Abstract
Prostate cancer (CaP) is the most common type of tumour disease in men. Early diagnosis of cancer of the prostate is very important, because the sooner the cancer is detected, the better it is treated. According to that fact, there is great interest in the finding of new markers including amino acids, proteins or nucleic acids. Prostate specific antigen (PSA) is commonly used and is the most important biomarker of CaP. This marker can only be detected in blood and its sensitivity is approximately 80%. Moreover, early stages cannot be diagnosed using this protein. Currently, there does not exist a test for diagnosis of early stages of prostate cancer. This fact motivates us to find markers sensitive to the early stages of CaP, which are easily detected in body fluids including urine. A potential is therefore attributed to the non-protein amino acid sarcosine, which is generated by glycine-N-methyltransferase in its biochemical cycle. In this review, we summarize analytical methods for quantification of sarcosine as a CaP marker. Moreover, pathways of the connection of synthesis of sarcosine and CaP development are discussed.
1. Introduction
1.1. History
Sarcosine, also known as N-methylglycine with the chemical formula CH3NHCH2COOH, was firstly isolated and named by German chemist Justus von Liebig in 1847. It is a non-proteinogenic amino acid that occurs as an intermediate product in the synthesis and degradation of amino acid glycine [1]. According to the Web of Knowledge database, there is an increasing incidence of keyword sarcosine as well as an increasing number of citations since 2009. This phenomenon confirms growing interest in sarcosine as a potential marker of a prostate cancer. The first publication describing the relationship between changes in level of sarcosine and progression of a prostate cancer was published in Nature by Sreekumar et al. in 2009 [1]. Since then sarcosine has been investigated as a new marker of prostate cancer by Issaq et al. [2] and has been identified as a metabolite greatly increasing during progression of a prostate cancer and metastatic process; which can be detected in urine [3].
1.2. Background
According to data describing CaP mortality and incidence in US in year 2013 prostate cancer exhibits the highest incidence (238,590 estimated new cases) and second highest mortality (29,720 estimated deaths) of all cancer types diagnosed in males [4]. Thanks to more developed methods of diagnosis is improved also the prognosis of patients with CaP. Therefore the fact that sarcosine does not occur or occurs in negligible concentration in urine of healthy persons is critical for its evaluation of potential disease marker [1]. This phenomenon reduces the risk of false positive and false negative results [5]. The possibility to use sarcosine as a marker of early stages of development of prostate cancer has been discussed in few papers [6–8]. Thanks to the application of sarcosine as a tumour marker, low cost analytical methods for its determination in the urine, tissue and blood plasma samples are being searched and developed. Biosynthesis of sarcosine has been shown to be certainly affected by the cancerogenesis (Figure 1). This phenomenon was described by Mukherjee et al. whose research revealed an important role of glycine N-methyltransferase (GNMT) in the metabolism of tissues of prostate cancer [9]. It has been found that GNMT is involved in the metabolism of methionine as well as in the gluconeogenesis and transformation of sarcosine to glycine (Figure 1). However, the role of sarcosine in carcinogenesis has not been fully understood and remains unknown, as indicate the results of the study published by Hakimi et al. [10]. Modulating the function of GNMT can be used to develop new strategies for treatment of prostate cancer. In addition, GNMT could serve as a new tumour marker to diagnose malignant progression of prostate cancer [11]. The most frequently used CaP screening marker—prostate specific antigen (PSA) is organ specific but cannot