M.; Hoang, A.; Haeffele, C.; Vonk, A. Feminization of male frogs in the wild. Nature 2002, 419, 895–895. [Google Scholar]
- Rohr, J.; Palmer, B. Aquatic herbicide exposure increases salamander desiccation risk eight months later in a terrestrial environment. Environ. Toxicol. Chem. 2009, 24, 1253–1258. [Google Scholar] [CrossRef]
- Suzawa, M.; Ingraham, H. The herbicide atrazine activates endocrine gene networks via non-steroidal nr5a nuclear receptors in fish and mammalian cells. PLoS One 2008, 3, e2117. [Google Scholar] [CrossRef]
- Fan, W.; Yanase, T.; Morinaga, H.; Gondo, S.T.; Okabe, T.; Nomura, M.; Komatsu, T.; Morohashi, K.; Hayes, T.; Takayanagi, R.; et al. Atrazine-Induced aromatase expression is sf-1 dependent: Implications for endocrine disruption in wildlife and reproductive cancers in humans. Environ. Health Perspect. 2007, 115, 720–727. [Google Scholar] [CrossRef]
- Albanito, L.; Lappano, R.; Madeo, A.; Chimento, A.; Prossnitz, E.R.; Cappello, A.R.; Dolce, V.; Abonante, S.; Pezzi, V.; Maggiolini, M. G-Protein–Coupled receptor 30 and estrogen receptor-α are involved in the proliferative effects induced by atrazine in ovarian cancer cells. Environ. Health Perspect. 2008, 116, 1648–1655. [Google Scholar] [CrossRef]
- Cooper, R.L.; Laws, S.C.; Das, P.C.; Narotsky, M.G.; Goldman, J.M.; Lee Tyrey, E.; Stoker, T.E. Atrazine and reproductive function: Mode and mechanism of action studies. Birth Defects Res. Part B Dev. Reprod. Toxicol. 2007, 80, 98–112. [Google Scholar]
- Mizota, K.; Ueda, H. Endocrine disrupting chemical atrazine causes degranulation through gq/11 protein-coupled neurosteroid receptor in mast cells. Toxicol. Sci. 2006, 90, 362–368. [Google Scholar] [CrossRef]
- Tchounwou, T.B.; Wilson, B.A.; Ishaque, A.B.; Schneider, J. Atrazine potentiation of arsenic trioxide-induced cytotoxicity and gene expression in human liver carcinoma cells (hepg2). Mol. Cell. Biochem. 2001, 222, 49–59. [Google Scholar] [CrossRef]
- Mills, P.K. Correlation analysis of pesticide use data and cancer incidence rates in california counties. Arch. Environ. Health 1998, 53, 410–416. [Google Scholar] [CrossRef]
- Amistadi, J.K.; Hall, J.K.; Bogus, A.R.; Mumma, R.O. Comparison of gas chromatography and immunoassay methods for the detection of atrazine in water and soil. J. Environ. Sci. Health B 1997, 32, 45–60. [Google Scholar]
- Brzezicki, J.M.; Andersen, M.E.; Cranmer, B.K.; Tessari, J.D. Quantitative identification of atrazine and its chlorinated metabolites in plasma. J. Anal. Toxicol. 2003, 27, 569–573. [Google Scholar] [CrossRef]
- Panuwet, P.; Nguyen, J.V.; Kuklenyik, P.; Udunka, S.O.; Needham, L.L.; Barr, D.B. Quantification of atrazine and its metabolites in urine by on-line solid-phase extraction-high-performance liquid chromatography-tandem mass spectrometry. Anal. Bioanal. Chem. 2008, 391, 1931–1939. [Google Scholar] [CrossRef]
- Wortberg, M.; Goodrow, M.H.; Gee, S.J.; Hammock, B.D. Immunoassay for simazine and atrazine with low cross-reactivity for propazine. J. Agric. Food Chem. 1996, 44, 2210–2219. [Google Scholar] [CrossRef]
- Suri, C.R.; Boro, R.; Nangia, Y.; Gandhi, S.; Sharma, P.; Wangoo, N.; Rajesh, K.; Shekhawat, G.S. Immunoanalytical techniques for analyzing pesticides in the environment. TrAC-Trends Anal. Chem. 2009, 28, 29–39. [Google Scholar] [CrossRef]
- Sanchez, P.E. DNA Aptamer Development for Detection of Atrazine and Protective Antigen Toxin Using Fluorescence Polarization; University of California: Riverside, CA, USA, 2012. [Google Scholar]
- Huang, D.-W.; Niu, C.-G.; Qin, P.-Z.; Ruan, M.; Zeng, G.-M. Time-Resolved fluorescence aptamer-based sandwich assay for thrombin detection. Talanta 2010, 83, 185–189. [Google Scholar] [CrossRef]
- Ozsos, M.S.; Mascini, M.; Palchetti, I. Nucleic Acid Biosensors for Environmental Pollution Monitoring; Royal Society of Chemistry: London, UK, 2011. [Google Scholar]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment-rna ligands to bacteriophage t4 DNA-polymerase. Science 1990, 249, 505–510. [Google Scholar]
