Resistin Stimulates Expression of Chemokine Genes in Chondrocytes via Combinatorial Regulation of C/EBPβ and NF-κB
Abstract
:1. Introduction
2. Results and Discussion
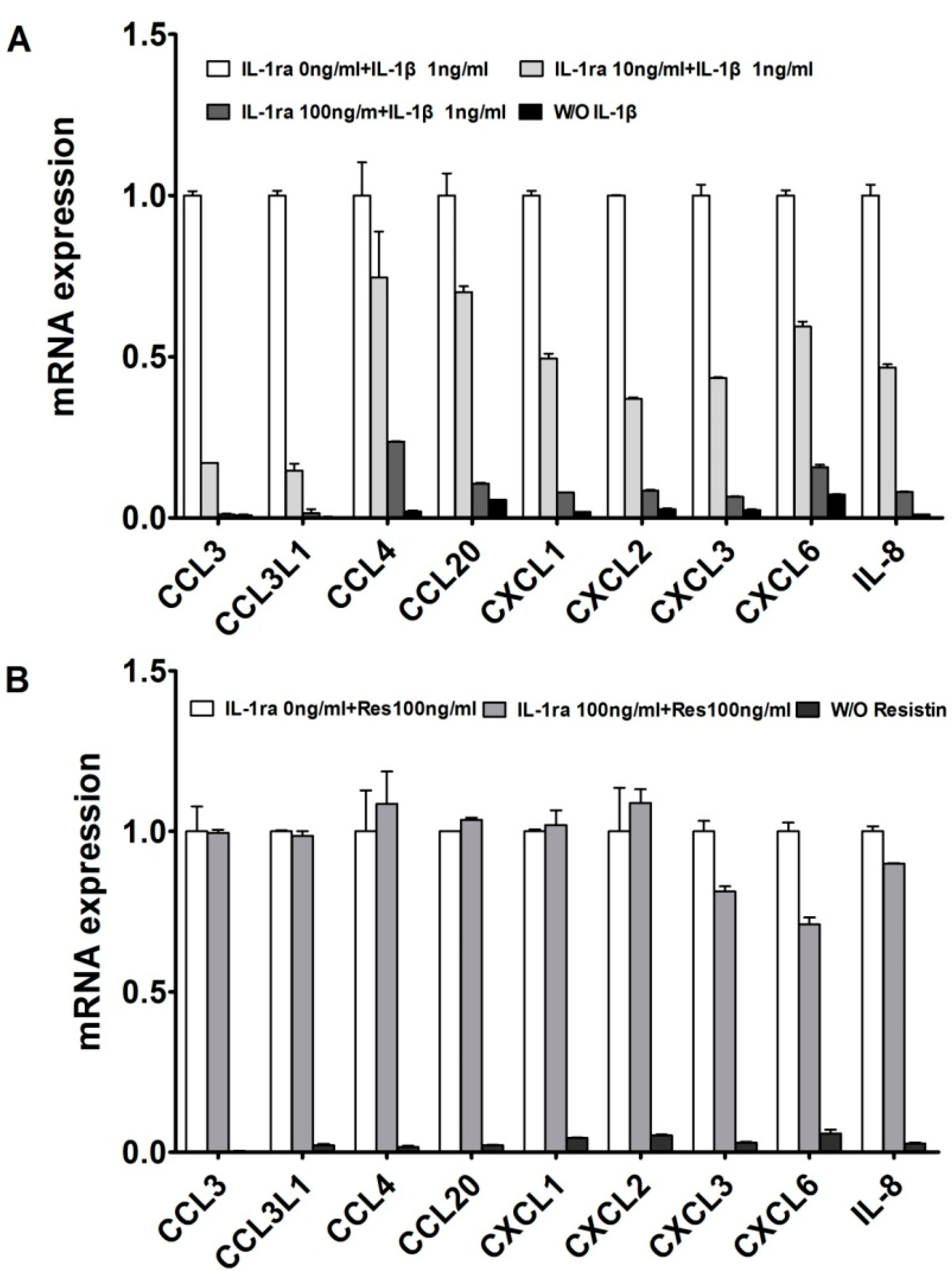
2.1. Resistin Independently Stimulates the Expression of Chemokine Genes in Normal Chondrocytes
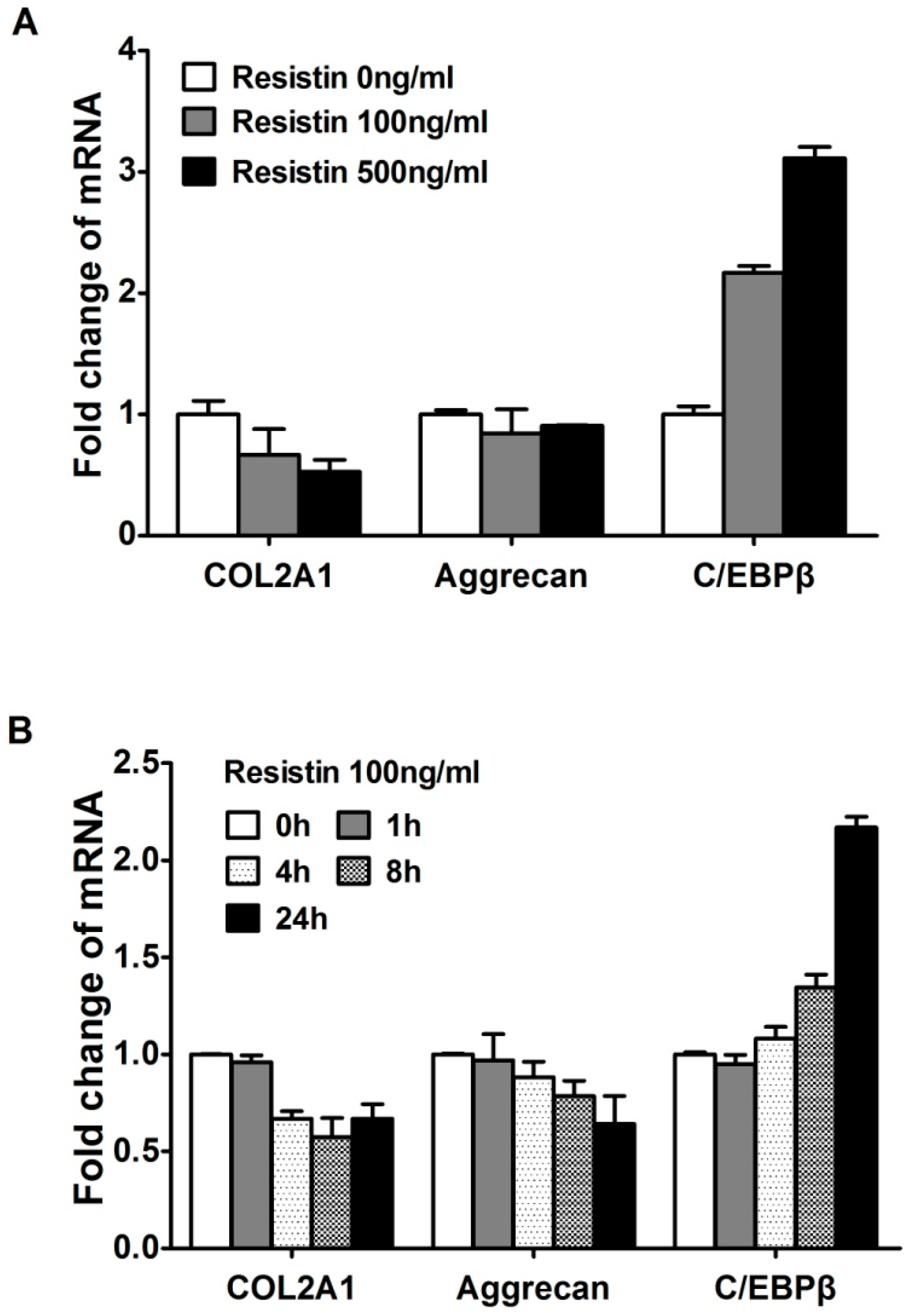
2.2. Resistin Stimulates the Expression of C/EBPβ in Chondrocytes
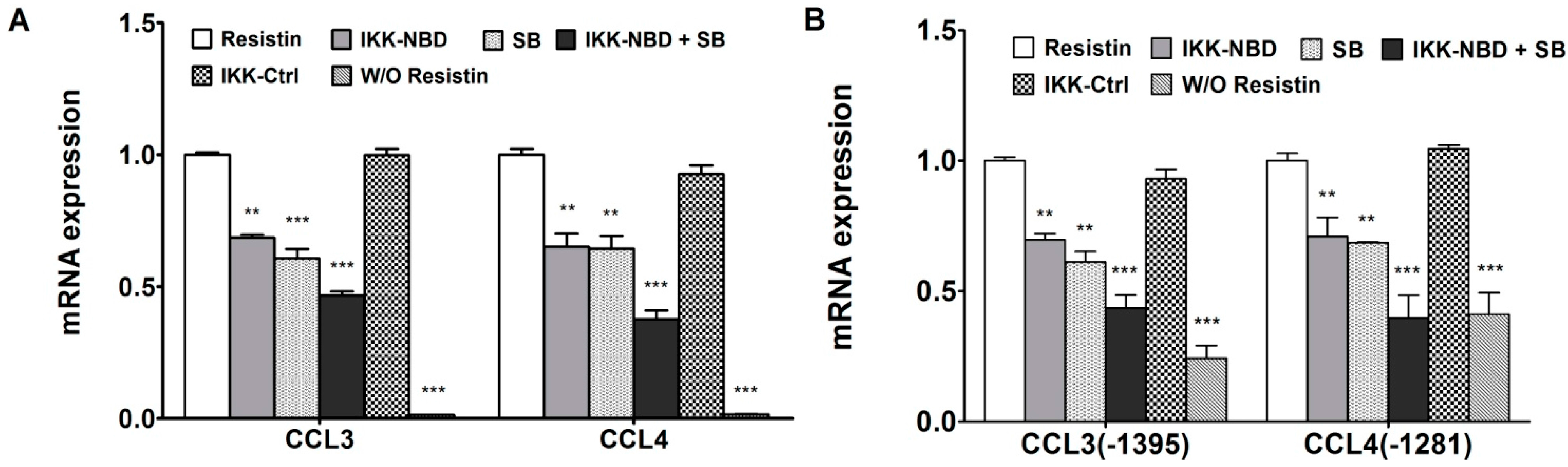
2.3. NF-κB Is Involved in the Up-Regulation of Chemokine Genes by Chondrocytes in Response to Resistin

2.4. C/EBPβ and NF-κB Co-Enhance the Expression of CCL3 and CCL4 in Human Chondrocytes in Response to Resistin
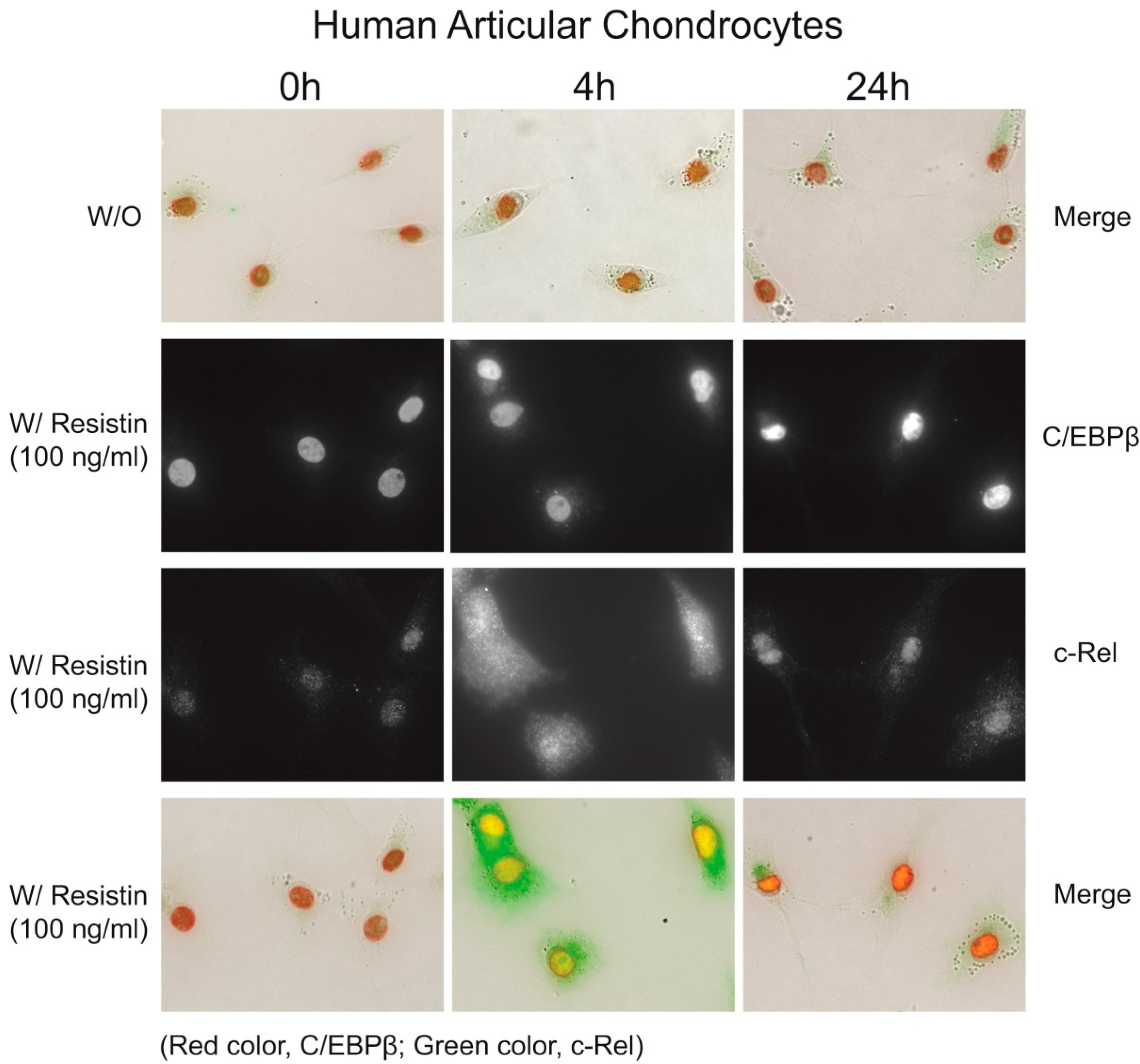
2.5. Immunohistochemistry of C/EBPβ and NF-κB
2.6. Discussion
3. Experimental Section
3.1. Materials
3.2. Cell Culture
3.3. RNA Isolation and Real Time Quantitative PCR
| Gene | Primer Sequence |
|---|---|
| GAPDH | F(5'-ACCCAGAAGACTGTGGATGG-3'), R(5'-GAGGCAGGGATGATGTTCTG-3') |
| COL2A1 | F(5'-CCCAGAGGTGACAAAGGAGA-3'), R(5'-CACCTTGGTCTCCAGAAGGA-3') |
| Aggrecan | F(5'-GGCACTAGTCAACCCTTTGG-3'), R(5'-CTGAACCCTGGTAACCCTGA-3') |
| C/EBPβ | F(5'-CTCGCAGGTCAAGAGCAAGG-3'), R(5'-TCGTCGCTGTGCTTGTCC-3') |
| CCL3 (MIP-1α) | F(5'-GCAACCAGTTCTCTGCATCA-3'), R(5'-TGGCTGCTCGTCTCAAAGTA-3') |
| CCL3L1 (LD78β) | F(5'-GTCCTCTCTGCACCACTTGC-3'), R(5'-GGAAGATGACACTGGGCTTG-3') |
| CCL4 (MIP-1β) | F(5'-GCTTTTCTTACACTGCGAGGA-3'), R(5'-CCAGGATTCACTGGGATCAG-3') |
| CCL20 (MIP-3α) | F(5'-GCTTTTCTTACACTGCGAGGA-3'), R(5'-CCAGGATTCACTGGGATCAG-3') |