specify the stage and the type of disease, so its use for prostate cancer screening is insufficient [12]. Therefore, nowadays prostatic antigen 3 (PCA 3) [13–15] and annexin (A3) [5,16–18] are the most widely used and generally accepted markers of non-invasive CaP in urine. There are also other newly discussed potential markers of CaP that may be useful in the diagnosis of progression of prostate cancer, which can be detected in urine, such as alpha-methylacyl-CoA racemase (AMACR) [19–21], which level is elevated in prostatic adenocarcinoma and high-grade intraepithelial neoplasia. TMPRSS2-ETS fusion gene conversions that is very common and specific in alterations in the prostate cancer cells. These genetic alterations lead the overexpression of ETS genes encoding the E26 family of transcription factors involved in cell proliferation [22]. Another used marker is the ratio of free to total PSA—f/tPSA. It was confirmed that this biomarker can better distinguish between patients with prostate cancer from patients with a benign hyperplasia of the prostate [23]. ProPSA—the most stable subform of PSA associated with cancer also exhibits potential, because several studies suggested the clinical usefulness of proPSA in the detection of prostate cancer using different non-commercial assays [24,25]. Some high potential serum biomarkers are: kallikrein 2, urokinase-type plasminogen activator/urokinase-type plasminogen activator receptor, interleukin-6/interleukin-6 receptor, pigment epithelium-derived factor (PEDF), fibronectin 1, chromogranin A ceruloplasmin and others [26,27], nevertheless their clinical utilization and their role in the active surveillance scenario needs to be studied further. The aim of this review is to describe the molecularly-biological aspects and biochemical pathways of biosynthesis of sarcosine and to summarize the basic data about suitability of chosen analytical techniques for determination of sarcosine in various matrixes.
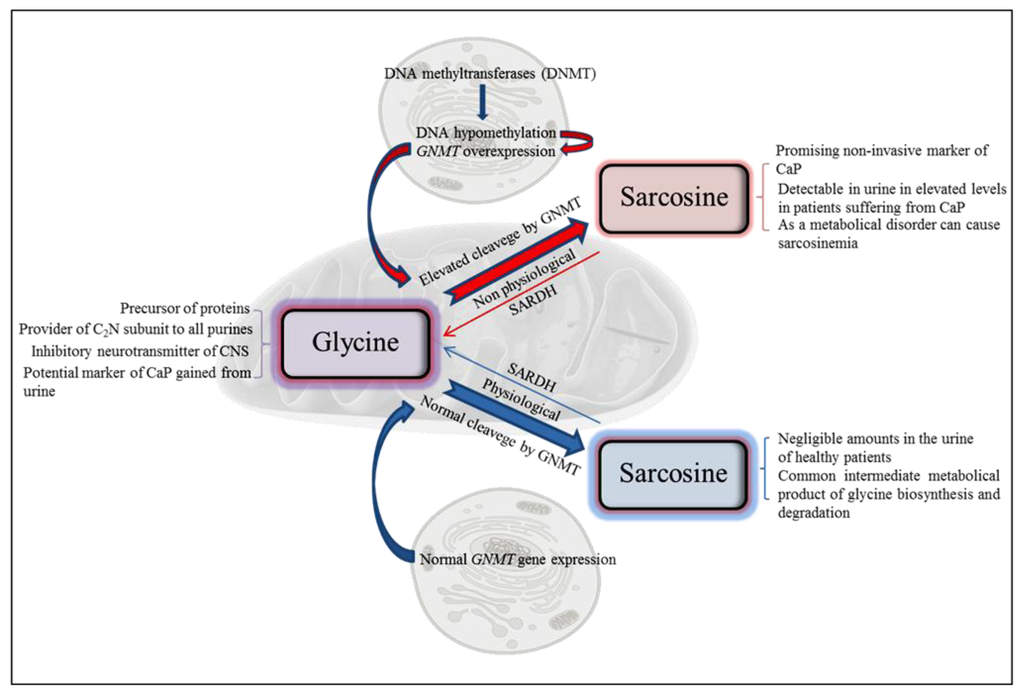
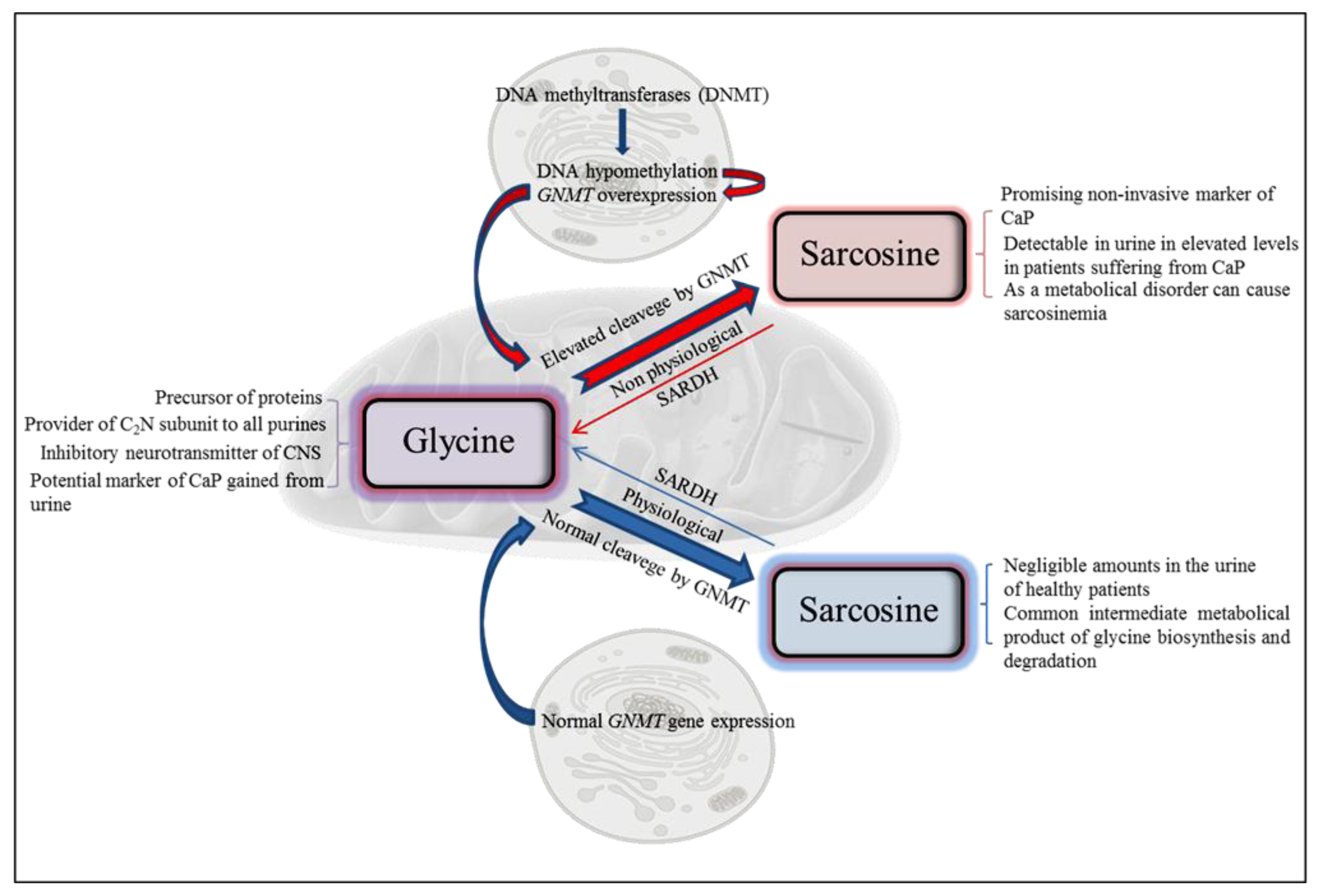
Figure 1.
Scheme of biological functions of sarcosine at both physiological and nonphysiological conditions. CaP—prostate carcinoma; GNMT—glycine N-methyltransferase; SARDH—sarcosine dehydrogenase.
2. Methods
A literature search covering the topic of the review was performed in following databases: Web of Science (Thomson Reuters, New York, NY, USA), PubMed (United States National Library of Medicine, Bethesda, MD, USA) Cochrane Library (Cochrane Collaboration, Baltimore, MD, USA), and Scopus (Elsevier, Amsterdam, Netherland) as it is shown in Figure 2. The presented figure shows a course of the process of the papers publications devoted to sarcosine. We looked for studies related to sarcosine and its metabolic properties, its molecularly-biological properties and possibilities of its analysis.

Figure 2.
Overview of the databases used for the evaluation of the papers related to the topic of the review.
The used keywords are shown in Figure 2. Keyword sarcosine in tumour tissue gave 77 results in all used databases. Keyword sarcosine in urine revealed 417 results in all databases and keyword sarcosine in serum provided 438 results within all the databases. Duplicating articles were subtracted for each term from these counts. Seven articles were subtracted for the keyword sarcosine in tumour tissue, 14 articles for sarcosine in serum and 7 articles for sarcosine in urine. The resulting number of articles was subjected to readout of articles that were not directly related to the review topic. We withdrew 65 articles that were not related to the keyword sarcosine in tumour tissue, 393 articles for the keyword sarcosine in serum and 332 articles for sarcosine in urine. The resulting number n is the sum of non-recurring articles in all the databases related to the searched keywords. The final numbers were as follows: 5 for sarcosine in tumour tissue, 31 for sarcosine in serum and 78 for sarcosine in urine. These numbers give clear evidence of the importance of sarcosine as a potential biomarker, whose possibility to be determined in urine represents the greatest potential. This fact is important, because urine is one of the most accessible and stable body fluids for analysis [28].