- Muller, S.R.; Berg, M.; Ulrich, M.M.; Schwarzenbach, R.P. Atrazine and its primary metabolites in swiss lakes: Input characteristics and long-term behavior in the water column. Environ. Sci. Technol. 1997, 31, 2104–2113. [Google Scholar] [CrossRef]
- Coupe, R.H.; Manning, M.A.; Foreman, W.T.; Goolsby, D.A.; Majewski, M.S. Occurrence of pesticides in rain and air in urban and agricultural areas of mississippi, april-september 1995. Sci. Total Environ. 2000, 248, 227–240. [Google Scholar] [CrossRef]
- Purcell, M.; Neault, J.F.; Malonga, H.; Arakawa, H.; Carpentier, R.; Tajmir-Riahi, H.A. Interactions of atrazine and 2,4-d with human serum albumin studied by gel and capillary electrophoresis, and ftir spectroscopy. Biochim. Biophys. Acta-Protein Struct. Mol. Enzym. 2001, 1548, 129–138. [Google Scholar]
- McCarthy, I.D.; Fuiman, L.A. Growth and protein metabolism in red drum (sciaenops ocellatus) larvae exposed to environmental levels of atrazine and malathion. Aquat. Toxicol. 2008, 88, 220–229. [Google Scholar] [CrossRef]
- Kucklick, J.R.; Bidleman, T.F. Organic contaminants in winyah bay, south-carolina. 1. Pesticides and polycyclic aromatic-hydrocarbons in subsurface and microlayer waters. Mar. Environ. Res. 1994, 37, 63–78. [Google Scholar] [CrossRef]
- Lin, Q.; Nguyen, T.; Pei, R.J.; Landry, D.W.; Stojanovic, M.N. Microfluidic aptameric affinity sensing of vasopressin for clinical diagnostic and therapeutic applications. Sens. Actuators B-Chem. 2011, 154, 59–66. [Google Scholar] [CrossRef]
- Yang, P.Y.; Wang, H.X.; Liu, Y.; Liu, C.C.; Huang, J.Y.; Liu, B.H. Microfluidic chip-based aptasensor for amplified electrochemical detection of human thrombin. Electrochem. Commun. 2010, 12, 258–261. [Google Scholar] [CrossRef]
- Soh, H.T.; Swensen, J.S.; Xiao, Y.; Ferguson, B.S.; Lubin, A.A.; Lai, R.Y.; Heeger, A.J.; Plaxco, K.W. Continuous, real-time monitoring of cocaine in undiluted blood serum via a microfluidic, electrochemical aptamer-based sensor. J. Am. Chem. Soc. 2009, 131, 4262–4266. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Ahmad, K.M.; Oh, S.S.; Kim, S.; McClellen, F.M.; Xiao, Y.; Soh, H.T. Probing the limits of aptamer affinity with a microfluidic selex platform. PLoS One 2011, 6, e27051. [Google Scholar]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. Selex—A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007, 24, 381–403. [Google Scholar] [CrossRef]
- Hamula, C.L.A.; Guthrie, J.W.; Zhang, H.; Li, X.; Le, X.C. Selection and analytical applications of aptamers. Trends Anal. Chem. 2006, 25, 681–691. [Google Scholar] [CrossRef]
- Hirao, I.; Spingola, M.; Peabody, D.; Ellington, A.D. The limits of specificity: An experimental analysis with rna aptamers to ms2 coat protein variants. Mol. Divers. 1998, 4, 75–89. [Google Scholar] [CrossRef]
- Hamula, C.L.; Le, X.C.; Li, X.-F. DNA aptamers binding to multiple prevalent m-types of streptococcus pyogenes. Anal. Chem. 2011, 83, 3640–3647. [Google Scholar] [CrossRef]
- Mehta, J.; Rouah-Martin, E.; van Dorst, B.; Maes, B.; Herrebout, W.; Scippo, M.-L.; Dardenne, F.; Blust, R.; Robbens, J. Selection and characterization of pcb-binding DNA aptamers. Anal. Chem. 2012, 84, 1669–1676. [Google Scholar] [CrossRef]
- Moore, P.D.; Yedjou, C.G.; Tchounwou, P.B. Malathion-Induced oxidative stress, cytotoxicity, and genotoxicity in human liver carcinoma (hepg2) cells. Environ. Toxicol. 2010, 25, 221–226. [Google Scholar]
- Pluth, J.M.; Nicklas, J.A.; O’Neill, J.P.; Albertini, R.J. Increased frequency of specific genomic deletions resulting from in vitro malathion exposure. Cancer Res. 1996, 56, 2393–2399. [Google Scholar]
- Reus, G.Z.; Valvassori, S.S.; Nuernberg, H.; Comim, C.M.; Stringari, R.B.; Padilha, P.T.; Leffa, D.D.; Tavares, P.; Dagostim, G.; Paula, M.M.; et al. DNA damage after acute and chronic treatment with malathion in rats. J. Agric. Food Chem. 2008, 56, 7560–7565. [Google Scholar] [CrossRef]
- Stojanovic, M.N.; de Prada, P.; Landry, D.W. Aptamer-Based folding fluorescent sensor for cocaine. J. Am. Chem. Soc. 2001, 123, 4928–4931. [Google Scholar] [CrossRef]
- Agency, U.S. E.P. Atrazine Updates. In Pesticides: Registration. Available online: http://www.epa.gov/oppsrrd1/reregistration/atrazine/atrazine_update.htm (accessed on 11 August 2014).
