Expression of Root-Related Transcription Factors Associated with Flooding Tolerance of Soybean (Glycine max)
Abstract
:1. Introduction
2. Results and Discussion
2.1. Physiological, Morphological and Anatomical Mechanism of Flooding Tolerance
2.1.1. Root Growth
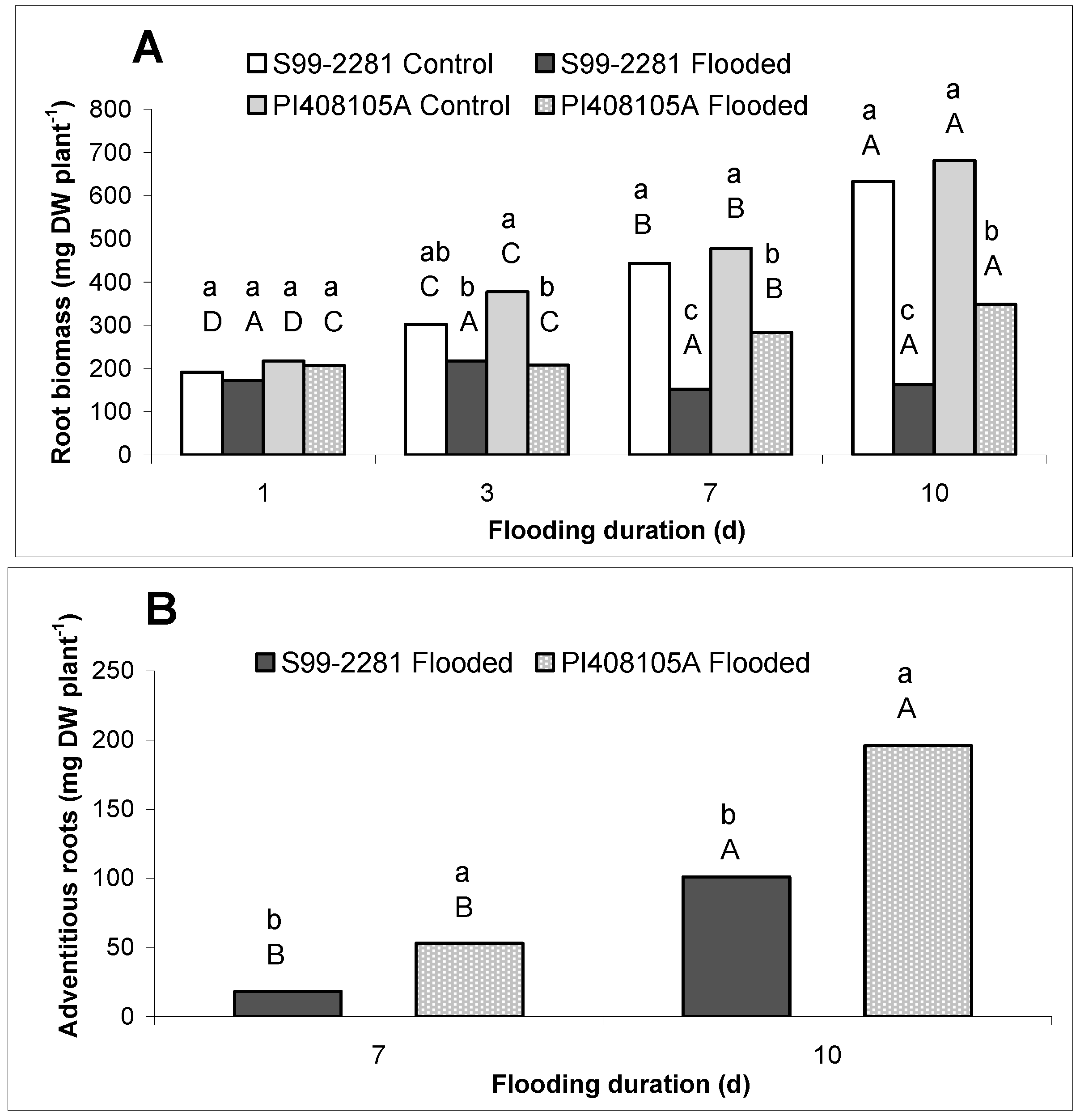
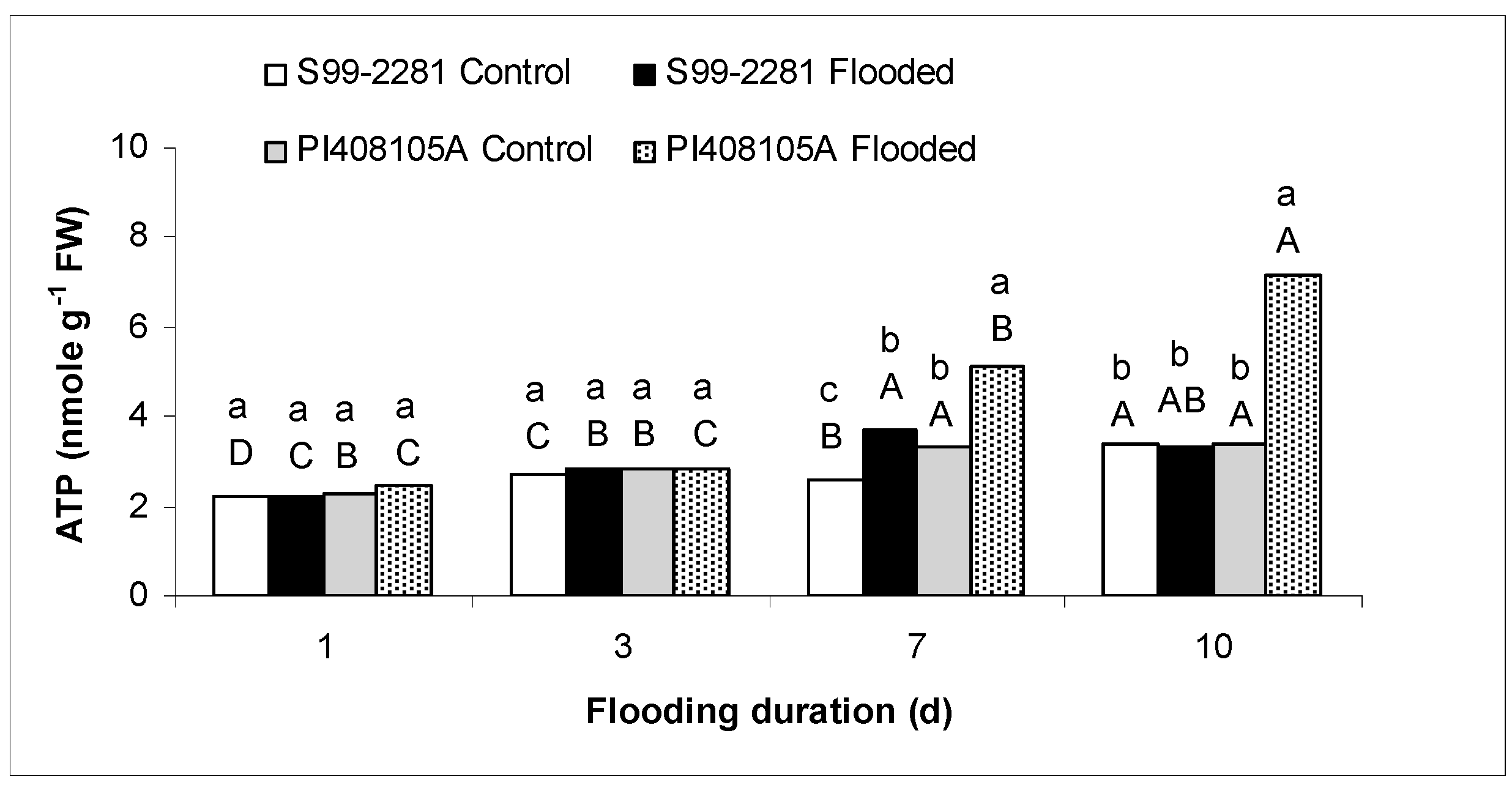
2.1.2. Root ATP Concentration
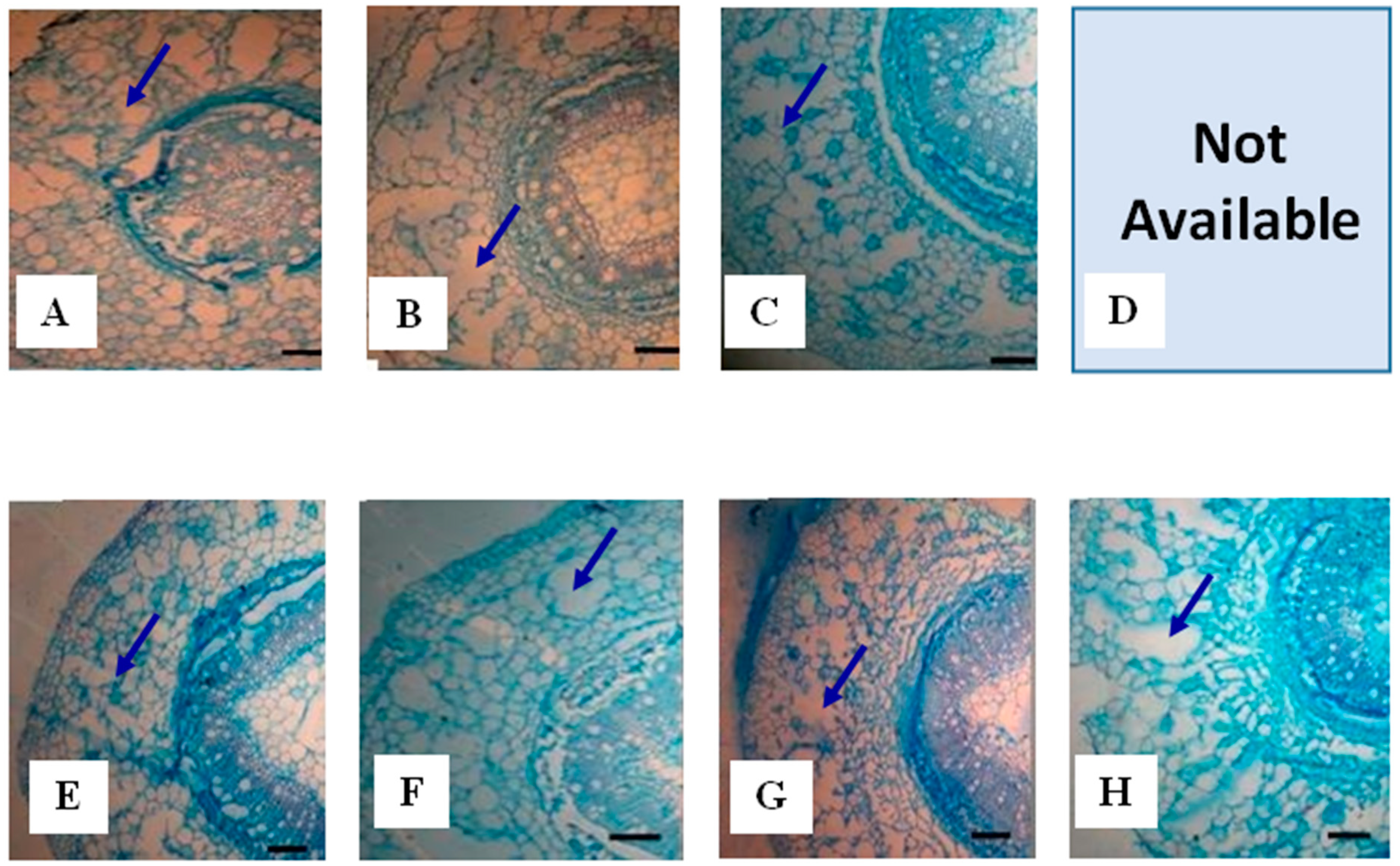
2.1.3. Root Aerenchyma
| Genotype | FD (day) | TRA (mm2) | CCA (mm2) | COA (mm2) | TAA (mm2) | PAA (%) | PAC (%) |
|---|---|---|---|---|---|---|---|
| PI408105A | 1 | 102.5 d * | 11.2 d | 91.3 c | 15.39 c | 15.1 b | 16.9 b |
