Association of CDKN2BAS Polymorphism rs4977574 with Coronary Heart Disease: A Case-Control Study and a Meta-Analysis
Abstract
:1. Introduction
2. Results
| Characteristics | Case (590) | Control (482) | p Value a |
|---|---|---|---|
| Age (years mean ± SD) | 61.73 ± 7.83 | 58.17 ± 8.79 | 0.0001 |
| Sex (male) | 418 | 254 | 0.003 |
| Smoking (n) | 144 | 81 | 0.018 |
| Hypertension (n) | 172 | 114 | 0.152 |
| Diabetes (n) | 58 | 33 | 0.124 |
| Family history (n) | 28 | 16 | 0.281 |
| TG (mmol/L) | 2.23 ± 1.02 | 2.28 ± 1.02 | 0.441 |
| TC (mmol/L) | 4.37 ± 1.08 | 4.31 ± 0.98 | 0.425 |
| HDL-C (mmol/L) | 1.07 ± 0.25 | 1.12 ± 0.26 | 0.001 |
| LDL-C (mmol/L) | 1.95 ± 1.15 | 1.75 ± 0.97 | 0.003 |
| Gender | Group | Genotype (Counts) | χ2 | p (df = 2) a | HWE | Allele (Counts) | χ2 | p (df = 1) | OR (95% CI) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | A | G | ||||||||
| All | Case (n = 590) | 122 | 305 | 163 | 0.36 | 547 | 631 | |||||
| Control (n = 482) | 138 | 267 | 77 | 23.41 | <0.0001 | 0.007 | 543 | 423 | 20.30 | <0.0001 | 1.48 (1.25–1.75) | |
| Male | Case (n = 418) | 86 | 220 | 112 | 0.28 | 392 | 444 | |||||
| Control (n = 254) | 70 | 144 | 40 | 12.32 | 0.014 | 0.02 | 284 | 224 | 10.27 | 0.001 | 1.44 (1.15–1.79) | |
| Female | Case (n = 172) | 36 | 85 | 51 | 1.00 | 157 | 187 | |||||
| Control (n = 228) | 68 | 123 | 37 | 11.4 | 0.002 | 0.18 | 259 | 197 | 9.78 | 0.002 | 1.57 (1.18–2.08) | |
| Gender | Group | Dominant | χ2 | p (df = 2) | OR (95% CI) | Recessive | χ2 | p (df = 1) | OR (95% CI) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | GG + GA | GA + AA | GG | ||||||||
| Female | Case | 36 | 136 | 121 | 51 | ||||||
| Control | 68 | 160 | 4.03 | 0.051 | 1.59 (0.89–2.62) | 191 | 37 | 10.29 | 0.003 | 2.14 (1.31–2.77) | |
| Gender | Age | Group | Genotype (Counts) | χ2 | p (df = 2) a | HWE | Allele (Counts) | χ2 | p (df = 1) | OR (95% CI) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs4977574 | AA | GA | GG | A | G | ||||||||
| Male | <65 | Case (n = 254) | 50 | 136 | 68 | 0.26 | 236 | 272 | 1.45 (1.11–1.90) | ||||
| Control (n = 191) | 53 | 107 | 31 | 8.63 | 0.04 | 0.08 | 213 | 169 | 7.55 | 0.006 | |||
| ≥65 | Case (n = 161) | 36 | 84 | 41 | 0.64 | 156 | 166 | 1.35 (0.88–2.05) | |||||
| Control (n = 60) | 15 | 37 | 8 | 3.75 | 0.30 | 0.07 | 67 | 53 | 1.91 | 0.16 | |||
| Female | <65 | Case (n = 89) | 18 | 39 | 32 | 0.39 | 75 | 103 | 1.87 (1.30–2.70) | ||||
| Control (n = 169) | 52 | 91 | 26 | 14.64 | 0.0002 | 0.20 | 195 | 143 | 11.31 | 0.0008 | |||
| ≥65 | Case (n = 81) | 18 | 46 | 17 | 0.27 | 82 | 80 | 1.16 (0.72–1.88) | |||||
| Control (n = 57) | 15 | 32 | 10 | 0.44 | 0.63 | 0.42 | 62 | 52 | 0.38 | 0.54 | |||