- Anderson, B.; Corbin, M.; Erickson, R.; Frankenberry, M.; Irene, S.; Pease, A.; Thawley, M.; Thurman, N. The ecological significance of atrazine effects on primary producers in surface water streams in the corn and sorghum growing region of the united states (part ii). In Off. Pestic. Programs Environ. Fate Effects Div.; 2009. Available online: http://www.epa.gov/scipoly/sap/meetings/2009/may/051209minutes.pdf (accessed on 14 August 2014). [Google Scholar]
- Reinemann, C.; Linkorn, S.; Stoltenburg, R. Aptamers for pharmaceuticals and their application in environmental analytics. Bioanal. Rev. 2012, 4, 1–30. [Google Scholar]
- Yildirim, N.; Long, F.; Gao, C.; He, M.; Shi, H.C.; Gu, A.Z. Aptamer-Based optical biosensor for rapid and sensitive detection of 17beta-estradiol in water samples. Environ. Sci. Technol. 2012, 46, 3288–3294. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, L.; Zhang, H.; Boey, F.; Song, S.; Fan, C. Aptamer-Based multicolor fluorescent gold nanoprobes for multiplex detection in homogeneous solution. Small 2010, 6, 201–204. [Google Scholar] [CrossRef]
- Saltmiras, D.A.; Lemley, A.T. Atrazine degradation by anodic fenton treatment. Water Res. 2002, 36, 5113–5119. [Google Scholar] [CrossRef]
- Mandelbaum, R.T.; Allan, D.L.; Wackett, L.P. Isolation and characterization of a pseudomonas sp that mineralizes the s-triazine herbicide atrazine. Appl. Environ. Microbiol. 1995, 61, 1451–1457. [Google Scholar]
- Herzberg, M.; Dosoretz, C.G.; Tarre, S.; Michael, B.; Dror, M.; Green, M. Simultaneous removal of atrazine and nitrate using a biological granulated activated carbon (bgac) reactor. J. Chem. Technol. Biotechnol. 2004, 79, 626–631. [Google Scholar] [CrossRef]
- Qian, J.; Lou, X.; Zhang, Y.; Xiao, Y.; Soh, H.T. Generation of highly specific aptamers via micromagnetic selection. Anal. Chem. 2009, 81, 5490–5495. [Google Scholar] [CrossRef]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. Flumag-Selex as an advantageous method for DNA aptamer selection. Anal. Bioanal. Chem. 2005, 383, 83–91. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Williams, R.M.; Crihfield, C.L.; Gattu, S.; Holland, L.A.; Sooter, L.J. In Vitro Selection of a Single-Stranded DNA Molecular Recognition Element against Atrazine. Int. J. Mol. Sci. 2014, 15, 14332-14347. https://doi.org/10.3390/ijms150814332
Williams RM, Crihfield CL, Gattu S, Holland LA, Sooter LJ. In Vitro Selection of a Single-Stranded DNA Molecular Recognition Element against Atrazine. International Journal of Molecular Sciences. 2014; 15(8):14332-14347. https://doi.org/10.3390/ijms150814332
Chicago/Turabian StyleWilliams, Ryan M., Cassandra L. Crihfield, Srikanth Gattu, Lisa A. Holland, and Letha J. Sooter. 2014. "In Vitro Selection of a Single-Stranded DNA Molecular Recognition Element against Atrazine" International Journal of Molecular Sciences 15, no. 8: 14332-14347. https://doi.org/10.3390/ijms150814332
APA StyleWilliams, R. M., Crihfield, C. L., Gattu, S., Holland, L. A., & Sooter, L. J. (2014). In Vitro Selection of a Single-Stranded DNA Molecular Recognition Element against Atrazine. International Journal of Molecular Sciences, 15(8), 14332-14347. https://doi.org/10.3390/ijms150814332