| PI408105A | 3 | 129.9 d | 24.5 c | 105.5 c | 26.64 b | 20.5 a | 25.2 a |
| PI408105A | 7 | 171.0 b | 32.9 b | 138.2 b | 30.90 a | 18.2 a | 22.6 a |
| PI408105A | 10 | - | - | No results * | - | - | - |
| S99-2281 | 1 | 150.1 c | 28.2 b | 121.9 b | 13.28 c | 8.9 c | 11.0 c |
| S99-2281 | 3 | 166.9 b | 29.6 b | 137.2 b | 25.07 b | 15.1 b | 18.3 b |
| S99-2281 | 7 | 213.9 a | 48.7 a | 165.2 a | 33.94 a | 15.9 b | 20.6 b |
| S99-2281 | 10 | 180.8 b | 25.2 c | 155.6 a | 23.01 b | 12.9 b | 14.9 b |
2.2. Expression of Root-Related Transcription Factors and Anaerobic Genes
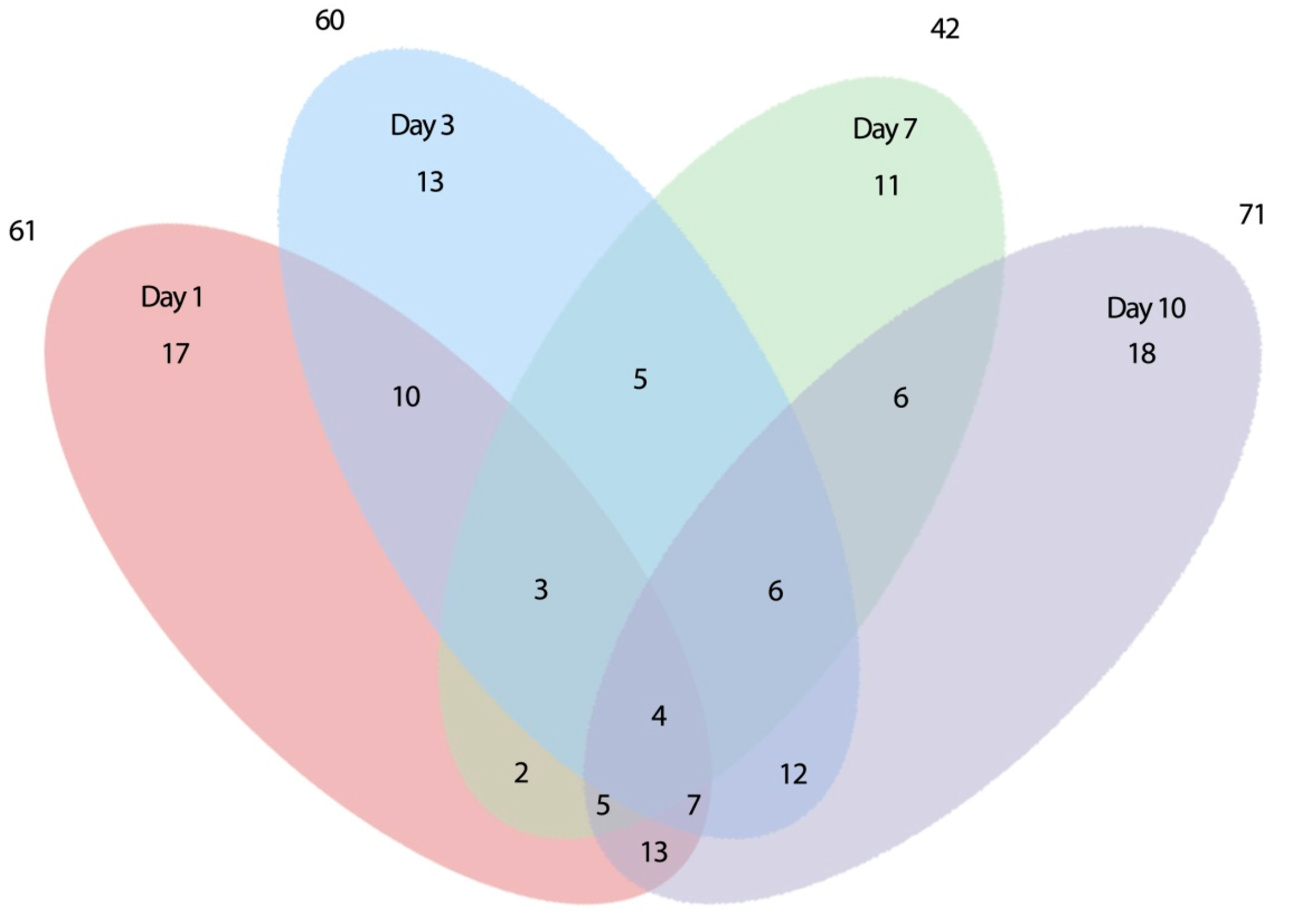
2.2.1. Early Differentially Expressed Genes between the Two Genotypes
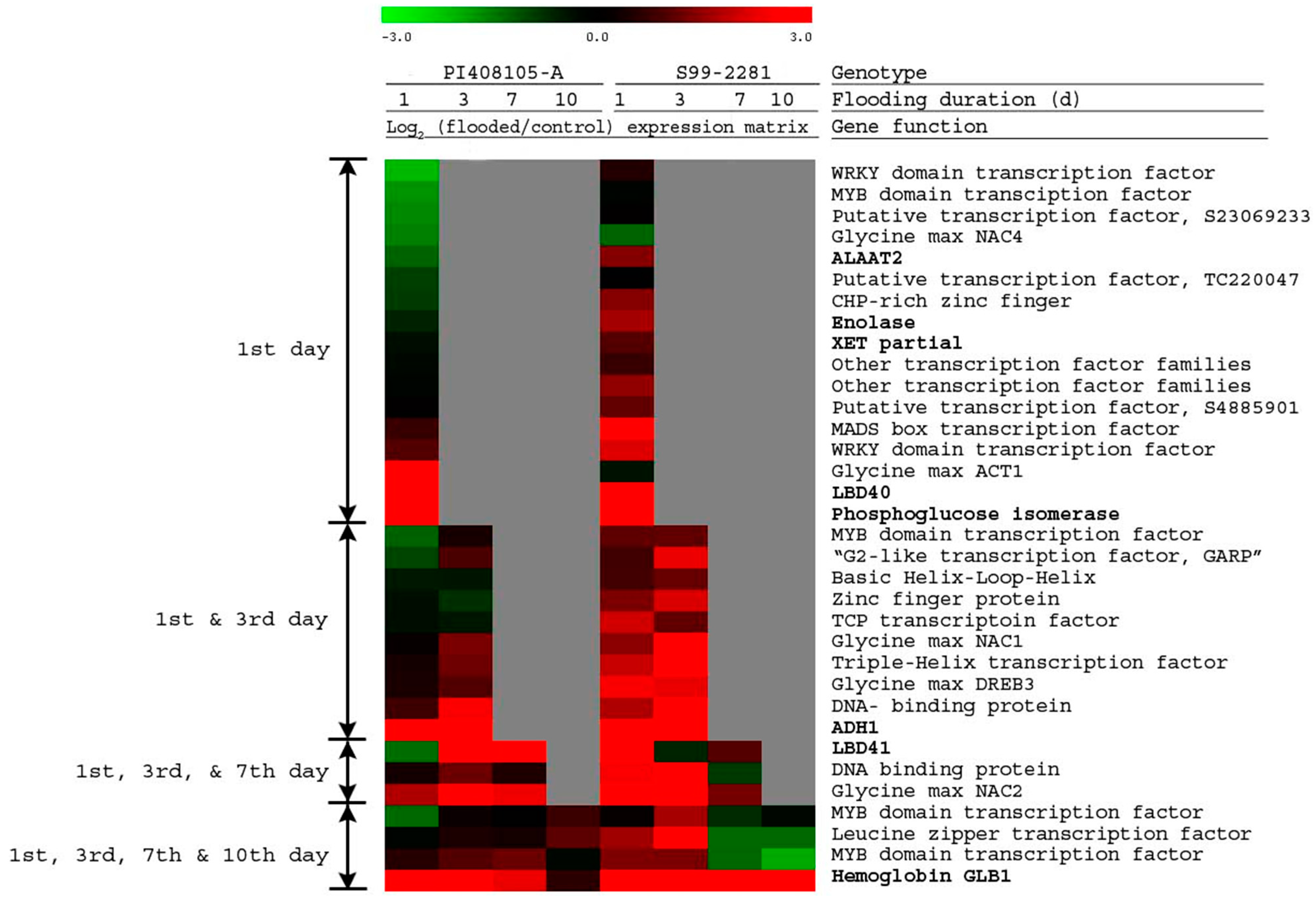
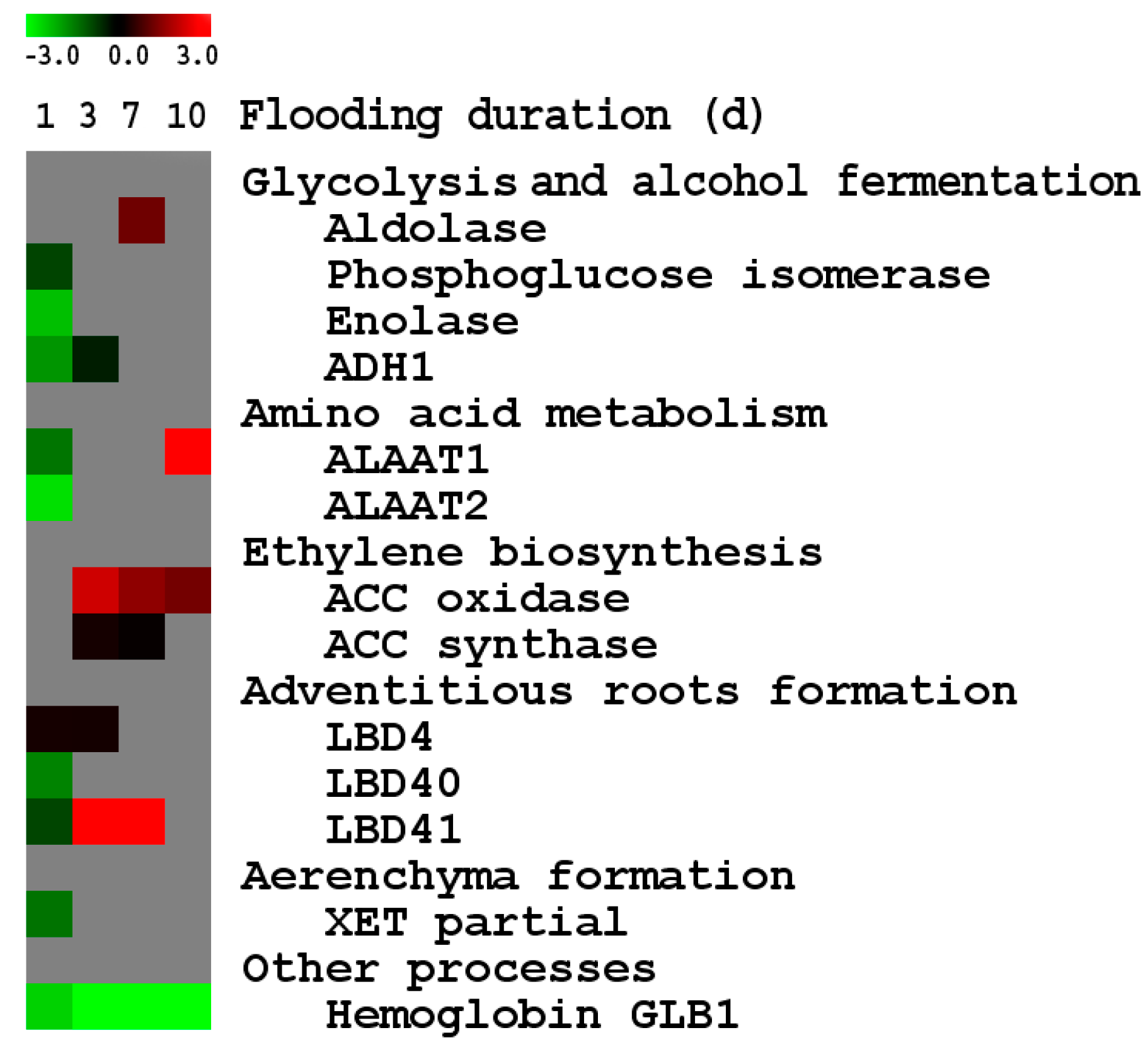
2.2.2. Differential Expression of Known Anaerobic Genes between the Two Genotypes
2.2.3. Gene Expression Associated with Flooding Tolerance
2.2.4. Expression of Known Anaerobic Genes
2.2.5. Flooding-Tolerance Candidate Genes
3. Experimental Section
3.1. Plant Materials and Flooding Treatment
3.2. Phenotypic Analysis of Responses to Flooding
3.2.1. Determination of Root Biomass
3.2.2. Analysis of Root ATP Content
3.2.3. Analysis of Aerenchyma
3.2.4. Statistical Analysis
3.3. High Throughput Quantitative Reverse-Transcription PCR (qRT-PCR)
3.3.1. RNA Isolation, DNase Treatment and cDNA Synthesis
3.3.2. Primers of Soybean Transcription Factors for qRT-PCR
3.3.3. qRT-PCR
3.3.4. Statistical Analysis and Data Presentation
4. Conclusions
Supplementary Materials
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fausey, N.R. Subirrigation/drainage systems for water table management in the Midwest. In Proceedings of the International Irrigation Exposition and Technical Conference, Atlanta, GA, USA, 20 November 1994; pp. 242–246.