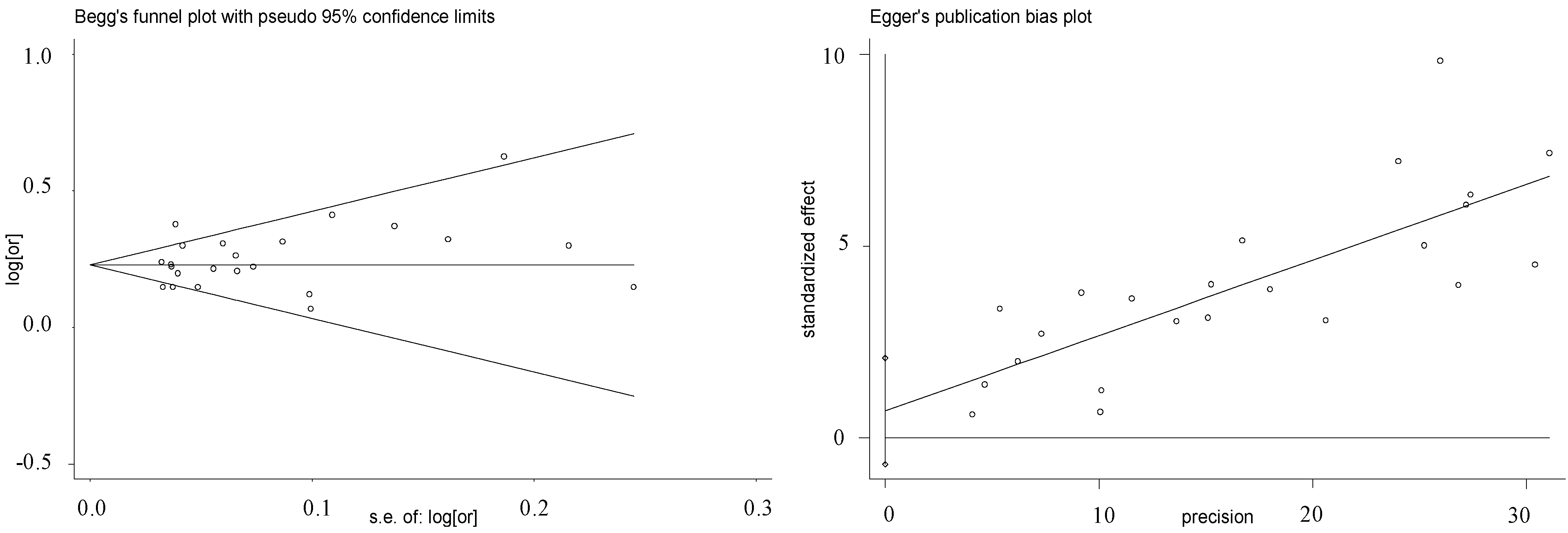
| Ethnic Group | Studies/Stages (n) | OR (95% CI) | Weight (%) | Z | p(z) | I2 | P | τ2 | Heterogeneity Statistic |
|---|---|---|---|---|---|---|---|---|---|
| Caucasians | 16 | 1.28 (1.23–1.32) | 80.72 | 13.11 | <0.0001 | 48.6% | 0.015 | 0.0024 | 29.17 |
| Asians | 7 | 1.23 (1.13–1.34) | 19.28 | 4.74 | <0.0001 | 41.0% | 0.118 | 0.0040 | 10.16 |
| Overall | 23 | 1.27 (1.22–1.31) | 100.00 | 12.85 | <0.0001 | 54.2% | 0.001 | 0.0034 | 48.01 |
3. Discussion
4. Experimental Section
4.1. Sample Collection
4.2. Biochemical Analysis
4.3. SNP Genotyping
4.4. Retrieval of Published Studies and Selection of Studies for the Meta-Analysis
4.5. Statistical Analyses
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Owan, T.E.; Roe, M.T.; Messenger, J.C.; Dai, D.; Michaels, A.D. Contemporary use of adjunctive thrombectomy during primary percutaneous coronary intervention for ST-elevation myocardial infarction in the United States. Catheter. Cardiovasc. Interv. 2012, 80, 1173–1180. [Google Scholar]
- Chien, K.L.; Hsu, H.C.; Su, T.C.; Chang, W.T.; Chen, P.C.; Sung, F.C.; Lin, H.J.; Chen, M.F.; Lee, Y.T. Constructing a point-based prediction model for the risk of coronary artery disease in a Chinese community: A report from a cohort study in Taiwan. Int. J. Cardiol. 2012, 157, 263–268. [Google Scholar]
- Tada, N.; Maruyama, C.; Koba, S.; Tanaka, H.; Birou, S.; Teramoto, T.; Sasaki, J. Japanese dietary lifestyle and cardiovascular disease. J. Atheroscler. Thromb. 2011, 18, 723–734. [Google Scholar]
- Kangas-Kontio, T.; Huotari, A.; Ruotsalainen, H.; Herzig, K.H.; Tamminen, M.; Ala-Korpela, M.; Savolainen, M.J.; Kakko, S. Genetic and environmental determinants of total and high-molecular weight adiponectin in families with low HDL-cholesterol and early onset coronary heart disease. Atherosclerosis 2010, 210, 479–485. [Google Scholar]
- Madjid, M.; Willerson, J.T. Inflammatory markers in coronary heart disease. Br. Med. Bull. 2010, 100, 23–38. [Google Scholar]
- Khurana, R.; Simons, M.; Martin, J.F.; Zachary, I.C. Role of angiogenesis in cardiovascular disease: A critical appraisal. Circulation 2005, 112, 1813–1824. [Google Scholar]
- Emanueli, C.; Madeddu, P. Angiogenesis gene therapy to rescue ischaemic tissues: Achievements and future directions. Br. J. Pharmacol. 2001, 133, 951–958. [Google Scholar]
- Jarinova, O.; Stewart, A.F.; Roberts, R.; Wells, G.; Lau, P.; Naing, T.; Buerki, C.; McLean, B.W.; Cook, R.C.; Parker, J.S.; et al. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler. Thromb. Vasc. Biol. 2009, 29, 1671–1677. [Google Scholar]