| CXCL1 (GRO-α) | F(5'-GGGAATTCACCCCAAGAAC-3'), R(5'-GATGCAGGATTGAGGCAAG-3') |
| CXCL2 (GRO-β) | F(5'-TGCAGGGAATTCACCTCAAG-3'), R(5'-TCTTAACCATGGGCGATGC-3') |
| CXCL3 (GRO-γ) | F(5'-ACCGAAGTCATAGCCACACTC-3'), R(5'-GGTGCTCCCCTTGTTCAGTA-3') |
| CXCL6 (GCP-2) | F(5'-GTTTACGCGTTACGCTGAGAG-3'), R(5'-ACTTCCACCTTGGAGCACTG-3') |
| IL-8 | F(5'-GAAGGTGCAGTTTTGCCAAG-3'), R(5'-TGTGGTCCACTCTCAATCACTC-3') |
| IL-1α | F(5'-TGCCTGAGATACCCAAAACC-3'), R(5'-AACAAGTTTGGATGGGCAAC-3') |
| IL-1β | F(5'-TCCAGGAGAATGACCTGAGC-3'), R(5'-GTGATCGTACAGGTGCATCG-3') |
| NF-κB1 (p50) | F(5'-CCTGGATGACTCTTGGGAAA-3'), R(5'-TCAGCCAGCTGTTTCATGTC-3') |
| NF-κB2 (p52) | F(5'-GAACAGCCTTGCATCTAGCC-3'), R(5'-TTTTCAGCAT GGATGTCAGC-3') |
| p65 (RelA) | F(5'-TCTGCTTCCAGGTGACAGTG-3'), R(5'-GCCAGAGTTTCGGTTCACTC-3') |
| c-Rel | F(5'-CGAACCCAATTTATGACAAC-3'), R(5'-TTTTGTTTCTTTGCTTTATTGC-3') |
| RelB | F(5'-CTGCTTCCAGGCCTCATATC-3'), R(5'-CGCAGCTCTGATGTGTTTGT-3') |
| IκBα | F(5'-GATCCGCCAGGTGAAGGG-3'), R(5'-GCAATTTCTGGCTGGTTGG-3') |
3.4. Plasmid Constructs
3.5. Transient Transfection and Luciferase Assay
3.6. Immunofluorescence
3.7. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lee, J.H.; Ort, T.; Ma, K.; Picha, K.; Carton, J.; Marsters, P.A.; Lohmander, L.S.; Baribaud, F.; Song, X.Y.; Blake, S. Resistin is elevated following traumatic joint injury and causes matrix degradation and release of inflammatory cytokines from articular cartilage in vitro. Osteoarthr. Cartil. 2009, 17, 613–620. [Google Scholar]