- VanToai, T.; Beuerlein, J.E.; Schmitthenner, A.F; Martin, S.K. Genetic variability for flooding tolerance in soybeans. Crop Sci. 1994, 6, 1112–1115. [Google Scholar]
- Sullivan, M.; VanToai, T.T.; Fausey, N.; Beuerlein, J.; Parkinson, R.; Soboyejo, A. Evaluating on-farm flooding impacts on soybean. Crop Sci. 2001, 41, 93–100. [Google Scholar]
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [PubMed]
- Rosenzweig, C.; Tubiello, F.N.; Goldberg, R.; Mills, E.; Bloomfield, J. Increased crop damage in the US from excess precipitation under climate change. Glob. Environ. Chang. 2002, 12, 197–202. [Google Scholar]
- Kennedy, R.A.; Rumpho, M.E.; Fox, T.C. Anaerobic metabolism in plants. Plant Physiol. 1992, 84, 1204–1209. [Google Scholar]
- Perata, V.M.; Alpi, A. Plant responses to anaerobiosis. Plant Sci. 1993, 93, 1–17. [Google Scholar]
- Ricard, B.; Couee, I.; Raymond, P.; Saglio, P.H.; Saint-Ges, V.; Pradet, A. Plant metabolism under hypoxia and anoxia. Plant Biochem. 1994, 32, 1–10. [Google Scholar]
- Sachs, M.M.; Subbaiah, C.C.; Sabb, I.N. Anaerobic gene expression and flooding tolerance in maize. J. Exp. Bot. 1996, 47, 1–15. [Google Scholar]
- Klok, E.J.; Wilson, I.W.; Wilson, D.; Chapman, S.C.; Ewing, R.M.; Somerville, S.C.; Peacock, W.J.; Dolferus, R.; Dennis, E.S. Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell 2002, 14, 2481–2494. [Google Scholar] [PubMed]
- Ismond, K.P.; Dolferus, R.; de Pauw, M.; Dennis, E.S.; Good, A.G. Enhanced low oxygen survival in Arabidopsis through increased metabolic flux in the fermentative pathway. Plant Physiol. 2003, 132, 1292–1302. [Google Scholar] [PubMed]
- Liu, F.; van Toai, T.T.; Moy, L.; Bock, G.; Linford, L.D.; Quackenbush, J. Global transcription profiling reveals novel insights into hypoxic response in Arabidopsis. Plant Physiol. 2005, 137, 1115–1120. [Google Scholar] [PubMed]
- Van Dongen, J.T.; Frohlich, A.; Ramirez-Aguilar, S.J.; Schauer, N.; Fernie, A.R.; Erban, A.; Kopka, J.; Clark, J.; Langer, A.; Geigenberger, P. Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of Arabidopsis plants. Ann. Bot. 2009, 103, 269–280. [Google Scholar] [PubMed]
- Christianson, J.A.; Wilson, I.W.; Llewellyn, D.J.; Dennis, E.S. The low-oxygen-induced NAC domain transcription factor ANAC102 affects viability of Arabidopsis seeds following low-oxygen treatment. Plant Physiol. 2009, 149, 1724–1738. [Google Scholar] [PubMed]
- Chang, W.W.; Huang, L.; Shen, M.; Webster, C.; Burlingame, A.L.; Roberts, J.K. Patterns of protein synthesis and tolerance of anoxia in root tips of maize seedlings acclimated to a low-oxygen environment and identification of proteins by mass spectrometry. Plant Physiol. 2000, 122, 295–318. [Google Scholar] [PubMed]
- Niroula, R.K.; Pucciariello, C.; Ho, V.T.; Novi, G.; Fukao, T.; Perata, P. SUB1A-dependent and -independent mechanisms are involved in the flooding tolerance of wild rice species. Plant J. 2012, 72, 282–293. [Google Scholar] [PubMed]
- Komatsu, S.; Nanjo, Y.; Nishimura, M. Proteomic analysis of the flooding tolerance mechanism in mutant soybean. J. Proteomics 2013, 79, 231–250. [Google Scholar] [PubMed]
- Fennoy, S.; Nong, T.; Bailey-Serres, J. Transcriptional and post-transcriptional processes regulate gene expression in oxygen-deprived roots of maize. Plant J. 1998, 15, 727–735. [Google Scholar]
- Russell, D.A.; Sachs, M.M. Protein synthesis in maize during anaerobic and heat Stress. Plant Physiol. 1992, 99, 615–620. [Google Scholar] [PubMed]
- Sachs, M.M. Molecular response to anoxic stress in maize. In Plant Life under Oxygen Deprivation; Jackson, M.B., Davies, D.D., Lambers, H., Eds.; SPB Academic Publishing: Hague, The Netherlands, 1991. [Google Scholar]
- Kelley, P.M.; Freeling, M. Anaerobic expression of maize fructose-1,6-diphosphate aldolase. J. Biol. Chem. 1984, 259, 14180–14183. [Google Scholar] [PubMed]
- Kelley, P.M.; Freeling, M. Anaerobic expression of maize glucose phosphate isomerase I. J. Biol. Chem. 1984, 259, 673–677. [Google Scholar] [PubMed]
- Kelley, P. Maize pyruvate decarboxylase mRNA is induced anaerobically. Plant Mol. Biol. 1989, 13, 213–222. [Google Scholar] [PubMed]
- Dennis, E.S.; Dolferus, R.; Ellis, M.; Rahman, M.; Wu, Y.; Hoeren, F.U.; Grover, A.; Ismond, K.P.; Good, A.G.; Peacock, W.J. Molecular strategies for improving waterlogging tolerance in plants. J. Exp. Bot. 2000, 51, 89–97. [Google Scholar] [PubMed]
- Voesenek, L.; Bailey-Serres, J. Flooding tolerance: O2 sensing and survival strategies. Curr. Opin. Plant Biol. 2013, 16, 647–653. [Google Scholar] [PubMed]
- De Vetten, N.C.; Ferl, R.J. Characterization of a maize G-box binding factor that is induced by hypoxia. Plant J. 1995, 7, 589–601. [Google Scholar] [PubMed]