- Liu, Y.; Sanoff, H.K.; Cho, H.; Burd, C.E.; Torrice, C.; Mohlke, K.L.; Ibrahim, J.G.; Thomas, N.E.; Sharpless, N.E. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS One 2009, 4, e5027. [Google Scholar]
- Broadbent, H.M.; Peden, J.F.; Lorkowski, S.; Goel, A.; Ongen, H.; Green, F.; Clarke, R.; Collins, R.; Franzosi, M.G.; Tognoni, G.; et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum. Mol. Genet. 2008, 17, 806–814. [Google Scholar]
- Liu, Y.; Sanoff, H.K.; Cho, H.; Burd, C.E.; Torrice, C.; Ibrahim, J.G.; Thomas, N.E.; Sharpless, N.E. Expression of p16(INK4a) in peripheral blood T–cells is a biomarker of human aging. Aging Cell 2009, 8, 439–448. [Google Scholar]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar]
- Pasmant, E.; Sabbagh, A.; Vidaud, M.; Bieche, I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011, 25, 444–448. [Google Scholar]
- Holdt, L.M.; Beutner, F.; Scholz, M.; Gielen, S.; Gabel, G.; Bergert, H.; Schuler, G.; Thiery, J.; Teupser, D. ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 620–627. [Google Scholar]
- Cunnington, M.S.; Santibanez Koref, M.; Mayosi, B.M.; Burn, J.; Keavney, B. Chromosome 9p21 SNPs associated with multiple disease phenotypes correlate with ANRIL expression. PLoS Genet. 2010, 6, e1000899. [Google Scholar]
- Psaty, B.M.; O’Donnell, C.J.; Gudnason, V.; Lunetta, K.L.; Folsom, A.R.; Rotter, J.I.; Uitterlinden, A.G.; Harris, T.B.; Witteman, J.C.; Boerwinkle, E.; et al. Cohorts for heart and aging research in genomic epidemiology (CHARGE) consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ. Cardiovasc. Genet. 2009, 2, 73–80. [Google Scholar]
- McPherson, R.; Pertsemlidis, A.; Kavaslar, N.; Stewart, A.; Roberts, R.; Cox, D.R.; Hinds, D.A.; Pennacchio, L.A.; Tybjaerg-Hansen, A.; Folsom, A.R.; et al. A common allele on chromosome 9 associated with coronary heart disease. Science 2007, 316, 1488–1491. [Google Scholar]
- Lotta, L.A. Genome-wide association studies in atherothrombosis. Eur. J. Int. Med. 2010, 21, 74–78. [Google Scholar]
- Burton, P.R.; Clayton, D.G.; Cardon, L.R.; Craddock, N.; Deloukas, P.; Duncanson, A.; Kwiatkowski, D.P.; McCarthy, M.I.; Ouwehand, W.H.; Samani, N.J.; et al. Genome-wide association study of 14,000 cases of seven common diseases and 3000 shared controls. Nature 2007, 447, 661–678. [Google Scholar]
- Zeggini, E.; Weedon, M.N.; Lindgren, C.M.; Frayling, T.M.; Elliott, K.S.; Lango, H.; Timpson, N.J.; Perry, J.R.; Rayner, N.W.; Freathy, R.M.; et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 2007, 316, 1336–1341. [Google Scholar]
- Scott, L.J.; Mohlke, K.L.; Bonnycastle, L.L.; Willer, C.J.; Li, Y.; Duren, W.L.; Erdos, M.R.; Stringham, H.M.; Chines, P.S.; Jackson, A.U.; et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 2007, 316, 1341–1345. [Google Scholar]