- Staikos, C.; Ververidis, A.; Drosos, G.; Manolopoulos, V.G.; Verettas, D.A.; Tavridou, A. The association of adipokine levels in plasma and synovial fluid with the severity of knee osteoarthritis. Rheumatology 2013, 52, 1077–1083. [Google Scholar]
- Senolt, L.; Housa, D.; Vernerova, Z.; Jirasek, T.; Svobodova, R.; Veigl, D.; Anderlova, K.; Muller-Ladner, U.; Pavelka, K.; Haluzik, M. Resistin in rheumatoid arthritis synovial tissue, synovial fluid and serum. Ann. Rheum. Dis. 2007, 66, 458–463. [Google Scholar]
- Presle, N.; Pottie, P.; Dumond, H.; Guillaume, C.; Lapicque, F.; Pallu, S.; Mainard, D.; Netter, P.; Terlain, B. Differential distribution of adipokines between serum and synovial fluid in patients with osteoarthritis. Contribution of joint tissues to their articular production. Osteoarthr. Cartil. 2006, 14, 690–695. [Google Scholar]
- Tilg, H.; Moschen, A.R. Adipocytokines: Mediators linking adipose tissue, inflammation and immunity. Nat. Rev. Immunol. 2006, 6, 772–783. [Google Scholar]
- Koerner, A.; Kratzsch, J.; Kiess, W. Adipocytokines: Leptin—The classical, resistin—The controversical, adiponectin—The promising, and more to come. Best Pract. Res. Clin. Endocrinol. Metab. 2005, 19, 525–546. [Google Scholar]
- Bokarewa, M.; Nagaev, I.; Dahlberg, L.; Smith, U.; Tarkowski, A. Resistin, an adipokine with potent proinflammatory properties. J. Immunol. 2005, 174, 5789–5795. [Google Scholar]
