Bistable Switch in let-7 miRNA Biogenesis Pathway Involving Lin28
Abstract
:1. Introduction
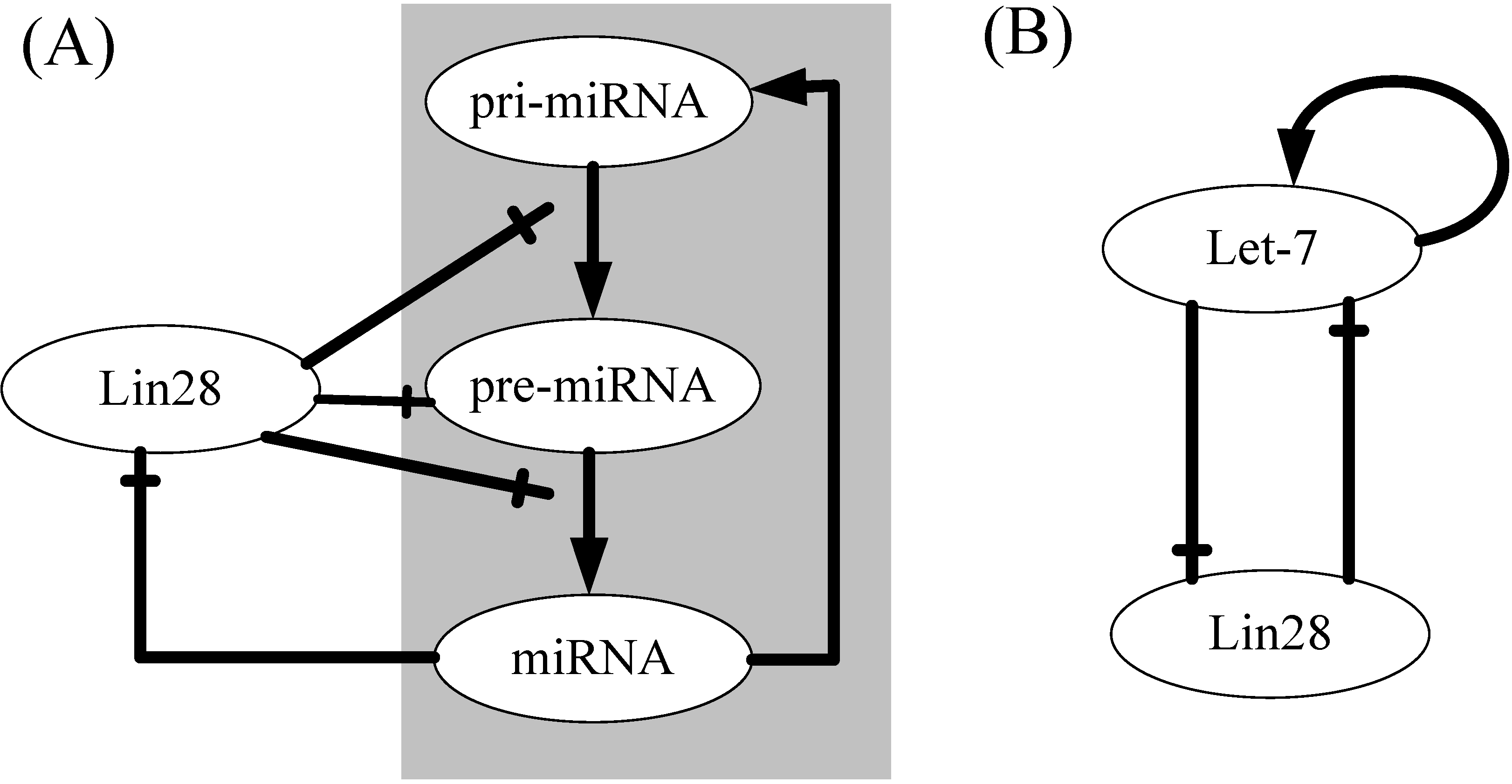
2. Results and Discussion
2.1. Model Introduction
2.2. Steady States of the Model
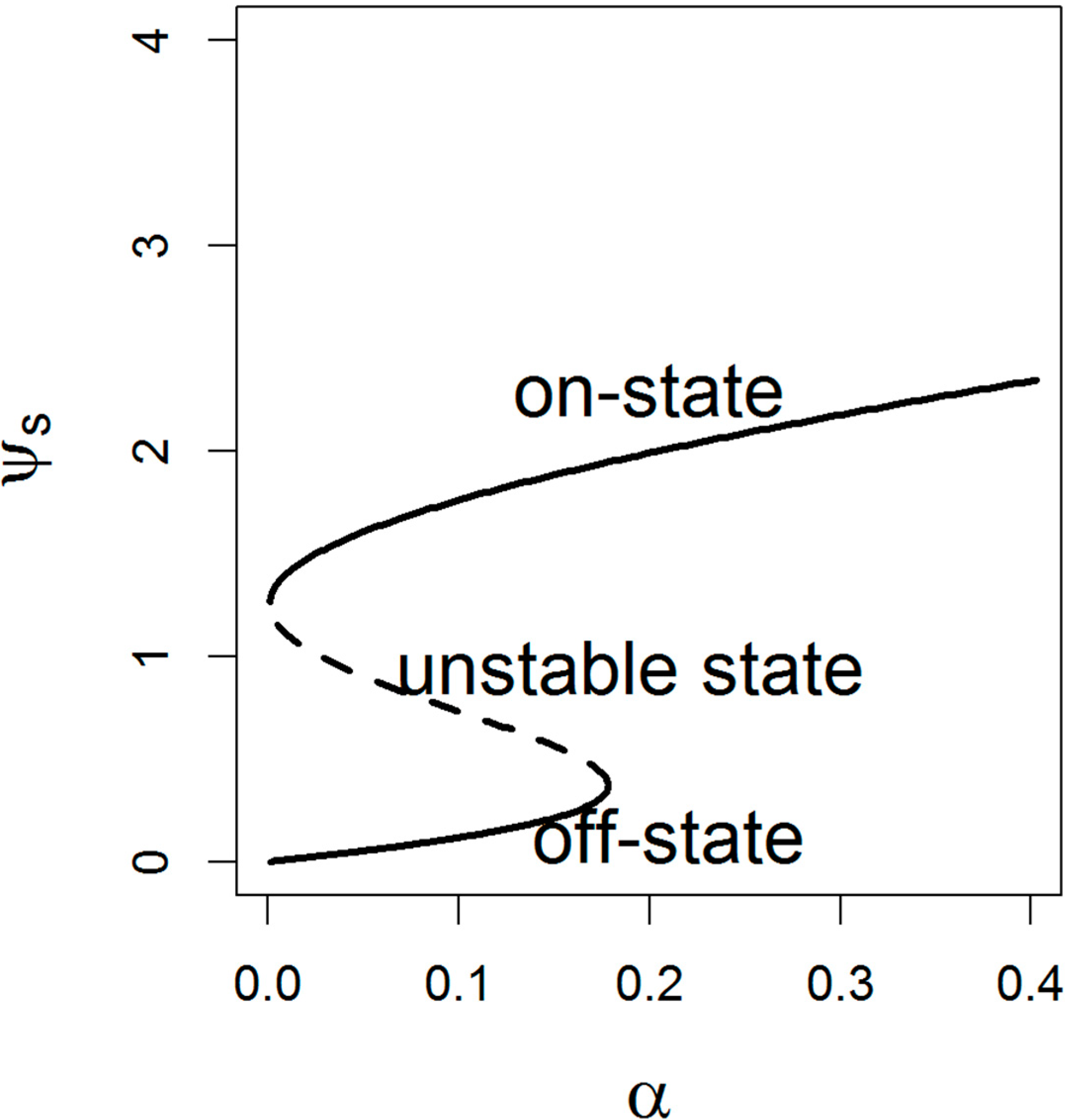
2.3. Effects of the Positive Feedback of let-7 on Switching Behavior

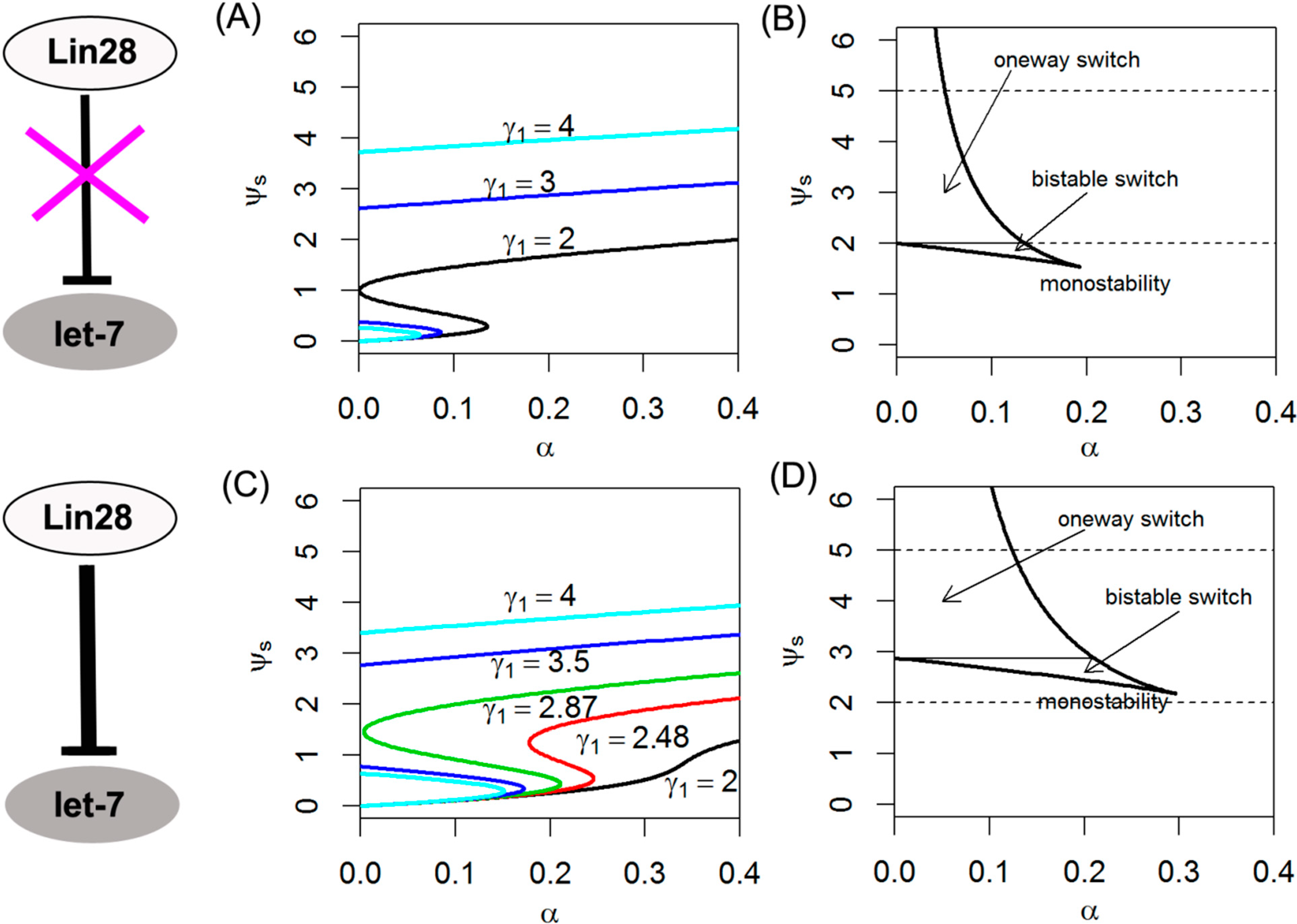
2.4. Effects of Dual Negative Feedback Regulation of Lin28 and let-7 on Switching Behavior
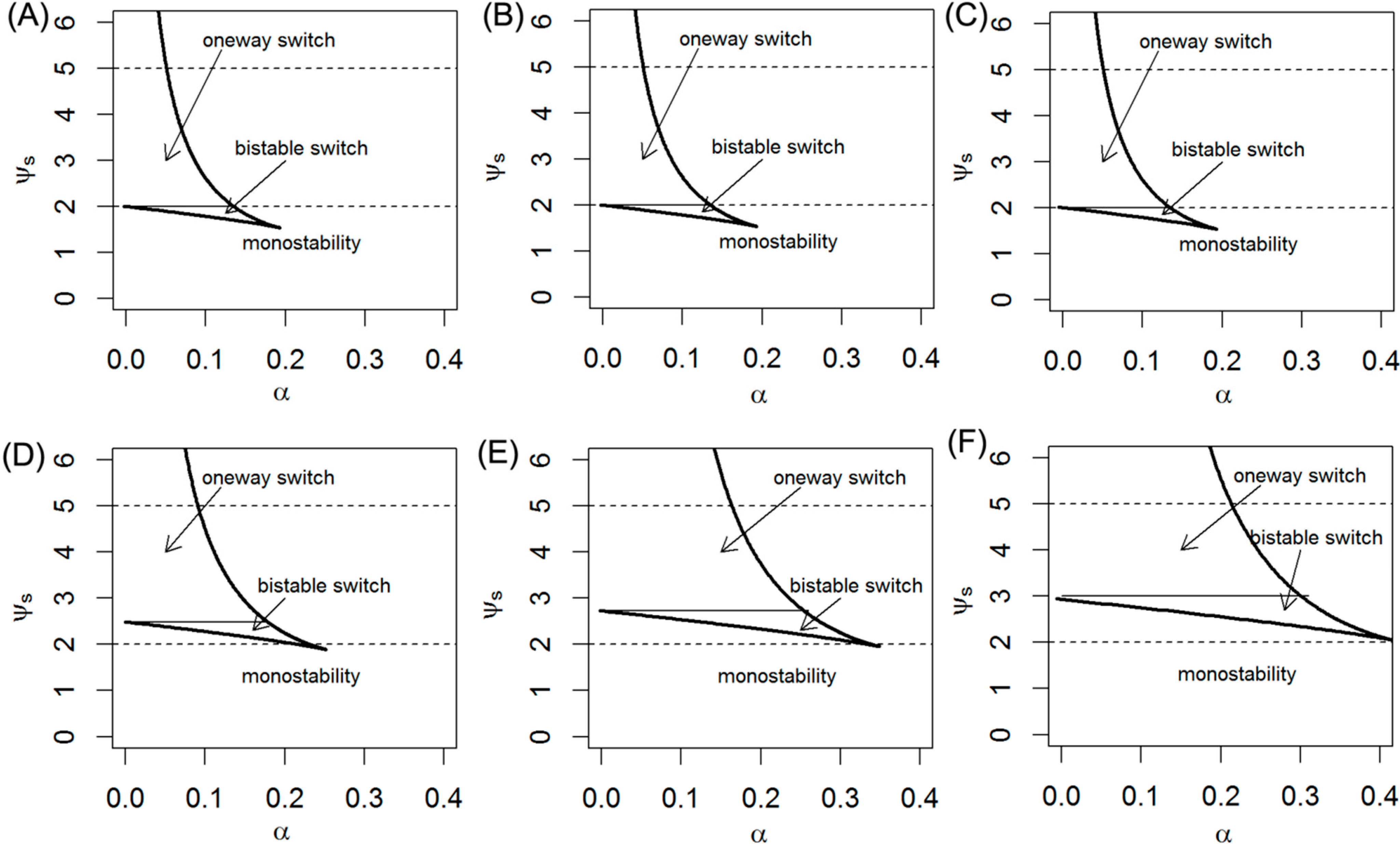
2.5. Effects of Expression of Lin28 and let-7 on Switching Behavior
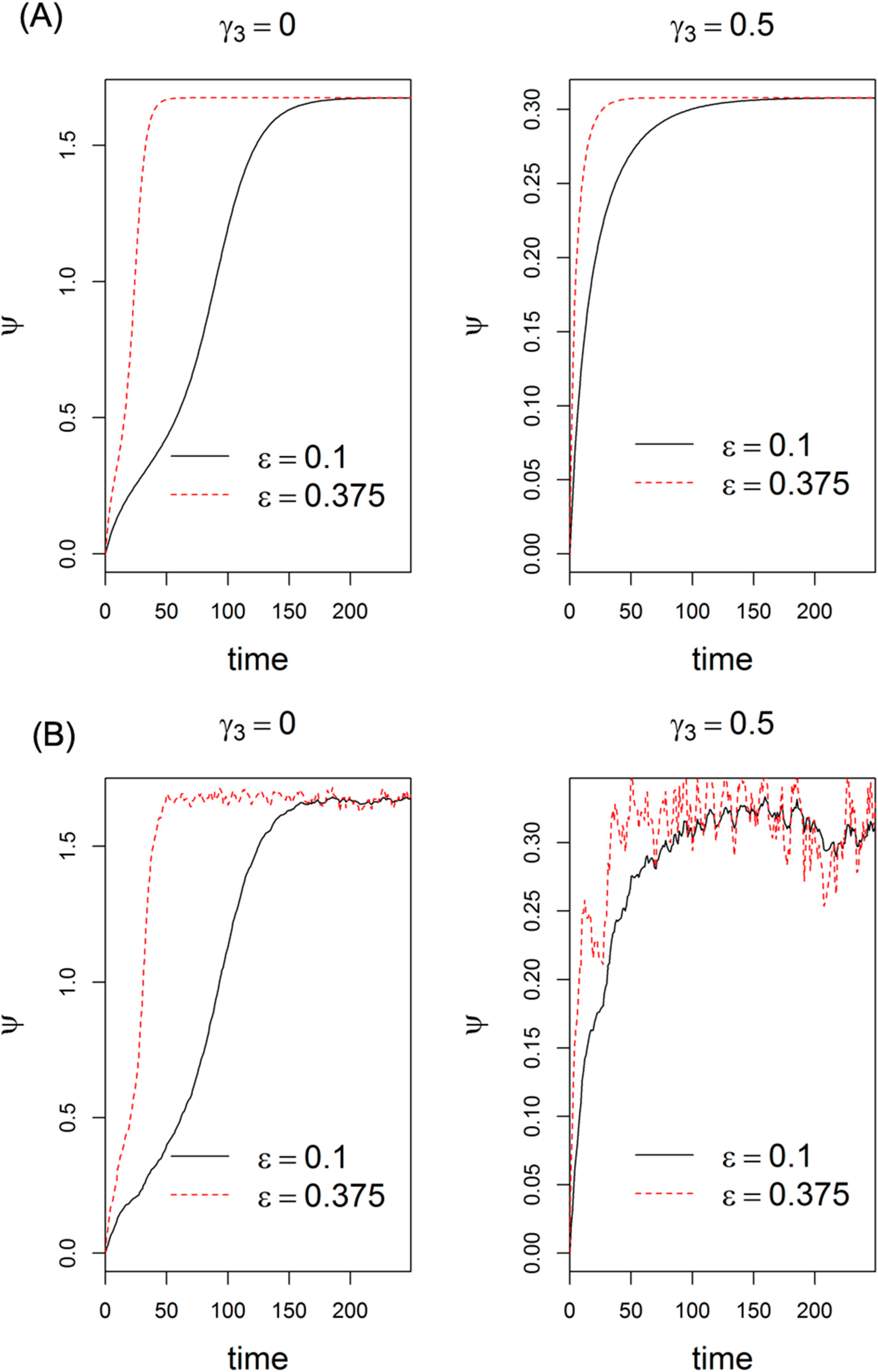
2.6. Implications of Lin28/let-7 Axis in Cancer Treatment
3. Experimental Section
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Suh, N.; Blelloch, R. Small RNAs in early mammalian development: From gametes to gastrulation. Development 2011, 138, 1653–1661. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.N.; Hata, A. Regulation of MicroRNA Biogenesis: A miRiad of mechanisms. Cell Commun. Signal. 2009, 7, 18. [Google Scholar] [CrossRef] [PubMed]
- Roush, S.; Slack, F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [PubMed]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef] [PubMed]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Muller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef]
- Thornton, J.E.; Gregory, R.I. How does Lin28 let-7 control development and disease? Trends Cell Biol. 2012, 22, 474–482. [Google Scholar] [CrossRef]