- Matarin, M.; Brown, W.M.; Singleton, A.; Hardy, J.A.; Meschia, J.F. Whole genome analyses suggest ischemic stroke and heart disease share an association with polymorphisms on chromosome 9p21. Stroke 2008, 39, 1586–1589. [Google Scholar]
- Schaefer, A.S.; Richter, G.M.; Dommisch, H.; Reinartz, M.; Nothnagel, M.; Noack, B.; Laine, M.L.; Folwaczny, M.; Groessner-Schreiber, B.; Loos, B.G.; et al. CDKN2BAS is associated with periodontitis in different European populations and is activated by bacterial infection. J. Med. Genet. 2011, 48, 38–47. [Google Scholar]
- Zhang, W.; Chen, Y.; Liu, P.; Chen, J.; Song, L.; Tang, Y.; Wang, Y.; Liu, J.; Hu, F.B.; Hui, R. Variants on chromosome 9p21.3 correlated with ANRIL expression contribute to stroke risk and recurrence in a large prospective stroke population. Stroke 2012, 43, 14–21. [Google Scholar]
- Helgadottir, A.; Thorleifsson, G.; Manolescu, A.; Gretarsdottir, S.; Blondal, T.; Jonasdottir, A.; Jonasdottir, A.; Sigurdsson, A.; Baker, A.; Palsson, A. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 2007, 316, 1491–1493. [Google Scholar]
- Samani, N.J.; Erdmann, J.; Hall, A.S.; Hengstenberg, C.; Mangino, M.; Mayer, B.; Dixon, R.J.; Meitinger, T.; Braund, P.; Wichmann, H.-E.; et al. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 2007, 357, 443–453. [Google Scholar]
- Qi, L.; Ma, J.; Qi, Q.; Hartiala, J.; Allayee, H.; Campos, H. Genetic risk score and risk of myocardial infarction in Hispanics. Circulation 2011, 123, 374–380. [Google Scholar]
- Wen, J.; Ronn, T.; Olsson, A.; Yang, Z.; Lu, B.; Du, Y.; Groop, L.; Ling, C.; Hu, R. Investigation of type 2 diabetes risk alleles support CDKN2A/B, CDKAL1, and TCF7L2 as susceptibility genes in a Han Chinese cohort. PLoS One 2010, 5, e9153. [Google Scholar]
- Lin, Y.; Li, P.; Cai, L.; Zhang, B.; Tang, X.; Zhang, X.; Li, Y.; Xian, Y.; Yang, Y.; Wang, L.; et al. Association study of genetic variants in eight genes/loci with type 2 diabetes in a Han Chinese population. BMC Med. Genet. 2010, 11, 97. [Google Scholar] [CrossRef]
- Hu, W.L.; Li, S.J.; Liu, D.T.; Wang, Y.; Niu, S.Q.; Yang, X.C.; Zhang, Q.; Yu, S.Z.; Jin, L.; Wang, X.F. Genetic variants on chromosome 9p21 and ischemic stroke in Chinese. Brain Res. Bull. 2009, 79, 431–435. [Google Scholar]
- Yang, X.C.; Zhang, Q.; Chen, M.L.; Li, Q.; Yang, Z.S.; Li, L.; Cao, F.F.; Chen, X.D.; Liu, W.J.; Jin, L.; et al. MTAP and CDKN2B genes are associated with myocardial infarction in Chinese Hans. Clin. Biochem. 2009, 42, 1071–1075. [Google Scholar]