- Zhang, Z.; Xing, X.; Hensley, G.; Chang, L.W.; Liao, W.; Abu-Amer, Y.; Sandell, L.J. Resistin induces expression of proinflammatory cytokines and chemokines in human articular chondrocytes via transcription and messenger RNA stabilization. Arthritis Rheumatol. 2010, 62, 1993–2003. [Google Scholar]
- Imamura, T.; Imamura, C.; McAlinden, A.; Davies, S.R.; Iwamoto, Y.; Sandell, L.J. A novel tumor necrosis factor alpha-responsive CCAAT/enhancer binding protein site regulates expression of the cartilage-derived retinoic acid-sensitive protein gene in cartilage. Arthritis Rheumatol. 2008, 58, 1366–1376. [Google Scholar]
- Brass, A.; Brenndorfer, E.D. The role of chemokines in hepatitis C virus-mediated liver disease. Int. J. Mol.Sci. 2014, 15, 4747–4779. [Google Scholar]
- Van der Kraan, P.M. Age-related alterations in TGFβ signaling as a causal factor of cartilage degeneration in osteoarthritis. Biomed. Mater. Eng. 2014, 24, 75–80. [Google Scholar]
- Huang, J.; Zhu, M.; Tao, Y.; Wang, S.; Chen, J.; Sun, W.; Li, S. Therapeutic properties of quercetin on monosodium urate crystal-induced inflammation in rat. J. Pharm. Pharmacol. 2012, 64, 1119–1127. [Google Scholar]
- Sandell, L.J.; Xing, X.; Franz, C.; Davies, S.; Chang, L.W.; Patra, D. Exuberant expression of chemokine genes by adult human articular chondrocytes in response to IL-1β. Osteoarthr. Cartil. 2008, 16, 1560–1571. [Google Scholar]
- Zhang, Z.; Bryan, J.L.; DeLassus, E.; Chang, L.W.; Liao, W.; Sandell, L.J. CCAAT/enhancer-binding protein beta and NF-κB mediate high level expression of chemokine genes CCL3 and CCL4 by human chondrocytes in response to IL-1β. J. Biol. Chem. 2010, 285, 33092–33103. [Google Scholar]