3. Molecular Biology of Sarcosine
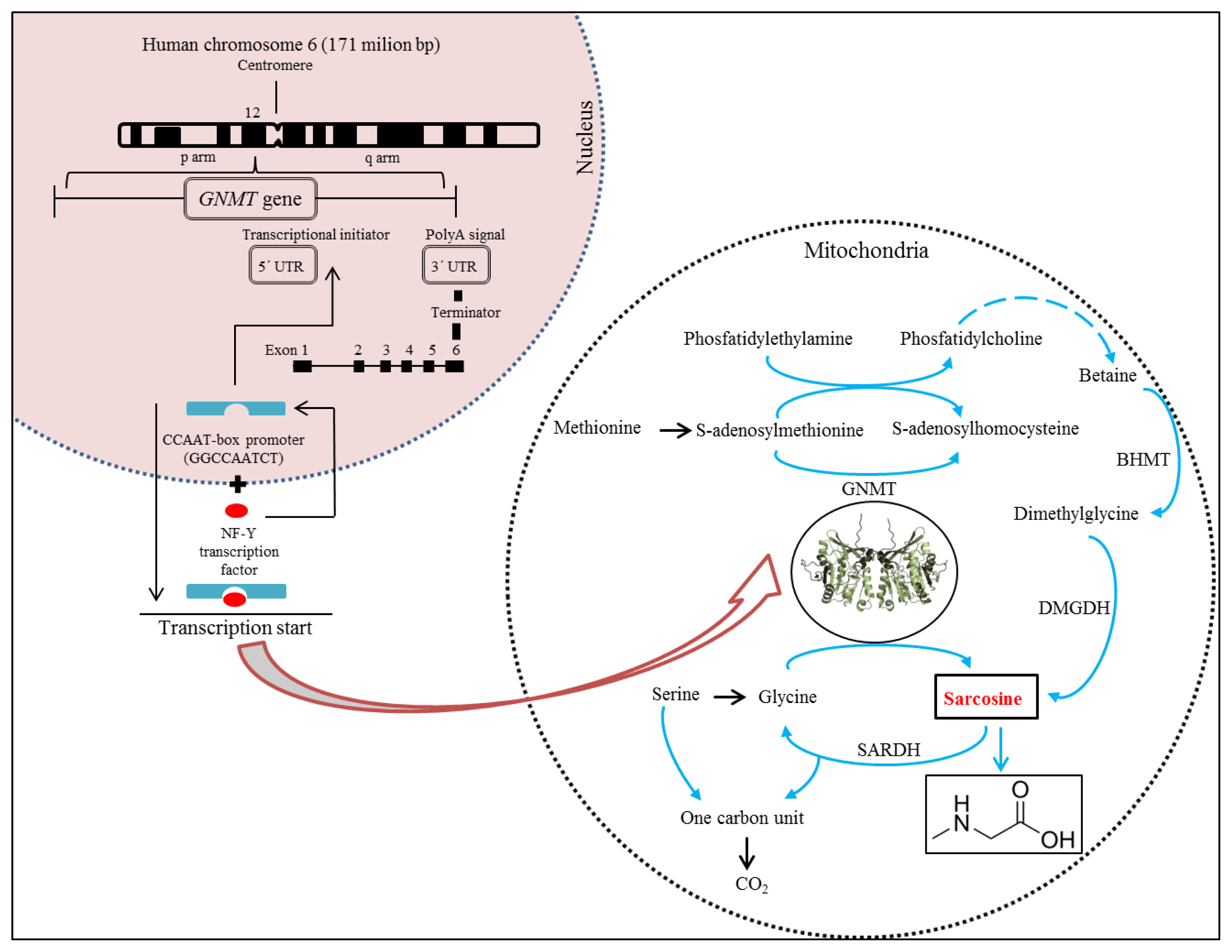
Glycine N-methyltransferase (GNMT) acts as an essential component that influences synthesis of sarcosine [29–32]. Synthesis of GNMT is controlled by the same gene named GNMT. It has recently been reported that the GNMT gene is located on the short (p) arm of chromosome 6 at position 12 (Figure 3) and acts as a tumour-susceptible gene [33]. According to the study by Ianni et al., T allele of the rs9462856 SNP in the promoter region of the GNMT gene is overexpressed in patients suffering from CaP and its overexpression significantly increases the risk of the disease [34]. Phenotypic analysis of three GNMT haplotypes (A, B, and C) indicated that haplotype C had the highest promoter activity and haplotype B had significantly higher activity compared to the haplotype A. The difference between the haplotypes B and C is due to the T allele of SNP1 that exerts a strong disequilibrium [35]. The GNMT gene contains in TATA-less core promoter region the Sp1 site and a CCAAT-box [36] that is one of the most ubiquitous elements being present in 30% of all eukaryotic promoters [37–39]. This region represents a binding site for the transcriptional factor, NF-Y, a trimer with histone-like subunits NF-YB/NF-YC and the sequence-specific NF-YA [40,41]. NF-Y is a sequence-specific transcription factor with nucleosome-like properties in nonspecific DNA binding that helps to establish permissive chromatin modifications at CCAAT promoters [38].
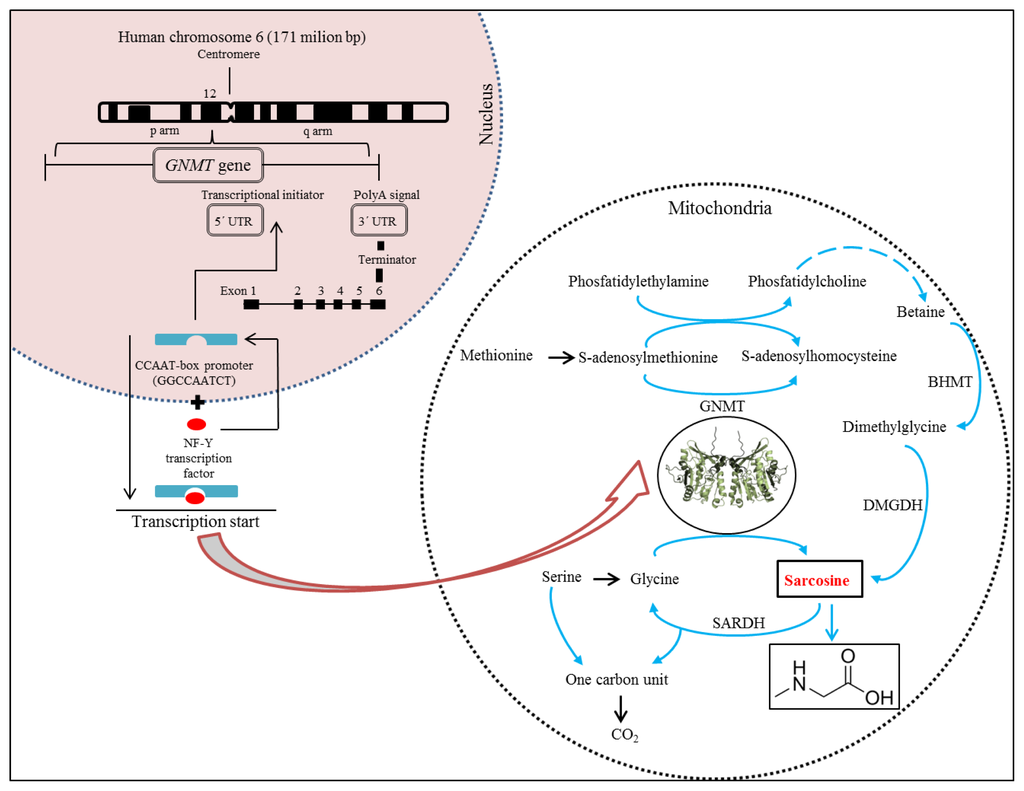
Figure 3.
Structure of human cine-N-methyltransferase (GNMT) gene in connection with scheme of sarcosine genesis in biochemical pathway.
The expression of the GNMT induced in this manner leads to synthesis of GNMT that contributes to the regulation of the level of S-adenosylmethionine (SAM) and influences gene expression by affecting the DNA methylation [34]. DNA methylation is an essential process in the body. Methyl groups transferred by SAM are used to synthesize many essential compounds including creatine and/or phosphatidylcholine. In addition, DNA methylation is essential for regulation of gene expression [42]. Deficiency of donors of methyl group (e.g., choline and methionine) or coenzymes of metabolism of methyl group (e.g., folate and vitamin B12) disturbs the intracellular levels of S-adenosylmethionine, triggers the DNA hypomethylation, and promotes cancers of the liver, prostate, and other organs [35,43]. Patients suffering from CaP have been revealed to show a decreased methylation of DNA, whereas patients positive for the GNMT T allele had a lower level of methylated DNA than controls with the same allele [34]. GNMT also binds a number of polycyclic aromatic hydrocarbons and inhibits the formation of DNA adducts [30].
Due to properties of GNMT, its excessive production causes a cleavage of glycine to sarcosine and elevated the presence of sarcosine in urine (Figures 1 and 3). Stabler et al. reported that increased flux through of GNMT results in the increased formation of homocysteine and sarcosine through increased utilization of SAM [44]. This makes sarcosine interesting in the field of non-invasive cancer biomarkers. Elevated levels of sarcosine correlated with progression of prostate cancer and metastatic process. Supplementation of sarcosine to prostate cancer cell lines induced a selection of invasive phenotype in culture [19]. Dahl et al. reported for the first time on a significant upregulation of a potent oncoprotein human epidermal growth factor receptor 2 (HER2/neu) in androgen-dependent prostate cancer cells upon exposure to exogenous sarcosine. That indicates that sarcosine may be involved in regulation of HER2/neu. However, information about these cellular mechanisms will be needed for detailed clarification [45].
