Combinatorial Measurement of CDKN1A/p21 and KIF20A Expression for Discrimination of DNA Damage-Induced Clastogenicity
Abstract
:1. Introduction
2. Results and Discussion
2.1. Gene Expression Profile of a Newly Selected Candidate Biomarker, KIF20A

| DNA Damage Class | Compounds a | Mechanism | Concentration (μg/mL) | RCG b (%) |
|---|---|---|---|---|
| Positive | MMC | Cross-link [6] | 1 | 47.5 |
| CP | Cross-link [7] | 10 | 52.7 | |
| MMS | Alkylate [8] | 40 | 42.9 | |
| EMS | Alkylate [8] | 1200 | 47.8 | |
| ETOP | Topoisomerase II inhibitor [9] | 1 | 40.5 | |
| HU | Antimetabolite in DNA synthesis [10] | 2500 | 41.3 | |
| ENU | Alkylate [8] | 125 | 44.6 | |
| CCNU | Cross-link [11] | 10 | 54.4 | |
| CAMP | Topoisomerase I inhibitor [12] | 0.005 | 57.0 | |
| 5-FU | Antimetabolite in DNA synthesis [13] | 25 | 46.0 | |
| A | Positive in the CA test c | 204 | 50.3 | |
| Negative | COLCH | Mitotic spindle inhibitor [14] | 0.04 | 42.7 |
| ADE | DNA precursor [15] | 1400 | 44.2 | |
| TXL | Mitotic spindle inhibitor [16] | 0.25 | 54.5 | |
| COLCE | Mitotic spindle inhibitor [17] | 10 | 37.9 | |
| 2-DA | DNA precursor [15] | 4000 | 58.5 | |
| CHX | Inhibitor of protein synthesis [18] | 800 | 52.3 | |
| NaCl d | Osmolality change [19] | 0.80% | 47.1 | |
| B | Negative in the CA test c | 346 | 48.5 | |
| C | Negative in the CA test c | 346 | 49.8 |
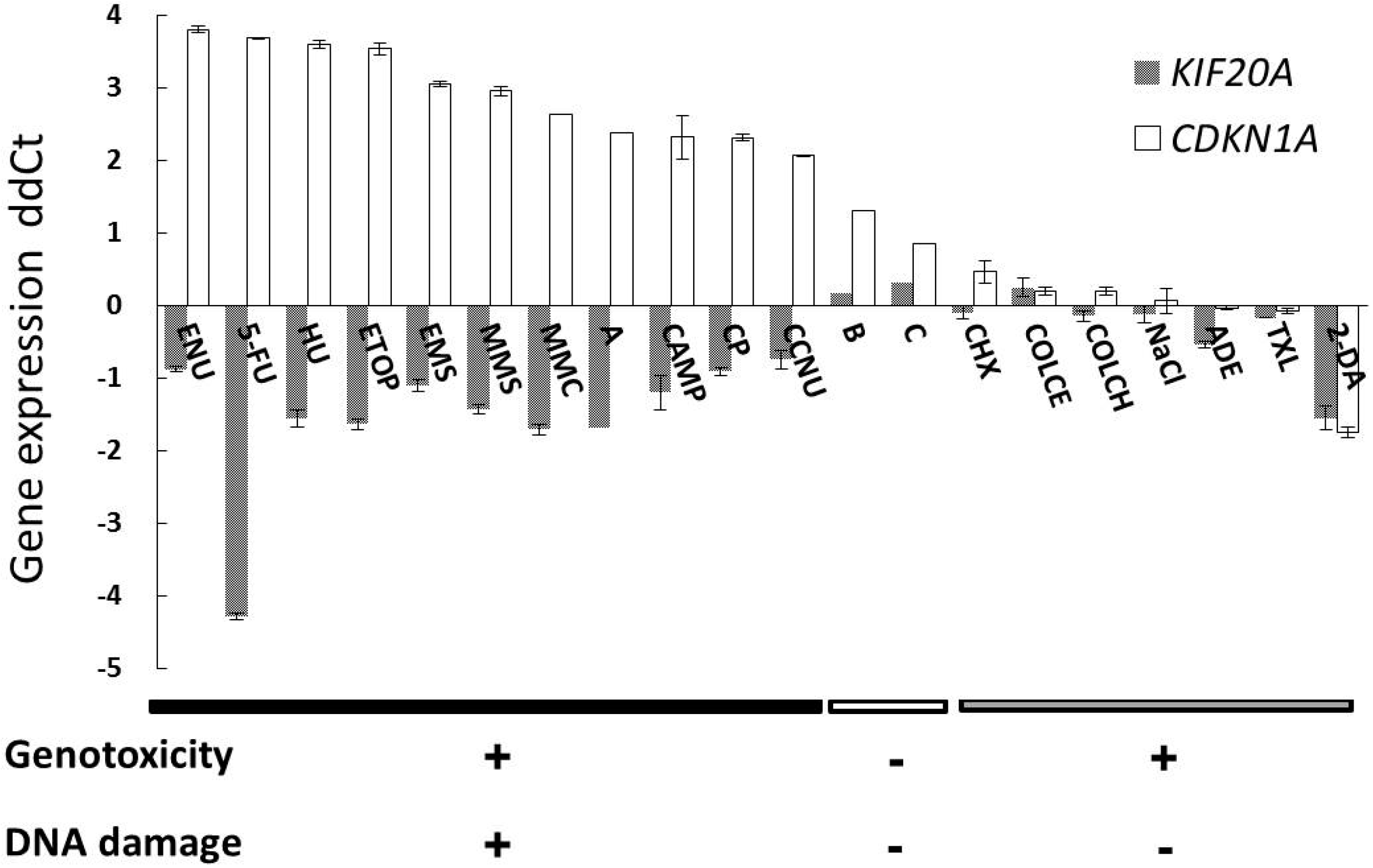
2.2. Expression Changes of Retinoblastoma (RB)/E2 Promoter Binding Factor (E2F) Regulatory Genes
| Gene Symbol | Probe Set ID | log2 (MMC/D) | log2 (MMS/D) | log2 (ETOP/D) | log2 (HU/D) | log2 (EMS/D) | log2 (CP/D) | log2 (ADE/D) |
|---|---|---|---|---|---|---|---|---|
| CCNA2 | 203418_at | −0.345 | −0.463 | −0.602 | −0.635 | −0.680 | −0.305 | −0.771 |
| CCNA2 | 213226_at | −0.358 | −0.334 | −0.671 | −0.602 | −0.429 | −0.221 | −0.540 |
| CDC25C | 205167_s_at | −0.961 | −0.403 | −0.696 | −0.657 | −0.354 | −0.735 | −1.963 |
| CDC25C | 217010_s_at | −0.506 | −0.411 | −0.615 | −0.402 | −0.597 | −0.509 | −1.454 |
| CDCA8 | 221520_s_at | −0.653 | −0.415 | −0.930 | −0.728 | −0.498 | −0.547 | −1.130 |
| CENPE | 205046_at | −1.389 | −1.107 | −1.968 | −1.414 | −0.917 | −0.637 | −1.610 |
| ECT2 | 219787_s_at | −0.607 | −0.651 | −1.033 | −0.881 | −0.574 | −0.642 | −1.526 |