- Hoeren, F.U.; Dolferus, R.; Wu, Y.; Peacock, J.W.; Dennis, E.S. Evidence for a role for ATMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (Adh1) by low oxygen. Genetics 1998, 149, 479–490. [Google Scholar] [PubMed]
- Baxter-Burrell, A.; Yang, A.Z.; Springer, P.S.; Bailey-Serres, J. RopGAP4-dependent rop GTPase rheostat control of Arabidopsis oxygen deprivation tolerance. Science 2002, 296, 2026–2028. [Google Scholar] [PubMed]
- Dordas, C.; Hasinoff, B.B.; Igamberdiev, A.U.; Manac'h, N.; Rivoal, J.; Hill, R.D. Expression of a stress-induced hemoglobin affects NO levels produced by alfalfa root cultures under hypoxic stress. Plant J. 2003, 35, 763–770. [Google Scholar] [PubMed]
- Dordas, C.; Hasinoff, B.B.; Rivoal, J.; Hill, R.D. Class-1 hemoglobins, nitrate and NO levels in anoxic maize cell-suspension cultures. Planta 2004, 219, 66–72. [Google Scholar] [PubMed]
- Vriezen, W.H.; Hulzink, R.; Mariani, C.; Voesenek, L.A.C.J. 1-Aminocyclopropane-1-carboxylate oxidase activity limits ethylene biosynthesis in Rumex palustris during submergence. Plant Physiol. 1999, 121, 189–196. [Google Scholar] [PubMed]
- Nie, X.; Singh, R.P.; Tai, G.C. Molecular characterization and expression analysis of 1-aminocyclopropane-1-carboxylate oxidase homologs from potato under abiotic and biotic stresses. Genome 2002, 45, 905–913. [Google Scholar] [PubMed]
- Saab, I.N.; Sachs, M.M. Complete cDNA and genomic sequence encoding a flooding-responsive gene from maize (Zea mays L.) homologous to xyloglucan endotransglycosylase. Plant Physiol. 1995, 108, 439–440. [Google Scholar] [PubMed]
- Saab, I.N.; Sachs, M.M. A flooding-induced xyloglucan endo-transglycosylase homolog in maize is responsive to ethylene and associated with aerenchyma. Plant Physiol. 1996, 112, 385–391. [Google Scholar] [PubMed]
- Loreti, E.; Poggi, A.; Novi, G.; Alpi, A.; Perata, P. A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol. 2005, 137, 1130–1138. [Google Scholar] [PubMed]
- Branco-Price, C.; Kawaguchi, R.; Ferreira, R.B.; Bailey-Serres, J. Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation. Ann. Bot. 2005, 96, 647–660. [Google Scholar] [PubMed]
- Lasanthi-Kudahettige, R.; Magneschi, L.; Loreti, E.; Gonzali, S.; Licausi, F.; Novi, G.; Beretta, O.; Vitulli, F.; Alpi, A.; Perata, P. Transcript profiling of the anoxic rice coleoptile. Plant Physiol. 2007, 144, 218–231. [Google Scholar] [PubMed]
- Narsai, R.; Howell, K.A.; Carroll, A.; Ivanova, A.; Millar, A.H.; Whelan, J. Defining core metabolic and transcriptomic responses to oxygen availability in rice embryos and young seedlings. Plant Physiol. 2009, 151, 306–322. [Google Scholar] [PubMed]
- Jung, K-H.; Seo, Y-S.; Walia, H.; Cao, P.; Fukao, T.; Canlas, P.E.; Amonpant, F.; Bailey-Serres, J.; Ronald, P.C. The submergence tolerance regulator Sub1a mediates stress-responsive expression of AP2/ERF transcription factors. Plant Physiol. 2010, 152, 1674–1692. [Google Scholar] [PubMed]
- Mustroph, A.; Lee, S.C.; Oosumi, T.; Zanetti, M.E.; Yang, H.; Ma, K.; Yaghoubi-Masihi, A.; Fukao, T.; Bailey-Serres, J. Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol. 2010, 152, 1484–1500. [Google Scholar] [PubMed]
- Kreuzwieser, J.; Hauberg, J.; Howell, K.A.; Carroll, A.; Rennenberg, H.; Millar, A.H.; Whelan, J. Differential response of gray poplar leaves and roots underpins stress adaptation during hypoxia. Plant Physiol. 2009, 149, 461–473. [Google Scholar] [PubMed]
- Oosterhuis, D.M.; Scott, H.D.; Hampton, R.E.; Wullschleger, S.D. Physiological response of two soybean [Glycine max, L. Merr] cultivars to short-term flooding. Environ. Exp. Bot. 1990, 30, 85–92. [Google Scholar]
- Gizlice, Z.; Carter, T.E., Jr.; Burton, J.W. Genetic base for North American public soybean cultivars released between 1947 and 1988. Crop Sci. 1994, 34, 1143–1151. [Google Scholar]
- Gizlice, Z.; Carter, T.E., Jr.; Burton, J.W. Genetic diversity patterns in North American public soybean cultivars based on coefficient of parentage. Crop Sci. 1996, 36, 753–765. [Google Scholar]
- Song, Q.; Hyten, D.L.; Jia, G.; Quigley, C.V.; Fickus, E.W.; Nelson, R.L.; Cregan, P.B. Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PLoS One 2013, 8, e54985. [Google Scholar] [PubMed]
- VanToai, T.T.; Nurjani, N. Screening for flooding tolerance of soybean. Soybean Genet. Newsl. 1996, 23, 210–213. [Google Scholar]
- Hou, F.F.; Thseng, F.S. Studies on the flooding tolerance of soybean seed: Varietal differences. Euphytica 1991, 57, 169–173. [Google Scholar]
- Shannon, J.G.; Stevens, W.E.; Wiebold, W.J.; McGraw, R.L.; Sleper, D.A.; Nguyen, H.T. Breeding soybeans for improved tolerance to flooding. In Proceedings of the 30th Soybean Research Conference, American Seed Trade Association, Chicago, IL, USA, 7 December 2005.