4. Metabolism of Sarcosine
Metabolites play essential role in an understanding the biological reactions and thereby the changes in their levels contribute to the development of new diagnostic and therapeutic methods to diagnose specific diseases [46,47]. Biochemical pathways of formation and oxidation of sarcosine occur in mitochondria and are provided by two basic pathways (Figure 3). Phosfatidylethylamine is methylated repeatedly by S-adenosylmethionine (SAM) during transulfuration to phosphatidylcholine in the first pathway with the resulting intermediate product betaine. This reaction forms dimethylglycine and regenerates methionine from homocysteine [44]. Dimethylglycine is subsequently converted to sarcosine via dimethylglycine dehydrogenase (DMGDH) [46,48]. The second metabolic pathway creates sarcosine during the transformation of the methyl group of S-adenosylmethionine catalysed by the enzyme glycine-N-methyltransferase (GNMT), that is a tetramer of identical 32 kDa subunits [49,50]. These two reactions ultimately produce 5, 10-methylenetetrahydrofolate and are dependent on oxidized flavoproteins [e.g., flavin adenine dinucleotide (FAD+)] [46]. This pathway also includes an oxidative phase, which removes the methyl group from sarcosine to create glycine and an active one-carbon unit by sarcosine dehydrogenase (SARDH), a mitochondrial flavoprotein [51] which, in a study by Montrose et al., was observed within the tumour tissue with GNMT and other enzymes and can explain the increases in sarcosine levels [48]. The absence of the activity of SARDH involves also conversion of choline to glycine in humans; this is transmitted genetically and represents a disorder in metabolism of amino acid that is manifested as sarcosinemia, a surplus of sarcosine accompanied by high concentrations of sarcosine in blood and urine [52].
5. Metabolomic of Prostate Cancer
In the last decade, advances in nuclear magnetic resonance spectroscopy (NMR) and mass spectrometry (MS) have been applied to identify metabolomics of prostate cancer that may show at clinically useful biomarkers [53]. NMR exploits the behaviour of molecules when placed in a magnetic field, allowing the identification of different nuclei based on their resonant frequency. MS determines the composition of molecules based on the mass-to-charge ratio in charged particles. The resultant metabolite detection and quantification is acquired as a data set called a spectrum [54]. Tumour metabolomic profile is highly dependent on its organ of origin, and exhibits unique patterns dependent on cancer type as well as differentiation status [55]. Prostate cancer has a distinct metabolic profile characterized by high levels of total choline (tCho) and phosphocholine, together with increasing amounts of the glycolytic products lactate and alanine [56]. In the studyof Swanson et al., healthy glandular tissues demonstrated significantly higher concentrations of citrate and polyamines than healthy stromal and prostate cancer tissues, while healthy glandular and stromal tissues demonstrated lower concentrations of choline, phosphocholine plus glycerophosphocholine and total cholin (tCho) than prostate cancer tissues [57]. The results in cancer metabolomic achieved through in vivo MRSI and ex vivo NMR investigations during the first 11 years of the 21st century are illustrating the areas where these technologies can be best translated into clinical practice [58].
6. Analytical Techniques for Detection of Sarcosine
Currently, new methods for reliable detection of sarcosine have been developed to confirm or contradict sarcosine as a possible marker of prostate cancer [2,7,59].
6.1. Microarray-Based Analysis
Microarray technology is based on the assumption that the amount of mRNA for each gene correlates with proteins amount. However, it is well know that a correlation between RNA expression and protein expression rarely holds because of posttranslational modification, protein degradation and feedback of protein on RNA expression [9]. Studying of gene expression provides useful information for both clinical and basic researches [60]. Microarray-based multiplex biomarker analysis of fusion genes in urine samples has been described [5]. Some promising non-invasive biomarkers with potential for detection in the urine were determined as PCA3, fusion genes, e.g., TMPRSS2:ETS, TERT, AMACR, GSTP1, MMP9, VEGF, ANXA3 and sarcosine [5]. The new urine tests should be useful in early diagnosis of prostate cancer and detection of aggressive tumours with the goal of radically improving treatment of CaP.
6.2. Chromatography
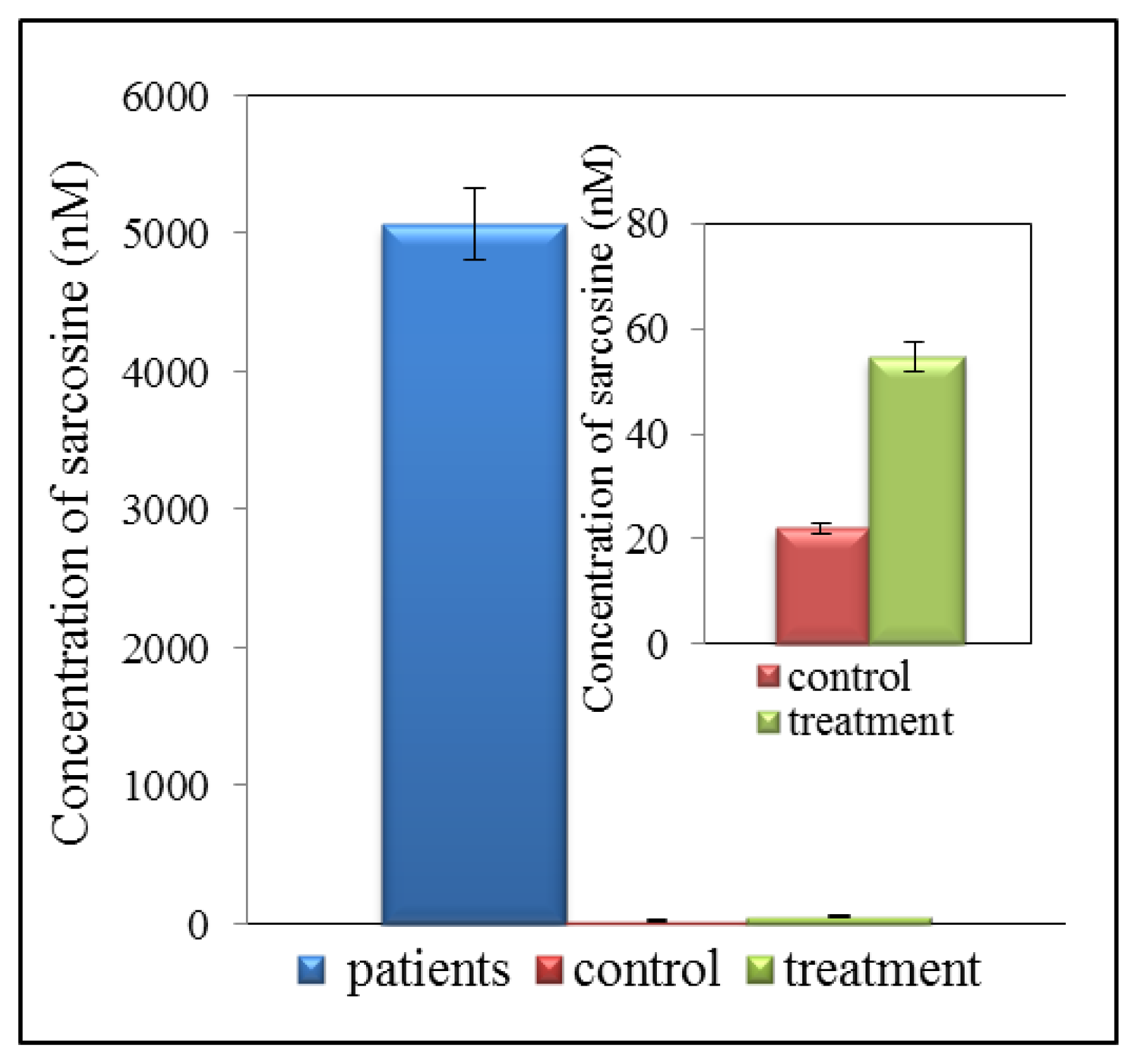
Detection of biomarkers using liquid chromatography (LC) is currently one of the most widely used analytical techniques. Advantage of LC application consists in repeatable and high throughput detection of low molecular mass compounds. It is therefore possible to detect and quantify very low concentrations of analytes in the presence of various interfering components. Very good results have been obtained by using chromatography in tandem with mass spectrometry used for determination of sarcosine in urine samples. This method is highly sensitive and allows determining sarcosine in very low concentrations. However, there is one great advantage, this method is able to separate sarcosine from its isomers and alanine, which may be responsible for a false-positive and false-negative results. Jentzmik et al. used gas chromatography with tandem mass spectrometry to determine sarcosine [7]. It was detected in the samples of both malignant and non-malignant tissues after radical prostatectomy from 92 patients suffering from prostate cancer. Significant differences in concentrations of sarcosine in malignant and non-malignant tissues were found. Concentration of sarcosine was more than 7% higher in malignant tissues compared to non-malignant. Based on these results it was concluded that sarcosine could not be considered as a suitable predictor of CaP [7]. In another published study gas chromatography (GC) and mass spectrometry (MS) were used. The results showed that the sarcosine/alanine ratios in patients with early and advanced prostate cancer were fairly constant showing no statistically significant differences between T-stages [61]. It was therefore concluded that sarcosine is not a suitable marker for prostate cancer. By comparing PSA with T-stages in the same group of patients it was found that PSA in the T1–T2 group of patients was significantly lower than in the T3–T4 group of patients confirming the well know merits and limitations of this marker [61]. Reverse phase high performance liquid chromatography (HPLC) is the most utilized technique for separation of amino acids [62,63] or peptides [64]. HPLC with tandem mass spectrometry, which is presently one of the most widely used analytical methods, was used to determine sarcosine and six other metabolites in urine [65]. This method is able to determine sarcosine in much lower levels than gas chromatography. The technique developed in this study is very simple, fast, sensitive, robust and repeatable. Cernei et al. showed that electrochemical detection is also a suitable method to detect sarcosine in very low concentration. However, this method can be used to analyse real samples only after application of suitable pre-treatment [21,66]. Application of ion-exchange liquid chromatography is also suitable to separate amino acids [21]. It can be used to analyse all organic compounds that contain amino acids [67]. Ion-exchange chromatography method developed by Cernei et al. revealed that level of sarcosine in urine of patients suffering from CaP is several times higher than that of cured patient (Figure 4). The level of sarcosine in healthy patients is only negligible [21]. Table 1 summarizes the method used for sarcosine detection.