| KIF11 | 204444_at | −0.486 | −0.325 | −0.729 | −0.035 | −0.349 | −0.409 | −0.951 |
| KIF18A | 221258_s_at | −1.037 | −0.571 | −1.125 | −0.395 | −0.750 | −0.873 | −2.083 |
| NEK2 | 204641_at | −0.832 | −0.500 | −1.186 | −0.861 | −0.436 | −0.394 | −0.602 |
| NEK2 | 211080_s_at | −0.780 | −0.881 | −1.230 | −0.948 | −0.647 | −0.400 | −1.668 |
2.3. Biological Significance of Genes Differentially Regulated on Adenine (ADE) Treatment

| Gene Ontology ID | Biological Processes | p-Value (Benjamini) |
|---|---|---|
| GO:0016070 | RNA metabolic process | 6.15 × 10−17 |
| GO:0006396 | RNA processing | 4.28 × 10−13 |
| GO:0000279 | M phase | 6.26 × 10−8 |
| GO:0007067 | Mitosis | 7.05 × 10−8 |
| GO:0019219 | Regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 8.70 × 10−8 |
| GO:0010468 | Regulation of gene expression | 7.34 × 10−8 |
| GO:0000087 | M phase of mitotic cell cycle | 7.16 × 10−8 |
| GO:0010556 | Regulation of macromolecule biosynthetic process | 9.86 × 10−8 |
| GO:0045449 | Regulation of transcription | 1.45 × 10−7 |
| GO:0031326 | Regulation of cellular biosynthetic process | 1.67 × 10−6 |
| GO:0034470 | ncRNA processing | 2.09 × 10−5 |
| GO:0016071 | mRNA metabolic process | 4.97 × 10−5 |
| GO:0008380 | RNA splicing | 1.14 × 10−4 |
| GO:0006397 | mRNA processing | 1.63 × 10−4 |
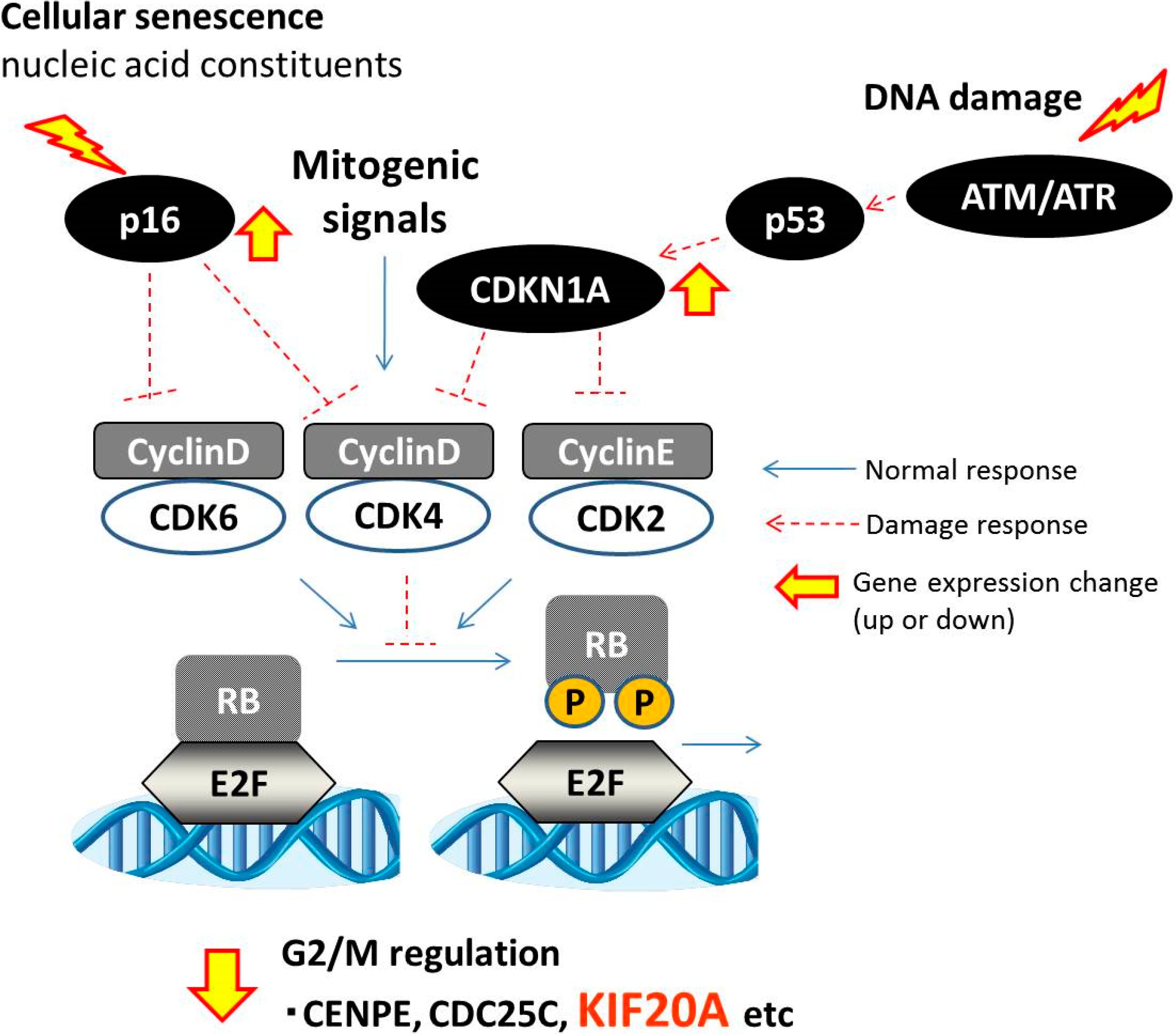
3. Experimental Section
3.1. In Vitro Cell Culture and Treatment
3.2. Microarray Analysis
3.3. Analysis of Biological Significance of Genes with Altered Expression on ADE Treatment
3.4. Quantitative RT-PCR Analysis (QPCR)
4. Conclusions
Supplementary Materials
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kirkland, D.J.; Müller, L. Interpretation of the biological relevance of genotoxicity test results: The importance of thresholds. Mutat. Res. 2000, 464, 137–147. [Google Scholar]
- Sakai, R.; Kondo, C.; Oka, H.; Miyajima, H.; Kubo, K.; Uehara, T. Utilization of CDKN1A/p21 gene for class discrimination of DNA damage-induced clastogenicity. Toxicology 2014, 315, 8–16. [Google Scholar]
- Martinez, L.A.; Yang, J.; Vazquez, E.S.; Rodriguez-Vargas Mdel, C.; Olive, M.; Hsieh, J.T.; Logothetis, C.J.; Navone, N.M. p21 modulates threshold of apoptosis induced by DNA-damage and growth factor withdrawal in prostate cancer cells. Carcinogenesis 2002, 23, 1289–1296. [Google Scholar]