- Urbach, A.; Yermalovich, A.; Zhang, J.; Spina, C.S.; Zhu, H.; Perez-Atayde, A.R.; Shukrun, R.; Charlton, J.; Sebire, N.; Mifsud, W.; et al. Lin28 sustains early renal progenitors and induces Wilms tumor. Genes Dev. 2014, 28, 971–982. [Google Scholar] [CrossRef]
- Zisoulis, D.G.; Kai, Z.S.; Chang, R.K.; Pasquinelli, A.E. Autoregulation of microRNA biogenesis by let-7 and Argonaute. Nature 2012, 486, 541–454. [Google Scholar] [PubMed]
- Nam, Y.; Chen, C.; Gregory, R.I.; Chou, J.J.; Sliz, P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 2011, 147, 1080–1091. [Google Scholar] [CrossRef] [PubMed]
- Newman, M.A.; Thomson, J.M.; Hammond, S.M. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 2008, 14, 1539–1549. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, S.R.; Daley, G.Q.; Gregory, R.I. Selective blockade of microRNA processing by Lin28. Science 2008, 320, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Hagan, J.P.; Piskounova, E.; Gregory, R.I. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2009, 16, 1021–1025. [Google Scholar] [CrossRef] [PubMed]
- Pillai, R.S.; Bhattacharyya, S.N.; Artus, C.G.; Zoller, T.; Cougot, N.; Basyuk, E.; Bertrand, E.; Filipowicz, W. Inhibition of translational initiation by let-7 MicroRNA in human cells. Science 2005, 309, 1573–1576. [Google Scholar] [CrossRef] [PubMed]