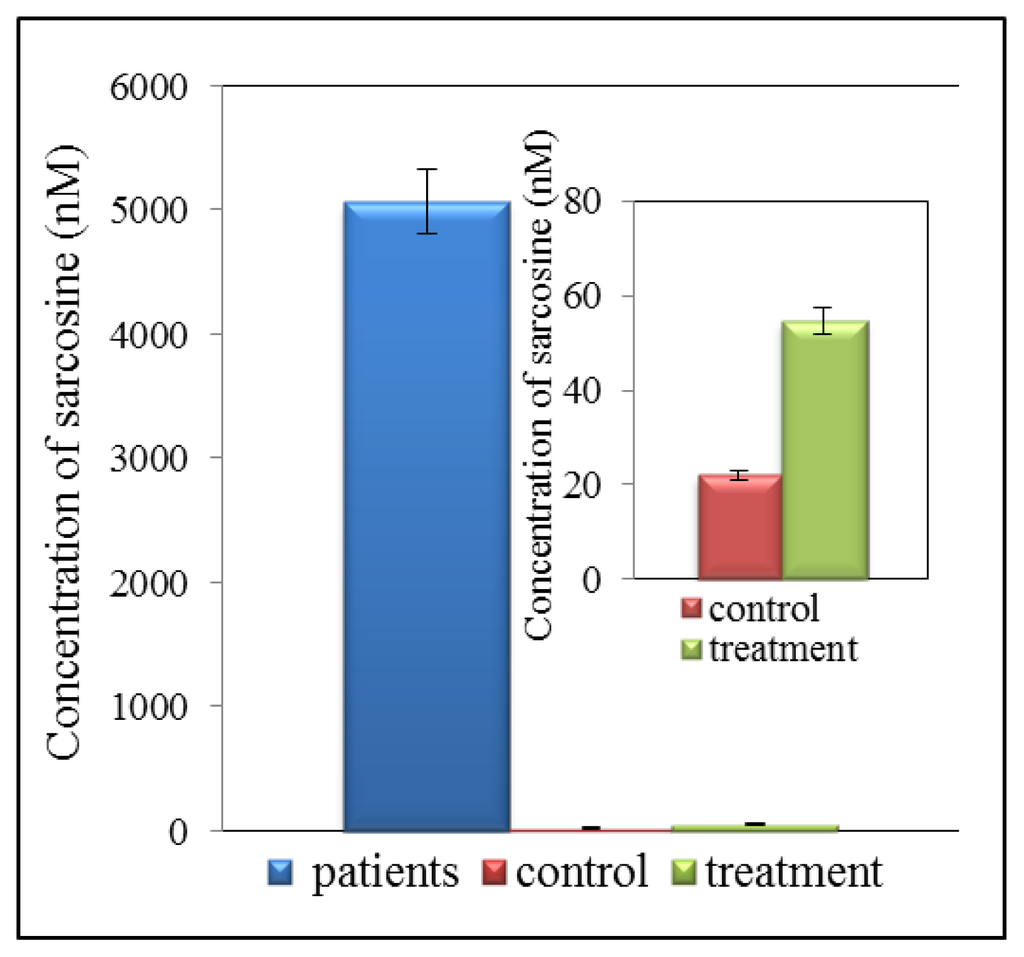
Figure 4.
Overview of concentration of sarcosine in urine of patients suffering from prostate cancer (CaP), treated patients and controls (healthy people).

Table 1.
Comparison of methods used for determination of sarcosine in different matrixes. In addition, the necessity to pre-treat samples is also indicated.
7. Summary of Results Obtained by Analysis of Samples from CaP Patients
Table 2 shows different types of prostate cancer diagnosed and published in the studies. Sarcosine has been determined in majority of presented works. Despite the fact that some publications do not claim sarcosine to be a potential marker for CaP, sarcosine was detected and quantified in almost all studies in amounts different from the controls. This statement corresponds with results by Khan et al. They validated sarcosine as an important oncometabolite using both in vivo and in vitro preclinical models. Moreover they confirmed that overexpression of GNMT in cells elevated also the sarcosine levels but had no effects on cell proliferation [68].

Table 2.
Overview of the published papers containing data about changes in level of sarcosine in samples of urine of patients with diagnosed prostate cancer.
Table 2 reveals the connection between sarcosine and prostate cancer. Sarcosine has been shown to be rapidly released into the urine supernatant [69]. Compared to study of Sreekumar et al. [1], Jentzmik et al. [69] suppose that the data by Sreekumar et al. [1] are more likely a result of cohort differences rather than true elevations of sarcosine levels in urine of patients suffering from prostate cancer [7]. Struys et al. [70] compared serum sarcosine concentrations between three groups of men, one of them was created by CaP patients, second were patients with increased PSA and the third one were individuals who have been assessed for vitamin B12 status. Results obtained by Struys and colleagues demonstrate that the mean of concentrations of sarcosine in serum does not discriminate between the three groups of men. The concentrations of sarcosine also did not demonstrate a correlation with tumour progression or PSA [70]. Despite this fact, other results shown in Table 2 more or less confirm the sarcosine as a potential CaP marker.