- Chinery, R.; Brockman, J.A.; Peeler, M.O.; Shyr, Y.; Beauchamp, R.D.; Coffey, R.J. Antioxidants enhance the cytotoxicity of chemotherapeutic agents in colorectal cancer: A p53-independent induction of p21WAF1/CIP1 via C/EBPβ. Nat. Med. 1997, 3, 1233–1241. [Google Scholar]
- Gartel, A.L.; Goufman, E.; Tevosian, S.G.; Shih, H.; Yee, A.S.; Tyner, A.L. Activation and repression of p21WAF1/CIP1 transcription by RB binding proteins. Oncogene 1998, 17, 3463–3469. [Google Scholar]
- Iyer, V.N.; Szybalski, W. A molecular mechanism of mitomycin action: Linking of complementary DNA strands. Proc. Natl. Acad. Sci. USA 1963, 50, 355–362. [Google Scholar]
- Zwelling, L.A.; Kohn, K.W. Mechanism of action of cis-dichlorodiammineplatinum(II). Cancer Treat. Rep. 1979, 63, 1439–1444. [Google Scholar]
- O’Neill, J.P.; Schenley, R.L.; Hsie, A.W. Cytotoxicity and mutagenicity of alkylating agents in cultured mammalian cells (CHO/HGPRT system): Mutagen treatment in the presence or absence of serum. Mutat. Res. 1979, 63, 381–385. [Google Scholar]
- Liu, L.F.; Liu, C.C.; Alberts, B.M. Type II DNA topoisomerases: Enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell 1980, 19, 697–707. [Google Scholar]
- Hendricks, S.P.; Mathews, C.K. Differential effects of hydroxyurea upon deoxyribonucleoside triphosphate pools, analyzed with vaccinia virus ribonucleotide reductase. J. Biol. Chem. 1998, 273, 29519–29523. [Google Scholar]
- Kohn, K.W. Interstrand cross-linking of DNA by 1,3-bis(2-chloroethyl)-1-nitrosourea and other 1-(2-haloethyl)-1-nitrosoureas. Cancer Res. 1977, 37, 1450–1454. [Google Scholar]
- Pommier, Y.; Redon, C.; Rao, V.A.; Seiler, J.A.; Sordet, O.; Takemura, H.; Antony, S.; Meng, L.; Liao, Z.; Kohlhagen, G.; et al. Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat. Res. 2003, 532, 173–203. [Google Scholar]
- McLeod, H.L.; Sludden, J.; Hardy, S.C.; Lock, R.E.; Hawksworth, G.M.; Cassidy, J. Autoregulation of 5-fluorouracil metabolism. Eur. J. Cancer 1998, 34, 1623–1627. [Google Scholar]
- Owellen, R.J.; Hartke, C.A.; Dickerson, R.M.; Hains, F. Inhibition of tubulin-microtubule polymerization by drugs of the Vinca alkaloid class. Cancer Res. 1976, 36, 1499–1502. [Google Scholar]