- Erdmann, J.; Grosshennig, A.; Braund, P.S.; Konig, I.R.; Hengstenberg, C.; Hall, A.S.; Linsel-Nitschke, P.; Kathiresan, S.; Wright, B.; Tregouet, D.-A.; et al. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat. Genet. 2009, 41, 280–282. [Google Scholar]
- Kathiresan, S.; Voight, B.F.; Purcell, S.; Musunuru, K.; Ardissino, D.; Mannucci, P.M.; Anand, S.; Engert, J.C.; Samani, N.J.; Schunkert, H.; et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Gene. 2009, 41, 334–341. [Google Scholar]
- Davies, R.W.; Dandona, S.; Stewart, A.F.; Chen, L.; Ellis, S.G.; Tang, W.H.; Hazen, S.L.; Roberts, R.; McPherson, R.; Wells, G.A. Improved prediction of cardiovascular disease based on a panel of single nucleotide polymorphisms identified through genome-wide association studies. Circ. Cardiovasc. Genet. 2010, 3, 468–474. [Google Scholar]
- Qi, L.; Parast, L.; Cai, T.; Powers, C.; Gervino, E.V.; Hauser, T.H.; Hu, F.B.; Doria, A. Genetic susceptibility to coronary heart disease in type 2 diabetes: 3 independent studies. J. Am. Coll. Cardiol. 2011, 58, 2675–2682. [Google Scholar]
- Peden, J.F.; Hopewell, J.C. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat. Genet. 2011, 43, 339–344. [Google Scholar]
- Erdmann, J.; Willenborg, C.; Nahrstaedt, J.; Preuss, M.; Konig, I.R.; Baumert, J.; Linsel-Nitschke, P.; Gieger, C.; Tennstedt, S.; Belcredi, P.; et al. Genome-wide association study identifies a new locus for coronary artery disease on chromosome 10p11.23. Eur. Heart J. 2011, 32, 158–168. [Google Scholar]
- Saade, S.; Cazier, J.B.; Ghassibe-Sabbagh, M.; Youhanna, S.; Badro, D.A.; Kamatani, Y.; Hager, J.; Yeretzian, J.S.; El-Khazen, G.; Haber, M.; et al. Large scale association analysis identifies three susceptibility loci for coronary artery disease. PLoS One 2011, 6, e29427. [Google Scholar]
- Burd, C.E.; Jeck, W.R.; Liu, Y.; Sanoff, H.K.; Wang, Z.; Sharpless, N.E. Expression of linear and novel circular forms of an INK4/ARF–associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010, 6, e1001233. [Google Scholar]
- Harismendy, O.; Notani, D.; Song, X.; Rahim, N.G.; Tanasa, B.; Heintzman, N.; Ren, B.; Fu, X.D.; Topol, E.J.; Rosenfeld, M.G.; et al. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response. Nature 2011, 470, 264–268. [Google Scholar]
- Hiramoto, J.S.; Katz, R.; Ix, J.H.; Wassel, C.; Rodondi, N.; Windham, B.G.; Harris, T.; Koster, A.; Satterfield, S.; Newman, A.; et al. Sex differences in the prevalence and clinical outcomes of subclinical peripheral artery disease in the health, aging, and body composition (Health ABC) study. Vascular 2014, 22, 142–148. [Google Scholar]
- Ober, C.; Loisel, D.A.; Gilad, Y. Sex-specific genetic architecture of human disease. Nat. Rev. Genet. 2008, 9, 911–922. [Google Scholar]