Moreover, Wu et al. [71] determined sarcosine in urine samples of patients suffering from CaP, and the authors finally conclude that value of sarcosine determined in urine has limited potential in the diagnostic algorithm of CaP. The authors suggested that the parameter sarcosine/creatinine ratio had not been accurate enough to diagnose CaP. In addition, it cannot reliably predict the histologic grade and behaviour of a tumour [71]. Lucarelli et al. [59] concluded that higher serum sarcosine levels were significantly associated with low- and intermediate-grade tumours in men with PSA < 4 ng/mL, nevertheless it is important to combine serum sarcosine with other tumour markers, such as PCA3 and [−2] pro PSA. Thereafter it could have an important role in selecting men with low grade/low risk PCa who should undergo active surveillance programs [59]. Bianchi et al. [72] concluded that sarcosine cannot be considered as a reliable marker for prostate cancer in urinary sediments. The analysis of a more extended number of samples taken from both healthy and CaP-diseased patients (localized and metastatic cancer) would be, according to extensiveness of issue, helpful to clarify definitively the role of sarcosine.
8. Conclusions
Non-proteinogenic amino acid sarcosine has a promising potential as a non-invasive marker of carcinoma of prostate as well as GNMT, which is closely connected with the presence and involvement of sarcosine in tissue of prostate tumours. Analytical demands on accurate and precise detection of low concentrations of sarcosine are high, and relatively expensive instrumentation is quite necessary. However, the limits of detection in units of nanograms can be achieved and reversed this fact. The finding that sarcosine does not occur in urine of healthy patients in detectable levels [1,5] and the necessity to eliminate negative results are very important for developing the low-cost, rapid, and reliable analytical methods.
Acknowledgments
Financial support from LPR 2012, CEITEC CZ.1.05/1.1.00/02.0068 and by MH CZ – DRO, University Hospital Motol, Prague, Czech Republic 00064203 is highly acknowledged.
Conflict of Interest
The authors declare no conflict of interest.
References
- Sreekumar, A.; Poisson, L.M.; Rajendiran, T.M.; Khan, A.P.; Cao, Q.; Yu, J.D.; Laxman, B.; Mehra, R.; Lonigro, R.J.; Li, Y.; et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 2009, 457, 910–914. [Google Scholar]
- Issaq, H.J.; Waybright, T.J.; Veenstra, T.D. Cancer biomarker discovery: Opportunities and pitfalls in analytical methods. Electrophoresis 2011, 32, 967–975. [Google Scholar]
- Cavaliere, B.; Macchione, B.; Monteleone, M.; Naccarato, A.; Sindona, G.; Tagarelli, A. Sarcosine as a marker in prostate cancer progression: A rapid and simple method for its quantification in human urine by solid-phase microextraction-gas chromatography-triple quadrupole mass spectrometry. Anal. Bioanal. Chem 2011, 400, 2903–2912. [Google Scholar]
- Siegel, R.; Naishadham, D.; Jemal, A. Cancer statistics, 2013. CA Cancer J. Clin 2013, 63, 11–30. [Google Scholar]
- Jamaspishvili, T.; Kral, M.; Khomeriki, I.; Student, V.; Kolar, Z.; Bouchal, J. Urine markers in monitoring for prostate cancer. Prostate Cancer Prostatic Dis 2010, 13, 12–19. [Google Scholar]
- Issaq, H.J.; Veenstra, T.D. Is sarcosine a biomarker for prostate cancer? J. Sep. Sci 2011, 34, 3619–3621. [Google Scholar]
- Jentzmik, F.; Stephan, C.; Lein, M.; Miller, K.; Kamlage, B.; Bethan, B.; Kristiansen, G.; Jung, K. Sarcosine in prostate cancer tissue is not a differential metabolite for prostate cancer aggressiveness and biochemical progression. J. Urol 2011, 185, 706–711. [Google Scholar]
- Palacios, D.A.; Rosser, C.J. Sarcosine, a biomarker for prostate cancer: Ready for prime time? Biomark. Med 2012, 6, 513–514. [Google Scholar]
- Mukherjee, S.; Cruz-Rodriguez, O.; Bolton, E.; Iniguez-Lluhi, J.A. The in vivo role of androgen receptor SUMOylation as revealed by androgen insensitivity syndrome and prostate cancer mutations targeting the proline/glycine residues of synergy control motifs. J. Biol. Chem 2012, 287, 31195–31206. [Google Scholar]
- Hakimi, J.M.; Schoenberg, M.P.; Rondinelli, R.H.; Piantadosi, S.; Barrack, E.R. Androgen receptor variants with short glutamine or glycine repeats may identify unique subpopulations of men with prostate cancer. Clin. Cancer Res 1997, 3, 1599–1608. [Google Scholar]
- Huang, J.; Jiang, L.M.; Ren, P.P.; Zhang, L.M.; Tang, H.R. Comprehensive solid-state NMR analysis reveals the effects of n-methylation on the molecular dynamics of glycine. J. Phys. Chem. B 2012, 116, 136–146. [Google Scholar]
- Patel, S.; Issa, M.M.; El-Galley, R. Evaluation of novel formula of PSA, age, prostate volume, and race in predicting positive prostate biopsy findings. Urology 2013, 81, 602–606. [Google Scholar]
- Morote, J.; Rigau, M.; Garcia, M.; Mir, C.; Ballesteros, C.; Planas, J.; Raventos, C.X.; Placer, J.; de Torres, I.M.; Reventos, J.; et al. Behavior of the PCA3 gene in the urine of men with high grade prostatic intraepithelial neoplasia. World J. Urol 2010, 28, 677–680. [Google Scholar]
- Goode, R.R.; Marshall, S.J.; Duff, M.; Chevli, E.; Chevli, K.K. Use of PCA3 in detecting prostate cancer in initial and repeat prostate biopsy patients. Prostate 2013, 73, 48–53. [Google Scholar]
- Shen, M.; Chen, W.; Yu, K.Y.; Chen, Z.G.; Zhou, W.; Lin, X.M.; Weng, Z.L.; Li, C.D.; Wu, X.L.; Tao, Z.H. The diagnostic value of PCA3 gene-based analysis of urine sediments after digital rectal examination for prostate cancer in a Chinese population. Exp. Mol. Pathol 2011, 90, 97–100. [Google Scholar]
- Wu, N.; Liu, S.Q.; Guo, C.M.; Hou, Z.J.; Sun, M.Z. The role of annexin A3 playing in cancers. Clin. Transl. Oncol 2013, 15, 106–110. [Google Scholar]
- Schostak, M.; Schwall, G.; Poznanovic, S.; Miller, K.; Messinger, D.; Klocker, H.; Stenzl, A.; Feyerabend, S.; Muller, M.; Schrattenholz, A. Annexin A3 quantification from supernatants of urine after dre provides a novel and clinically easy available biomarker for the non-invasive diagnosis of prostate cancer. J. Urol 2007, 177, 470–470. [Google Scholar]
- Schostak, M.; Schwall, G.P.; Poznanovic, S.; Groebe, K.; Muller, M.; Messinger, D.; Miller, K.; Krause, H.; Pelzer, A.; Horninger, W.; et al. Annexin A3 in urine: A highly specific noninvasive marker for prostate cancer early detection. J. Urol 2009, 181, 343–353. [Google Scholar]