- Scott, D.; Galloway, S.M.; Marshall, R.R.; Ishidate, M., Jr.; Brusick, D.; Ashby, J.; Myhr, B.C. International commission for protection against environmental mutagens and carcinogens—Genotoxicity under extreme culture conditions. A report from ICPEMC Task Group 9. Mutat. Res. 1991, 257, 147–205. [Google Scholar]
- Schiff, P.B.; Fant, J.; Horwitz, S.B. Promotion of microtubule assembly in vitro by taxol. Nature 1979, 277, 665–667. [Google Scholar]
- Hashimoto, K.; Todo, T. Mitotic slippage underlies the relationship between p53 dysfunction and the induction of large micronuclei by colcemid. Mutagenesis 2013, 28, 457–464. [Google Scholar]
- Schneider-Poetsch, T.; Ju, J.; Eyler, D.E.; Dang, Y.; Bhat, S.; Merrick, W.C.; Green, R.; Shen, B.; Liu, J.O. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 2010, 6, 209–217. [Google Scholar]
- Ashby, J.; Ishidate, M., Jr. lastogenicity in vitro of the Na, K, Ca and Mg salts of saccharin; and of magnesium chloride; consideration of significance. Mutat. Res. 1986, 163, 63–73. [Google Scholar]
- Eguchi, T.; Takaki, T.; Itadani, H.; Kotani, H. RB silencing compromises the DNA damage-induced G2/M checkpoint and causes deregulated expression of the ECT2 oncogene. Oncogene 2007, 26, 509–520. [Google Scholar]
- Ishida, S.; Huang, E.; Zuzan, H.; Spang, R.; Leone, G.; West, M.; Nevins, J.R. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 2001, 21, 4684–4699. [Google Scholar]
- Poon, R.Y.; Jiang, W.; Toyoshima, H.; Hunter, T. Cyclin-dependent kinases are inactivated by a combination of p21 and Thr-14/Tyr-15 phosphorylation after UV-induced DNA damage. J. Biol. Chem. 1996, 271, 13283–13291. [Google Scholar]
- Johnson, D.G.; Schneider-Broussard, R. Role of E2F in cell cycle control and cancer. Front. Biosci. 1998, 3, d447–d448. [Google Scholar]
- Xu, X.; Bieda, M.; Jin, V.X.; Rabinovich, A.; Oberley, M.J.; Green, R.; Farnham, P.J. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 2007, 17, 1550–1561. [Google Scholar]
- Van Oevelen, C.; Wang, J.; Asp, P.; Yan, Q.; Kaelin, W.G., Jr.; Kluger, Y.; Dynlacht, B.D. A role for mammalian Sin3 in permanent gene silencing. Mol. Cell 2008, 32, 359–370. [Google Scholar]
- Fontijn, R.D.; Goud, B.; Echard, A.; Jollivet, F.; van Marle, J.; Pannekoek, H; Horrevoets, A.J. The human kinesin-like protein RB6K is under tight cell cycle control and is essential for cytokinesis. Mol. Cell. Biol. 2001, 21, 2944–2955. [Google Scholar]