- Zhdanov, V. Two novel aspects of the kinetics of gene expression including miRNAs. Centr. Eur. J. Phys. 2013, 11, 448–456. [Google Scholar] [CrossRef]
- Zhdanov, V.P. Kinetic models of gene expression including non-coding RNAs. Phys. Rep. 2011, 500, 1–42. [Google Scholar] [CrossRef]
- Cai, S.; Zhou, P.; Liu, Z. Functional characteristics of a double negative feedback loop mediated by microRNAs. Cogn. Neurodyn. 2013, 7, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Zhdanov, V.P. Bistability in gene transcription: Interplay of messenger RNA, protein, and nonprotein coding RNA. Biosystems 2009, 95, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Price, J.; Ribeiro, A.S. Bistability in a stochastic RNA-mediated gene network. Phys. Rev. E Stat. Nonlin. Soft. Matter. Phys. 2013, 88, 032714. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Chang, X.; Liu, Z.; Chen, L.; Wang, R. Bistability and oscillations in gene regulation mediated by small noncoding RNAs. PLoS One 2011, 6, e17029. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Raghavachari, S. Quantifying negative feedback regulation by micro-RNAs. Phys. Biol. 2011, 8, 055002. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Y.; Xu, X.; Wang, Y.H. Toward a system-level understanding of microRNA pathway via mathematical modeling. Biosystems 2010, 100, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, X.; Ma, Z.; Huo, Y.; Xiao, Z.; Li, Y.; Wang, Y. Dynamic mechanisms for pre-miRNA binding and export by Exportin-5. RNA 2011, 17, 1511–1528. [Google Scholar] [CrossRef] [PubMed]