- Cao, D.L.; Ye, D.W.; Zhang, H.L.; Zhu, Y.; Wang, Y.X.; Yao, X.D. A multiplex model of combining gene-based, protein-based, and metabolite-based with positive and negative markers in urine for the early diagnosis of prostate cancer. Prostate 2011, 71, 700–710. [Google Scholar]
- Dabir, P.D.; Ottosen, P.; Hoyer, S.; Hamilton-Dutoit, S. Comparative analysis of three- and two-antibody cocktails to AMACR and basal cell markers for the immunohistochemical diagnosis of prostate carcinoma. Diagn. Pathol 2012, 7, 1–6. [Google Scholar]
- Cernei, N.; Zitka, O.; Skalickova, S.; Gumulec, J.; Masarik, M.; Hrabec, R.; Adam, V.; Kizek, R. Sarcosine in urine of patients with prostate carcinome (in Czech). Praktický Lékař 2012, 92, 444–448. [Google Scholar]
- Fernandez-Serra, A.; Rubio-Briones, J.; Garcia-Casado, Z.; Solsona, E.; Lopez-Guerrero, J.A. Prostate cancer: The revolution of the fusion genes. Actas Urol. Esp 2011, 35, 420–428. [Google Scholar]
- Strittmatter, F.; Stieber, P.; Nagel, D.; Fullhase, C.; Walther, S.; Stief, C.; Waidelich, R. Detection of prostate cancer with complexed PSA and complexed/total PSA ratio: Is there any advantage? Eur. J. Med. Res 2011, 16, 445–450. [Google Scholar]
- Filella, X.; Gimenez, N. Evaluation of [−2]proPSA and Prostate Health Index (phi) for the detection of prostate cancer: A systematic review and meta-analysis. Clin. Chem. Lab. Med 2013, 51, 729–739. [Google Scholar]
- Tosoian, J.J.; Loeb, S.; Feng, Z.Y.; Isharwal, S.; Landis, P.; Elliot, D.J.; Veltri, R.; Epstein, J.I.; Partin, A.W.; Carter, H.B.; et al. Association of [−2]proPSA with biopsy reclassification during active surveillance for prostate cancer. J. Urol 2012, 188, 1131–1136. [Google Scholar]
- Vesprini, D.; Liu, S.; Nam, R. Predicting high risk disease using serum and DNA biomarkers. Curr. Opin. Urol 2013, 23, 252–260. [Google Scholar]
- Pin, E.; Fredolini, C.; Petricoin, E.F. The role of proteomics in prostate cancer research: Biomarker discovery and validation. Clin. Biochem 2013, 46, 524–538. [Google Scholar]
- Ryan, D.; Robards, K.; Prenzler, P.D.; Kendall, M. Recent and potential developments in the analysis of urine: A review. Anal. Chim. Acta 2011, 684, 17–29. [Google Scholar]
- Wang, Y.C.; Chen, Y.M.; Lin, Y.J.; Liu, S.P.; Chiang, E.P.I. GNMT expression increases hepatic folate contents and folate-dependent methionine synthase-mediated homocysteine remethylation. Mol. Med 2011, 17, 486–494. [Google Scholar]
- Yen, C.H.; Lin, Y.T.; Chen, H.L.; Chen, S.Y.; Chen, Y.M.A. The multi-functional roles of GNMT in toxicology and cancer. Toxicol. Appl. Pharmacol 2013, 266, 67–75. [Google Scholar]
- Soriano, A.; Castillo, R.; Christov, C.; Andres, J.; Moliner, V.; Tunon, I. Catalysis in glycine N-methyltransferase: Testing the electrostatic stabilization and compression hypothesis. Biochemistry 2006, 45, 14917–14925. [Google Scholar]
- Velichkova, P.; Himo, F. Methyl transfer in glycine N-methyltransferase. A theoretical study. J. Phys. Chem. B 2005, 109, 8216–8219. [Google Scholar]
- Song, Y.H.; Shiota, M.; Kuroiwa, K.; Naito, S.; Oda, Y. The important role of glycine N-methyltransferase in the carcinogenesis and progression of prostate cancer. Mod. Pathol 2011, 24, 1272–1280. [Google Scholar]
- Ianni, M.; Porcellini, E.; Carbone, I.; Potenzoni, M.; Pieri, A.M.; Pastizzaro, C.D.; Benecchi, L.; Licastro, F. Genetic factors regulating inflammation and DNA methylation associated with prostate cancer. Prostate Cancer Prostatic Dis 2013, 16, 56–60. [Google Scholar]
- Huang, Y.C.; Lee, C.M.; Chen, M.; Chung, M.Y.; Chang, Y.H.; Huang, W.J.S.; Ho, D.M.T.; Pan, C.C.; Wu, T.T.; Yang, S.; et al. Haplotypes, loss of heterozygosity, and expression levels of glycine N-methyltransferase in prostate cancer. Clin. Cancer Res 2007, 13, 1412–1420. [Google Scholar]
- Lee, C.M.; Shih, Y.P.; Wu, C.H.; Chen, Y.M.A. Characterization of the 5′ regulatory region of the human Glycine N-methyltransferase gene. Gene 2009, 443, 151–157. [Google Scholar]
- Mantovani, R. The molecular biology of the CCAAT-binding factor NF-Y. Gene 1999, 239, 15–27. [Google Scholar]
- Nardini, M.; Gnesutta, N.; Donati, G.; Gatta, R.; Forni, C.; Fossati, A.; Vonrhein, C.; Moras, D.; Romier, C.; Bolognesi, M.; et al. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell 2013, 152, 132–143. [Google Scholar]
- Petroni, K.; Kumimoto, R.W.; Gnesutta, N.; Calvenzani, V.; Fornari, M.; Tonelli, C.; Holt, B.F.; Mantovani, R. The promiscuous life of plant nuclear factor Y transcription factors. Plant Cell 2012, 24, 4777–4792. [Google Scholar]
- Dolfini, D.; Minuzzo, M.; Pavesi, G.; Mantovani, R. The short isoform of NF-YA belongs to the embryonic stem cell transcription factor circuitry. Stem Cells 2012, 30, 2450–2459. [Google Scholar]
- Calvenzani, V.; Testoni, B.; Gusmaroli, G.; Lorenzo, M.; Gnesutta, N.; Petroni, K.; Mantovani, R.; Tonelli, C. Interactions and CCAAT-binding of arabidopsis thaliana NF-Y subunits. PLoS One 2012, 7, 1–11. [Google Scholar]
- Luka, Z.; Cerone, R.; Phillips, J.A.; Mudd, S.H.; Wagner, C. Mutations in human glycine N-methyltransferase give insights into its role in methionine metabolism. Hum. Genet 2002, 110, 68–74. [Google Scholar]
- Liao, Y.J.; Liu, S.P.; Lee, C.M.; Yen, C.H.; Chuang, P.C.; Chen, C.Y.; Tsai, T.F.; Huang, S.F.; Lee, Y.H.W.; Chen, Y.M.A. Characterization of a glycine N-methyltransferase gene knockout mouse model for hepatocellular carcinoma: Implications of the gender disparity in liver cancer susceptibility. Int. J. Cancer 2009, 124, 816–826. [Google Scholar]
- Stabler, S.; Koyama, T.; Zhao, Z.G.; Martinez-Ferrer, M.; Allen, R.H.; Luka, Z.; Loukachevitch, L.V.; Clark, P.E.; Wagner, C.; Bhowmick, N.A. Serum methionine metabolites are risk factors for metastatic prostate cancer progression. PLoS One 2011, 6, e22486. [Google Scholar]
- Dahl, M.; Bouchelouche, P.; Kramer-Marek, G.; Capala, J.; Nordling, J.; Bouchelouche, K. Sarcosine induces increase in HER2/neu expression in androgen-dependent prostate cancer cells. Mol. Biol. Rep 2011, 38, 4237–4243. [Google Scholar]
- Metallo, C.M. Expanding the reach of cancer metabolomics. Cancer Prev. Res 2012, 5, 1337–1340. [Google Scholar]
- Burton, C.; Gamagedara, S.; Ma, Y.F. A novel enzymatic technique for determination of sarcosine in urine samples. Anal. Methods 2012, 4, 141–146. [Google Scholar]