- Okazaki, K.; Yu, H.; Davies, S.R.; Imamura, T.; Sandell, L.J. A promoter element of the CD-RAP gene is required for repression of gene expression in non-cartilage tissues in vitro and in vivo. J. Cell Biochem. 2006, 97, 857–868. [Google Scholar]
- Lu, Y.C.; Kim, I.; Lye, E.; Shen, F.; Suzuki, N.; Suzuki, S.; Gerondakis, S.; Akira, S.; Gaffen, S.L.; Yeh, W.C.; et al. Differential role for c-Rel and C/EBPβ/δ in TLR-mediated induction of proinflammatory cytokines. J. Immunol. 2009, 182, 7212–7221. [Google Scholar]
- Venza, I.; Cucinotta, M.; Visalli, M.; de Grazia, G.; Oliva, S.; Teti, D. Pseudomonas aeruginosa induces interleukin-8 (IL-8) gene expression in human conjunctiva through the recruitment of both RelA and CCAAT/enhancer-binding protein beta to the IL-8 promoter. J. Biol. Chem. 2009, 284, 4191–4199. [Google Scholar]
- Hirata, M.; Kugimiya, F.; Fukai, A.; Ohba, S.; Kawamura, N.; Ogasawara, T.; Kawasaki, Y.; Saito, T.; Yano, F.; Ikeda, T.; et al. C/EBPβ promotes transition from proliferation to hypertrophic differentiation of chondrocytes through transactivation of p57. PLoS One 2009, 4, e4543. [Google Scholar]
- Basak, C.; Pathak, S.K.; Bhattacharyya, A.; Mandal, D.; Pathak, S.; Kundu, M. NF-κB- and C/EBPβ-driven interleukin-1β gene expression and PAK1-mediated caspase-1 activation play essential roles in interleukin-1β release from Helicobacter pylori lipopolysaccharide-stimulated macrophages. J. Biol. Chem. 2005, 280, 4279–4288. [Google Scholar]