- Aguda, B.D.; Kim, Y.; Piper-Hunter, M.G.; Friedman, A.; Marsh, C.B. MicroRNA regulation of a cancer network: Consequences of the feedback loops involving miR-17–92, E2F, and Myc. Proc. Natl. Acad. Sci. USA 2008, 105, 19678–19683. [Google Scholar] [CrossRef] [PubMed]
- Iliopoulos, D.; Hirsch, H.A.; Struhl, K. An epigenetic switch involving NF-κB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 2009, 139, 693–706. [Google Scholar] [CrossRef]
- Kim, C.W.; Vo, M.T.; Kim, H.K.; Lee, H.H.; Yoon, N.A.; Lee, B.J.; Min, Y.J.; Joo, W.D.; Cha, H.J.; Park, J.W.; et al. Ectopic over-expression of tristetraprolin in human cancer cells promotes biogenesis of let-7 by down-regulation of Lin28. Nucleic Acids Res. 2012, 40, 3856–3869. [Google Scholar] [CrossRef]
- Yao, G.; Lee, T.J.; Mori, S.; Nevins, J.R.; You, L. A bistable Rb-E2F switch underlies the restriction point. Nat. Cell Biol. 2008, 10, 476–482. [Google Scholar] [CrossRef]
- Li, Y.; Li, Y.; Zhang, H.; Chen, Y. MicroRNA-mediated positive feedback loop and optimized bistable switch in a cancer network Involving miR-17–92. PLoS One 2011, 6, e26302. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, M.; Miki, Y.; Masuda, M.; Hata, S.; Shibahara, Y.; Hirakawa, H.; Suzuki, T.; Sasano, H. LIN28: A regulator of tumor-suppressing activity of let-7 microRNA in human breast cancer. J. Steroid Biochem. Mol. Boil. 2012, 131, 101–106. [Google Scholar] [CrossRef]