- Fukao, T.; Xu, K.; Ronald, P.C.; Bailey-Serres, J. A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 2006, 18, 2021–2034. [Google Scholar] [PubMed]
- Licausi, F.; van Dongen, J.T.; Giuntoli, B.; Novi, G.; Santaniello, A.; Geigenberger, P.; Perata, P. HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J. 2010, 62, 302–315. [Google Scholar] [PubMed]
- Leyva-González, M.A.; Ibarra-Laclette, E.; Cruz-Ramírez, A.; Herrera-Estrella, L. Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS One 2012, 7, e48138. [Google Scholar] [PubMed]
- Banti, V.; Mafessoni, F.; Loreti, E.; Alpi, A.; Perata, P. The heat-inducible transcription factor HsfA2 enhances anoxia tolerance in Arabidopsis. Plant Physiol. 2010, 152, 1471–1483. [Google Scholar] [PubMed]
- VanToai, T.; Yang, Y.; Ling, P.; Boru, G.; Karica, M.; Roberts, V.; Hua, D.; Bishop, B. Monitoring soybean tolerance to flooding stress by image processing technique. In Digital Imaging and Spectral Techniques: Applications to Precision Agriculture and Crop Physiology; ASA Special Publication No 66; VanToai, T.T., Schepers, J., McDonald, M., Tarpley, L., Eds.; The American Society of Agronomy: Madison, WI, USA, 2003; pp. 43–51. [Google Scholar]
- Justin, S.; Armstrong, W. Evidence for the involvement of ethylene in aerenchyma formation in adventitious roots of rice (Oryza sativa). New Phytol. 1991, 118, 49–62. [Google Scholar]
- Parlanti, S.; Kudahettige, N.P.; Lombardi, L.; Mensuali-Sodi, A.; Alpi, A.; Perata, P.; Pucciariello, C. Distinct mechanisms for aerenchyma formation in leaf sheaths of rice genotypes displaying a quiescence or escape strategy for flooding tolerance. Ann. Bot. 2011, 107, 1335–1343. [Google Scholar] [CrossRef] [PubMed]
- He, C.J.; Finlayson, S.A.; Drew, M.C.; Jordan, W.R.; Morgan, P.W. Ethylene biosynthesis during aerenchyma formation in roots of maize subjected to mechanical impedance and hypoxia. Plant Physiol. 1996, 112, 1679–1685. [Google Scholar] [PubMed]
- Gunawardena, A.; Pearce, D.M.; Jackson, M.B.; Hawes, C.R.; Evans, D.E. Characterization of programmed cell death during aerenchyma formation induced by ethylene or hypoxia in roots of maize (Zea mays L.). Planta 2001, 212, 205–214. [Google Scholar] [PubMed]
- Trought, M.C.T.; Drew, M.C. The development of waterlogging damage in young wheat plants in anaerobic solution cultures. J. Exp. Bot. 1980, 31, 1573–1580. [Google Scholar]
- Takeda, K. Varietal variation of flooding tolerance in barley seedlings, and its diallel analysis. Jpn. J. Breed. 1989, 39 (Suppl. 1), 174–175. [Google Scholar]
- Stanca, A.M.; Romagosa, I.; Takeda, K.; Lundborg, T.; Terzi, V.; Cattivelli, L. Diversity in abiotic stress tolerances. In Diversity in Barley (Hordeum vulgare); Von Bothmer, R., van Hintum, T.H., Knüpffer, H., Sato, K., Eds.; Elsevier Science B.V.: Amsterdam, The Netherlands, 2003; pp. 280–291. [Google Scholar]
- Teutsch, C.D.; Sulc, R.M. Influence of seedling growth stage on flooding injury in alfalfa. Agron. J. 1997, 89, 970–975. [Google Scholar]
- Bacanamwo, M.; Purcell, L.C. Soybean root morphological and anatomical traits associated with acclimation to flooding. Crop Sci. 1999, 39, 143–149. [Google Scholar]
- Thomas, A.L.; Guerreiro, S.M.C.; Sodek, L. Aerenchyma formation and recovery from hypoxia of the flooded root system of nodulated soybean. Ann. Bot. 2005, 96, 1191–1198. [Google Scholar] [PubMed]
- Subbaiah, C.C.; Sachs, M.M. Molecular and cellular adaptations of maize to flooding stress. Ann. Bot. 2003, 91, 119–127. [Google Scholar] [PubMed]
- Johns, M.A.; Alleman, M.; Freeling, M. Differential regulation of the Adhl gene in maize: Facts and theories. In Genetic Engineering of Plants; Osuge, T.I., Meredith, C.P., Hollander, A., Eds.; Plenum Press: New York, NY, USA, 1983; pp. 61–79. [Google Scholar]
- Dolferus, R.; Klok, E.J.; Delessert, C.; Wilson, S.; Ismond, K.P.; Good, A.G.; Peacock, W.J.; Dennis, E.S. Enhancing the anaerobic response. Ann. Bot. 2003, 91, 111–117. [Google Scholar] [PubMed]
- Shuai, B.; Reynaga-Peña, C.G.; Springer, P.S. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002, 129, 747–761. [Google Scholar] [PubMed]
- Okushima, Y.; Fukaki, H.; Onoda, M.; Theologis, A.; Tasaka, M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 2007, 19, 118–130. [Google Scholar] [PubMed]