- Esteves, C.L.; Kelly, V.; Breton, A.; Taylor, A.I.; West, C.C.; Donadeu, F.X.; Peault, B.; Seckl, J.R.; Chapman, K.E. Proinflammatory cytokine induction of 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1) in human adipocytes is mediated by MEK.; C/EBPβ.; and NF-κB/RelA. J. Clin. Endocrinol. Metab. 2014, 99, E160–E168. [Google Scholar]
- Rahman, S.M.; Janssen, R.C.; Choudhury, M.; Baquero, K.C.; Aikens, R.M.; de la Houssaye, B.A.; Friedman, J.E. CCAAT/enhancer-binding protein beta (C/EBPβ) expression regulates dietary-induced inflammation in macrophages and adipose tissue in mice. J. Biol. Chem. 2012, 287, 34349–34360. [Google Scholar]
- Choi, S.J.; Oba, T.; Callander, N.S.; Jelinek, D.F.; Roodman, G.D. AML-1A and AML-1B regulation of MIP-1α expression in multiple myeloma. Blood 2003, 101, 3778–3783. [Google Scholar]
- Tseng, P.C.; Hsu, H.C.; Janmanchi, D.; Lin, C.H.; Kuo, Y.H.; Chou, C.K.; Yeh, S.F. Helioxanthin inhibits interleukin-1β-induced MIP-1 beta production by reduction of c-jun expression and binding of the c-jun/CREB1 complex to the AP-1/CRE site of the MIP-1 beta promoter in Huh7 cells. Biochem. Pharmacol. 2008, 76, 1121–1133. [Google Scholar]
- Otero, J.E.; Dai, S.; Alhawagri, M.A.; Darwech, I.; Abu-Amer, Y. IKKβ activation is sufficient for RANK-independent osteoclast differentiation and osteolysis. J. Bone Miner. Res. 2010, 25, 1282–1294. [Google Scholar]