- Safford, M.M.; Brown, T.M.; Muntner, P.M.; Durant, R.W.; Glasser, S.; Halanych, J.H.; Shikany, J.M.; Prineas, R.J.; Samdarshi, T.; Bittner, V.A.; et al. Association of race and sex with risk of incident acute coronary heart disease events. JAMA 2012, 308, 1768–1774. [Google Scholar]
- Silander, K.; Alanne, M.; Kristiansson, K.; Saarela, O.; Ripatti, S.; Auro, K.; Karvanen, J.; Kulathinal, S.; Niemela, M.; Ellonen, P.; et al. Gender differences in genetic risk profiles for cardiovascular disease. PLoS One 2008, 3, e3615. [Google Scholar]
- Zhou, J.; Huang, Y.; Huang, R.S.; Wang, F.; Xu, L.; Le, Y.; Yang, X.; Xu, W.; Huang, X.; Lian, J.; et al. A case-control study provides evidence of association for a common SNP rs974819 in PDGFD to coronary heart disease and suggests a sex-dependent effect. Thromb. Res. 2012, 130, 602–606. [Google Scholar]
- Huang, Y.; Lian, J.; Huang, R.S.; Wang, F.; Xu, L.; Le, Y.; Yang, X.; Xu, W.; Huang, X.; Ye, M.; et al. Positive association between rs10918859 of the NOS1AP gene and coronary heart disease in male Han Chinese. Genet. Test. Mol. Biomark. 2013, 17, 25–29. [Google Scholar]
- Peng, P.; Lian, J.; Huang, R.S.; Xu, L.; Huang, Y.; Ba, Y.; Yang, X.; Huang, X.; Dong, C.; Zhang, L.; et al. Meta-analyses of KIF6 Trp719Arg in coronary heart disease and statin therapeutic effect. PLoS One 2012, 7, e50126. [Google Scholar]
- Lian, J.; Huang, Y.; Huang, R.S.; Xu, L.; Le, Y.; Yang, X.; Xu, W.; Huang, X.; Ye, M.; Zhou, J.; et al. Meta-analyses of four eosinophil related gene variants in coronary heart disease. J. Thromb. Thrombolysis 2013, 36, 394–401. [Google Scholar]
- Zhang, L.; Yuan, F.; Liu, P.; Fei, L.; Huang, Y.; Xu, L.; Hao, L.; Qiu, X.; Le, Y.; Yang, X.; et al. Association between PCSK9 and LDLR gene polymorphisms with coronary heart disease: Case-control study and meta-analysis. Clin. Biochem. 2013, 46, 727–732. [Google Scholar]
- Xu, L.; Zhou, J.; Huang, S.; Huang, Y.; Le, Y.; Jiang, D.; Wang, F.; Yang, X.; Xu, W.; Huang, X.; et al. An association study between genetic polymorphisms related to lipoprotein-associated phospholipase A2 and coronary heart disease. Exp. Ther. Med. 2013, 5, 742–750. [Google Scholar]
- Zhou, L.T.; Qin, L.; Zheng, D.C.; Song, Z.K.; Ye, L. Meta-analysis of genetic association of chromosome 9p21 with early-onset coronary artery disease. Gene 2012, 510, 185–188. [Google Scholar]
- Nora, J.J.; Lortscher, R.H.; Spangler, R.D.; Nora, A.H.; Kimberling, W.J. Genetic–epidemiologic study of early-onset ischemic heart disease. Circulation 1980, 61, 503–508. [Google Scholar]