- Lin, Z.; Zhong, S.; Grierson, D. Recent advances in ethylene research. J. Exp. Bot. 2009, 60, 3311–3336. [Google Scholar] [PubMed]
- Hunt, P.W.; Klok, E.J.; Trevaskis, B.; Watts, R.A.; Ellis, M.H.; Peacock, W.J.; Dennis, E.S. Increased level of hemoglobin 1 enhances survival of hypoxic stress and promotes early growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2002, 99, 17197–17202. [Google Scholar] [PubMed]
- Ma, S.; Gong, Q.; Bohnert, H.J. Dissecting salt stress pathways. J. Exp. Bot. 2006, 57, 1097–1107. [Google Scholar] [PubMed]
- Tran, L.S.P.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [PubMed]
- Tran, L.S.; Quach, T.N.; Guttikonda, S.K.; Aldrich, D.L.; Kumar, R.; Neelakandan, A.; Valliyodan, B.; Nguyen, H.T. Molecular characterization of stress-inducible GmNAC genes in soybean. Mol. Genet. Genomics 2009, 281, 647–664. [Google Scholar] [PubMed]
- Liao, Y.; Zhang, J.S.; Chen, S.Y.; Zhang, W.K. Role of soybean GmbZIP132 under abscisic acid and salt stresses. J. Integr. Plant Biol. 2008, 50, 221–230. [Google Scholar]
- Kiba, T.; Naitou, T.; Koizumi, N.; Yamashino, T.; Sakakibara, H.; Mizuno, T. Combinatorial microarray analysis revealing Arabidopsis genes implicated in cytokinin responses through the His-Asp phosphorelay circuitry. Plant Cell Physiol. 2005, 46, 339–355. [Google Scholar] [PubMed]
- Ikeda, M.; Ohme-Takagi, M. A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol. 2009, 50, 970–975. [Google Scholar] [PubMed]
- Chen, Y.Y.X.; He, K.; Liu, M.; Li, J.; Gao, Z.; Lin, Z.; Zhang, Y.; Wang, X.; Qiu, X.; Shen, Y.; et al. The MYB transcription factor superfamily of Arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 2006, 60, 107–124. [Google Scholar] [PubMed]
- Fehr, W.R.; Caviness, C.E. Stages of Soybean Development; Cooperation Extension Service Special Report 80; Iowa State University: Ames, IA, USA, 1977. [Google Scholar]
- Johansen, D.A. Plant Microtechnique, 3rd ed.; Paul B. Hoeber, Inc.: New York, NY, USA, 1940; p. 790. [Google Scholar]
- Bukastsh, F. Bemerkungen zur Doppelfärbung astrablau-safrarin. Microkosmos 1972, 61, 255. [Google Scholar]
- Wang, Z.; Libault, M.; Joshi, T.; Valliyodan, B.; Nguyen, H.T.; Xu, D.; Stacey, G.; Cheng, J. SoyDB: A knowledge database of soybean transcription factors. BMC Plant Biol. 2010, 10. [Google Scholar] [CrossRef] [PubMed]
- Ramakers, C.; Ruijter, J.M.; LekAnnalse Deprez, R.H.; Moorman, A.F.M. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [PubMed]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.; Moorman, A.F. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Snesrud, E.C.; Haas, B.; Cheung, F.; Town, C.D.; Quackenbush, J. Gene expression analyses of Arabidopsis chromosome 2 using a genomic DNA amplicon microarray. Genome Res. 2003, 13, 327–340. [Google Scholar] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valliyodan, B.; Van Toai, T.T.; Alves, J.D.; De Fátima P. Goulart, P.; Lee, J.D.; Fritschi, F.B.; Rahman, M.A.; Islam, R.; Shannon, J.G.; Nguyen, H.T. Expression of Root-Related Transcription Factors Associated with Flooding Tolerance of Soybean (Glycine max). Int. J. Mol. Sci. 2014, 15, 17622-17643. https://doi.org/10.3390/ijms151017622
Valliyodan B, Van Toai TT, Alves JD, De Fátima P. Goulart P, Lee JD, Fritschi FB, Rahman MA, Islam R, Shannon JG, Nguyen HT. Expression of Root-Related Transcription Factors Associated with Flooding Tolerance of Soybean (Glycine max). International Journal of Molecular Sciences. 2014; 15(10):17622-17643. https://doi.org/10.3390/ijms151017622
Chicago/Turabian StyleValliyodan, Babu, Tara T. Van Toai, Jose Donizeti Alves, Patricia De Fátima P. Goulart, Jeong Dong Lee, Felix B. Fritschi, Mohammed Atiqur Rahman, Rafiq Islam, J. Grover Shannon, and Henry T. Nguyen. 2014. "Expression of Root-Related Transcription Factors Associated with Flooding Tolerance of Soybean (Glycine max)" International Journal of Molecular Sciences 15, no. 10: 17622-17643. https://doi.org/10.3390/ijms151017622
APA StyleValliyodan, B., Van Toai, T. T., Alves, J. D., De Fátima P. Goulart, P., Lee, J. D., Fritschi, F. B., Rahman, M. A., Islam, R., Shannon, J. G., & Nguyen, H. T. (2014). Expression of Root-Related Transcription Factors Associated with Flooding Tolerance of Soybean (Glycine max). International Journal of Molecular Sciences, 15(10), 17622-17643. https://doi.org/10.3390/ijms151017622