- Rybak, A.; Fuchs, H.; Smirnova, L.; Brandt, C.; Pohl, E.E.; Nitsch, R.; Wulczyn, F.G. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 2008, 10, 987–993. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, J.; Gao, L.; McClellan, S.; Finan, M.A.; Butler, T.W.; Owen, L.B.; Piazza, G.A.; Xi, Y. MiR-181 mediates cell differentiation by interrupting the Lin28 and let-7 feedback circuit. Cell Death Differ. 2012, 19, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Cimadamore, F.; Amador-Arjona, A.; Chen, C.; Huang, C.T.; Terskikh, A.V. SOX2-LIN28/let-7 pathway regulates proliferation and neurogenesis in neural precursors. Proc. Natl. Acad. Sci. USA 2013, 110, E3017–E3026. [Google Scholar] [CrossRef] [PubMed]
- Moss, E.G.; Tang, L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev. Biol. 2003, 258, 432–442. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, S.R.; Powers, J.T.; Einhorn, W.; Hoshida, Y.; Ng, T.L.; Toffanin, S.; O’Sullivan, M.; Lu, J.; Phillips, L.A.; Lockhart, V.L.; et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 2009, 41, 843–848. [Google Scholar] [CrossRef]
- Gantier, M.P.; McCoy, C.E.; Rusinova, I.; Saulep, D.; Wang, D.; Xu, D.; Irving, A.T.; Behlke, M.A.; Hertzog, P.J.; Mackay, F.; et al. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 2011, 39, 5692–5703. [Google Scholar] [CrossRef]