- Ichihara, S.; Yamamoto, K.; Asano, H.; Nakatochi, M.; Sukegawa, M.; Ichihara, G.; Izawa, H.; Hirashiki, A.; Takatsu, F.; Umeda, H.; et al. Identification of a glutamic acid repeat polymorphism of ALMS1 as a novel genetic risk marker for early-onset myocardial infarction by genome-wide linkage analysis. Circ. Cardiovasc. Genet. 2013, 6, 569–578. [Google Scholar] [CrossRef]
- Shen, G.Q.; Girelli, D.; Li, L.; Rao, S.; Archacki, S.; Olivieri, O.; Martinelli, N.; Park, J.E.; Chen, Q.; Topol, E.J.; et al. A novel molecular diagnostic marker for familial and early-onset coronary artery disease and myocardial infarction in the LRP8 Gene. Circ. Cardiovasc. Genet. 2014, 7, 514–520. [Google Scholar]
- Rac, M.; Kurzawski, G.; Safranow, K.; Sagasz-Tysiewicz, D.; Krzystolik, A.; Poncyljusz, W.; Olszewska, M.; Dawid, G.; Chlubek, D. Association of CD36 gene polymorphisms with echo- and electrocardiographic parameters in patients with early onset coronary artery disease. Arch. Med. Sci. 2013, 9, 640–650. [Google Scholar]
- Anderson, C.D.; Rosand, J. Genome-wide linkage approach yields novel early onset myocardial infarction locus in East Asians. Circ. Cardiovasc. Genet. 2013, 6, 531–532. [Google Scholar]
- Beckie, T.M.; Groer, M.W.; Beckstead, J.W. The relationship between polymorphisms on chromosome 9p21 and age of onset of coronary heart disease in black and white women. Genet. Test. Mol. Biomark. 2011, 15, 435–442. [Google Scholar]
- Lin, H.F.; Tsai, P.C.; Liao, Y.C.; Lin, T.H.; Tai, C.T.; Juo, S.H.; Lin, R.T. Chromosome 9p21 genetic variants are associated with myocardial infarction but not with ischemic stroke in a Taiwanese population. J. Investig. Med. 2011, 59, 926–930. [Google Scholar]
- Chen, Z.; Qian, Q.; Ma, G.; Wang, J.; Zhang, X.; Feng, Y.; Shen, C.; Yao, Y. A common variant on chromosome 9p21 affects the risk of early-onset coronary artery disease. Mol. Biol. Rep. 2009, 36, 889–893. [Google Scholar]
- Assimes, T.L.; Knowles, J.W.; Basu, A.; Iribarren, C.; Southwick, A.; Tang, H.; Absher, D.; Li, J.; Fair, J.M.; Rubin, G.D.; et al. Susceptibility locus for clinical and subclinical coronary artery disease at chromosome 9p21 in the multi-ethnic ADVANCE study. Hum. Mol. Genet. 2008, 17, 2320–2328. [Google Scholar]
- Lewandowski, M.; Szwed, H.; Kowalik, I. Searching for the optimal strategy for the diagnosis of stable coronary artery disease. Cost-effectiveness of the new algorithm. Cardiol. J. 2007, 14, 544–551. [Google Scholar]
- Higgs, Z.C.; Macafee, D.A.; Braithwaite, B.D.; Maxwell-Armbold, C.A. The Seldinger technique: 50 years on. Lancet 2005, 366, 1407–1409. [Google Scholar]
- Gabriel, S.; Ziaugra, L.; Tabbaa, D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr. Protoc. Hum. Genet. 2009, 2, 1–18. [Google Scholar]