- Montrose, D.C.; Zhou, X.K.; Kopelovich, L.; Yantiss, R.K.; Karoly, E.D.; Subbaramaiah, K.; Dannenberg, A.J. Metabolic profiling, a noninvasive approach for the detection of experimental colorectal neoplasia. Cancer Prev. Res 2012, 5, 1358–1367. [Google Scholar]
- Luka, Z.; Pakhomova, S.; Loukachevitch, L.V.; Newcomer, M.E.; Wagner, C. Differences in folate-protein interactions result in differing inhibition of native rat liver and recombinant glycine N-methyltransferase by 5-methyltetrahydrofolate. Bba Proteins Proteomics 2012, 1824, 286–291. [Google Scholar]
- Mitchell, A.D.; Benevenga, N.J. Importance of sarcosine formation in methionine methyl carbon oxidation in rat. J. Nutr 1976, 106, 1702–1713. [Google Scholar]
- Bar-joseph, I.; Pras, E.; Reznik-Wolf, H.; Marek-Yagel, D.; Abu-Horvitz, A.; Dushnitzky, M.; Goldstein, N.; Rienstein, S.; Dekel, M.; Pode-Shakked, B.; et al. Mutations in the sarcosine dehydrogenase gene in patients with sarcosinemia. Hum. Genet 2012, 131, 1805–1810. [Google Scholar]
- Bergeron, F.; Otto, A.; Blache, P.; Day, R.; Denoroy, L.; Brandsch, R.; Bataille, D. Molecular cloning and tissue distribution of rat sarcosine dehydrogenase. Eur. J. Biochem 1998, 257, 556–561. [Google Scholar]
- Trock, B.J. Application of metabolomics to prostate cancer. Urol. Oncol. Semin. Orig. Investig 2011, 29, 572–581. [Google Scholar]
- Spratlin, J.L.; Serkova, N.J.; Eckhardt, S.G. Clinical applications of metabolomics in oncology: A review. Clin. Cancer Res 2009, 15, 431–440. [Google Scholar]
- Kami, K.; Fujimori, T.; Sato, H.; Sato, M.; Yamamoto, H.; Ohashi, Y.; Sugiyama, N.; Ishihama, Y.; Onozuka, H.; Ochiai, A.; et al. Metabolomic profiling of lung and prostate tumor tissues by capillary electrophoresis time-of-flight mass spectrometry. Metabolomics 2013, 9, 444–453. [Google Scholar]
- Cheng, L.L.; Wu, C.; Smith, M.R.; Gonzalez, R.G. Non-destructive quantitation of spermine in human prostate tissue samples using HRMAS 1H-NMR spectroscopy at 9.4 T. FEBS Lett 2001, 494, 112–116. [Google Scholar]
- Swanson, M.G.; Zektzer, A.S.; Tabatabai, Z.L.; Simko, J.; Jarso, S.; Keshari, K.R.; Schmitt, L.; Carroll, P.R.; Shinohara, K.; Vigneron, D.B.; et al. Quantitative analysis of prostate metabolites using 1H HR-MAS spectroscopy. Magn. Reson. Med 2006, 55, 1257–1264. [Google Scholar]
- DeFeo, E.M.; Wu, C.L.; McDougal, W.S.; Cheng, L.L. A decade in prostate cancer: From NMR to metabolomics. Nat. Rev. Urol 2011, 8, 301–311. [Google Scholar]
- Lucarelli, G.; Fanelli, M.; Larocca, A.M.V.; Germinario, C.A.; Rutigliano, M.; Vavallo, A.; Selvaggi, F.P.; Bettocchi, C.; Battaglia, M.; Ditonno, P. Serum sarcosine increases the accuracy of prostate cancer detection in patients with total serum PSA less than 4.0 ng/mL. Prostate 2012, 72, 1611–1621. [Google Scholar]
- Shaughnessy, J.D.; Barlogie, B. Interpreting the molecular biology and clinical behavior of multiple myeloma in the context of global gene expression profiling. Immunol. Rev 2003, 194, 140–163. [Google Scholar]
- Bohm, L.; Serafin, A.M.; Fernandez, P.; van der Watt, G.; Bouic, P.J.D.; Harvey, J. Plasma sarcosine does not distinguish early and advanced stages of prostate cancer. SAMJ S. Afr. Med. J 2012, 102, 677–679. [Google Scholar]
- Chen, J.; Zhou, L.N.; Zhang, X.Y.; Lu, X.; Cao, R.; Xu, C.J.; Xu, G.W. Urinary hydrophilic and hydrophobic metabolic profiling based on liquid chromatography-mass spectrometry methods: Differential metabolite discovery specific to ovarian cancer. Electrophoresis 2012, 33, 3361–3369. [Google Scholar]
- Schuller, D.J.; Reisch, C.R.; Moran, M.A.; Whitman, W.B.; Lanzilotta, W.N. Structures of dimethylsulfoniopropionate-dependent demethylase from the marine organism Pelagabacter ubique. Protein Sci 2012, 21, 289–298. [Google Scholar]
- Cernei, N.; Masarik, M.; Gumulec, J.; Zitka, O.; Babula, P.; Kizek, R. Determination of sarcosine as possible tumour marker of prostate tumours. J. Biochem. Techol 2010, 2, S7–S8. [Google Scholar]
- Jiang, Y.Q.; Cheng, X.L.; Wang, C.A.; Ma, Y.F. Quantitative determination of sarcosine and related compounds in urinary samples by liquid chromatography with tandem mass spectrometry. Anal. Chem 2010, 82, 9022–9027. [Google Scholar]
- Cernei, N.; Zitka, O.; Ryvolova, M.; Adam, V.; Masarik, M.; Hubalek, J.; Kizek, R. Spectrometric and electrochemical analysis of sarcosine as a potential prostate carcinoma marker. Int. J. Electrochem. Sci 2012, 7, 4286–4301. [Google Scholar]
- LeBoucher, J.; Charret, C.; CoudrayLucas, C.; Giboudeau, J.; Cynober, L. Amino acid determination in biological fluids by automated ion-exchange chromatography: Performance of Hitachi L-8500A. Clin. Chem 1997, 43, 1421–1428. [Google Scholar]
- Khan, A.P.; Rajendiran, T.M.; Ateeq, B.; Asangani, I.A.; Athanikar, A.R.; Yocum, A.K.; Mehra, R.; Siddiqui, J.; Palapattu, G.; Wei, J.T.; et al. The role of sarcosine metabolism in prostate cancer progression. Neoplasia 2013, 15, 491–501. [Google Scholar]
- Jentzmik, F.; Stephan, C.; Miller, K.; Schrader, M.; Erbersdobler, A.; Kristiansen, G.; Lein, M.; Jung, K. Sarcosine in urine after digital rectal examination fails as a marker in prostate cancer detection and identification of aggressive tumours. Eur. Urol. 2010, 58, 12–18. [Google Scholar]
- Struys, E.A.; Heijboer, A.C.; van Moorselaar, J.; Jakobs, C.; Blankenstein, M.A. Serum sarcosine is not a marker for prostate cancer. Ann. Clin. Biochem 2010, 47, 282–282. [Google Scholar]
- Wu, H.; Liu, T.T.; Ma, C.G.; Xue, R.Y.; Deng, C.H.; Zeng, H.Z.; Shen, X.Z. GC/MS-based metabolomic approach to validate the role of urinary sarcosine and target biomarkers for human prostate cancer by microwave-assisted derivatization. Anal. Bioanal. Chem 2011, 401, 635–646. [Google Scholar]
- Bianchi, F.; Dugheri, S.; Musci, M.; Bonacchi, A.; Salvadori, E.; Arcangeli, G.; Cupelli, V.; Lanciotti, M.; Masieri, L.; Serni, S.; et al. Fully automated solid-phase microextraction-fast gas chromatography-mass spectrometry method using a new ionic liquid column for high-throughput analysis of sarcosine and N-ethylglycine in human urine and urinary sediments. Anal. Chim. Acta 2011, 707, 197–203. [Google Scholar]
- Cao, D.L.; Ye, D.W.; Zhu, Y.; Zhang, H.L.; Wang, Y.X.; Yao, X.D. Efforts to resolve the contradictions in early diagnosis of prostate cancer: A comparison of different algorithms of sarcosine in urine. Prostate Cancer Prostatic Dis 2011, 14, 166–172. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).