- Wang, L.X.; Wang, J.; Qu, T.T.; Zhang, Y.; Shen, Y.F. Reversible acetylation of Lin28 mediated by PCAF and SIRT1. Biochim. Biophys. Acta 2014, 1843, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Li, N.; Liang, S.; Huang, Q.; Coukos, G.; Zhang, L. Identification of microRNAs regulating reprogramming factor LIN28 in embryonic stem cells and cancer cells. J. Biol. Chem. 2010, 285, 41961–41971. [Google Scholar] [CrossRef]
- Rybak, A.; Fuchs, H.; Hadian, K.; Smirnova, L.; Wulczyn, E.A.; Michel, G.; Nitsch, R.; Krappmann, D.; Wulczyn, F.G. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat. Cell Biol. 2009, 11, 1411–1420. [Google Scholar] [CrossRef] [PubMed]
- Paulsson, J. Summing up the noise in gene networks. Nature 2004, 427, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Levine, E.; Zhang, Z.; Kuhlman, T.; Hwa, T. Quantitative characteristics of gene regulation by small RNA. PLoS Biol. 2007, 5, e229. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Cassidy, J.J.; Reinke, C.A.; Fischboeck, S.; Carthew, R.W. A microRNA imparts robustness against environmental fluctuation during development. Cell 2009, 137, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.J.; Zhang, X.P.; Liu, F.; Wang, W. Interlinking positive and negative feedback loops creates a tunable motif in gene regulatory networks. Phys. Rev. E Stat. Nonlin. Soft. Matter. Phys. 2009, 80, 011926. [Google Scholar] [CrossRef] [PubMed]