- Huang, Y.; Yu, X.; Wang, L.; Zhou, S.; Sun, J.; Feng, N.; Nie, S.; Wu, J.; Gao, F.; Fei, B.; et al. Four genetic polymorphisms of lymphotoxin-α gene and cancer risk: A systematic review and meta-analysis. PLoS One 2013, 8, e82519. [Google Scholar]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Sham, P.C.; Curtis, D. Monte Carlo tests for associations between disease and alleles at highly polymorphic loci. Ann. Hum. Genet. 1995, 59, 97–105. [Google Scholar]
- Yu, J.; Huang, J.; Liang, Y.; Qin, B.; He, S.; Xiao, J.; Wang, H.; Zhong, R. Lack of association between apolipoprotein C3 gene polymorphisms and risk of coronary heart disease in a Han population in East China. Lipids Health Dis. 2011, 10, 200. [Google Scholar]
- Dupont, W.D.; Plummer, W.D., Jr. Power and sample size calculations. A review and computer program. Control. Clin. Trials 1990, 11, 116–128. [Google Scholar]
- Stroup, D.F.; Berlin, J.A.; Morton, S.C.; Olkin, I.; Williamson, G.D.; Rennie, D.; Moher, D.; Becker, B.J.; Sipe, T.A.; Tracker, S.B.; et al. Meta-analysis of observational studies in epidemiology: A proposal for reporting. JAMA 2000, 283, 2008–2012. [Google Scholar]
- Egger, M.; Davey Smith, G.; Schneider, M.; Minder, C. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997, 315, 629–634. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Ye, H.; Hong, Q.; Xu, X.; Jiang, D.; Xu, L.; Dai, D.; Sun, J.; Gao, X.; Duan, S. Association of CDKN2BAS Polymorphism rs4977574 with Coronary Heart Disease: A Case-Control Study and a Meta-Analysis. Int. J. Mol. Sci. 2014, 15, 17478-17492. https://doi.org/10.3390/ijms151017478
Huang Y, Ye H, Hong Q, Xu X, Jiang D, Xu L, Dai D, Sun J, Gao X, Duan S. Association of CDKN2BAS Polymorphism rs4977574 with Coronary Heart Disease: A Case-Control Study and a Meta-Analysis. International Journal of Molecular Sciences. 2014; 15(10):17478-17492. https://doi.org/10.3390/ijms151017478
Chicago/Turabian StyleHuang, Yi, Huadan Ye, Qingxiao Hong, Xuting Xu, Danjie Jiang, Limin Xu, Dongjun Dai, Jie Sun, Xiang Gao, and Shiwei Duan. 2014. "Association of CDKN2BAS Polymorphism rs4977574 with Coronary Heart Disease: A Case-Control Study and a Meta-Analysis" International Journal of Molecular Sciences 15, no. 10: 17478-17492. https://doi.org/10.3390/ijms151017478
APA StyleHuang, Y., Ye, H., Hong, Q., Xu, X., Jiang, D., Xu, L., Dai, D., Sun, J., Gao, X., & Duan, S. (2014). Association of CDKN2BAS Polymorphism rs4977574 with Coronary Heart Disease: A Case-Control Study and a Meta-Analysis. International Journal of Molecular Sciences, 15(10), 17478-17492. https://doi.org/10.3390/ijms151017478