- Maier, P.J.; Zemoura, K.; Acuna, M.A.; Yevenes, G.E.; Zeilhofer, H.U.; Benke, D. Ischemia-like oxygen and glucose deprivation mediates down-regulation of cell surface γ-aminobutyric acid B receptors via the endoplasmic reticulum (ER) stress-induced transcription factor CCAAT/enhancer-binding protein (C/EBP)-homologous protein (CHOP). J. Biol. Chem. 2014, 289, 12896–12907. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Zhang, Z.; Kang, Y.; Hou, C.; Duan, X.; Sheng, P.; Sandell, L.J.; Liao, W. Resistin Stimulates Expression of Chemokine Genes in Chondrocytes via Combinatorial Regulation of C/EBPβ and NF-κB. Int. J. Mol. Sci. 2014, 15, 17242-17255. https://doi.org/10.3390/ijms151017242
Zhang Z, Zhang Z, Kang Y, Hou C, Duan X, Sheng P, Sandell LJ, Liao W. Resistin Stimulates Expression of Chemokine Genes in Chondrocytes via Combinatorial Regulation of C/EBPβ and NF-κB. International Journal of Molecular Sciences. 2014; 15(10):17242-17255. https://doi.org/10.3390/ijms151017242
Chicago/Turabian StyleZhang, Ziji, Zhiqi Zhang, Yan Kang, Changhe Hou, Xin Duan, Puyi Sheng, Linda J. Sandell, and Weiming Liao. 2014. "Resistin Stimulates Expression of Chemokine Genes in Chondrocytes via Combinatorial Regulation of C/EBPβ and NF-κB" International Journal of Molecular Sciences 15, no. 10: 17242-17255. https://doi.org/10.3390/ijms151017242
APA StyleZhang, Z., Zhang, Z., Kang, Y., Hou, C., Duan, X., Sheng, P., Sandell, L. J., & Liao, W. (2014). Resistin Stimulates Expression of Chemokine Genes in Chondrocytes via Combinatorial Regulation of C/EBPβ and NF-κB. International Journal of Molecular Sciences, 15(10), 17242-17255. https://doi.org/10.3390/ijms151017242