- Ren, B.; Cam, H.; Takahashi, Y.; Volkert, T.; Terragni, J.; Young, R.A.; Dynlacht, B.D. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes 2002, 16, 245–256. [Google Scholar]
- Van Gent, D.C.; Hoeijmakers, J.H.; Kanaar, R. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2001, 2, 196–206. [Google Scholar]
- Ohtani, N.; Yamakoshi, K.; Takahashi, A.; Hara, E. The p16INK4a-RB pathway: Molecular link between cellular senescence and tumor suppression. J. Med. Investig. 2004, 51, 146–153. [Google Scholar]
- Lundberg, A.S.; Hahn, W.C.; Gupta, P.; Weinberg, R.A. Genes involved in senescence and immortalization. Curr. Opin. Cell. Biol. 2000, 12, 705–709. [Google Scholar]
- Scholpa, N.E.; Zhang, X.; Kolli, R.T.; Cummings, B.S. Epigenetic changes in p21 expression in renal cells after exposure to bromate. Toxicol. Sci. 2014. [Google Scholar] [CrossRef]
- DAVID. Available online: http://david.abcc.ncifcrf.gov/ (accessed on 27 February 2014).
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sakai, R.; Morikawa, Y.; Kondo, C.; Oka, H.; Miyajima, H.; Kubo, K.; Uehara, T. Combinatorial Measurement of CDKN1A/p21 and KIF20A Expression for Discrimination of DNA Damage-Induced Clastogenicity. Int. J. Mol. Sci. 2014, 15, 17256-17269. https://doi.org/10.3390/ijms151017256
Sakai R, Morikawa Y, Kondo C, Oka H, Miyajima H, Kubo K, Uehara T. Combinatorial Measurement of CDKN1A/p21 and KIF20A Expression for Discrimination of DNA Damage-Induced Clastogenicity. International Journal of Molecular Sciences. 2014; 15(10):17256-17269. https://doi.org/10.3390/ijms151017256
Chicago/Turabian StyleSakai, Rina, Yuji Morikawa, Chiaki Kondo, Hiroyuki Oka, Hirofumi Miyajima, Kihei Kubo, and Takeki Uehara. 2014. "Combinatorial Measurement of CDKN1A/p21 and KIF20A Expression for Discrimination of DNA Damage-Induced Clastogenicity" International Journal of Molecular Sciences 15, no. 10: 17256-17269. https://doi.org/10.3390/ijms151017256
APA StyleSakai, R., Morikawa, Y., Kondo, C., Oka, H., Miyajima, H., Kubo, K., & Uehara, T. (2014). Combinatorial Measurement of CDKN1A/p21 and KIF20A Expression for Discrimination of DNA Damage-Induced Clastogenicity. International Journal of Molecular Sciences, 15(10), 17256-17269. https://doi.org/10.3390/ijms151017256