- Tsai, T.Y.; Choi, Y.S.; Ma, W.; Pomerening, J.R.; Tang, C.; Ferrell, J.E., Jr. Robust, tunable biological oscillations from interlinked positive and negative feedback loops. Science 2008, 321, 126–129. [Google Scholar] [CrossRef] [PubMed]
- Oscill8. Available online: http://oscill8.sourceforge.net (accessed on 20 October 2014).
- The R Project for Statistical Computing. Available online: http://www.R-project.org (accessed on 20 October 2014).
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, F.; Yu, W.; Wang, X. Bistable Switch in let-7 miRNA Biogenesis Pathway Involving Lin28. Int. J. Mol. Sci. 2014, 15, 19119-19133. https://doi.org/10.3390/ijms151019119
Shi F, Yu W, Wang X. Bistable Switch in let-7 miRNA Biogenesis Pathway Involving Lin28. International Journal of Molecular Sciences. 2014; 15(10):19119-19133. https://doi.org/10.3390/ijms151019119
Chicago/Turabian StyleShi, Fei, Wenbao Yu, and Xia Wang. 2014. "Bistable Switch in let-7 miRNA Biogenesis Pathway Involving Lin28" International Journal of Molecular Sciences 15, no. 10: 19119-19133. https://doi.org/10.3390/ijms151019119
APA StyleShi, F., Yu, W., & Wang, X. (2014). Bistable Switch in let-7 miRNA Biogenesis Pathway Involving Lin28. International Journal of Molecular Sciences, 15(10), 19119-19133. https://doi.org/10.3390/ijms151019119



