Metasin—An Intra-Operative RT-qPCR Assay to Detect Metastatic Breast Cancer in Sentinel Lymph Nodes
Abstract
:1. Introduction
2. Results
2.1. Patient Demographics
2.2. Characteristics of the PCR Assay
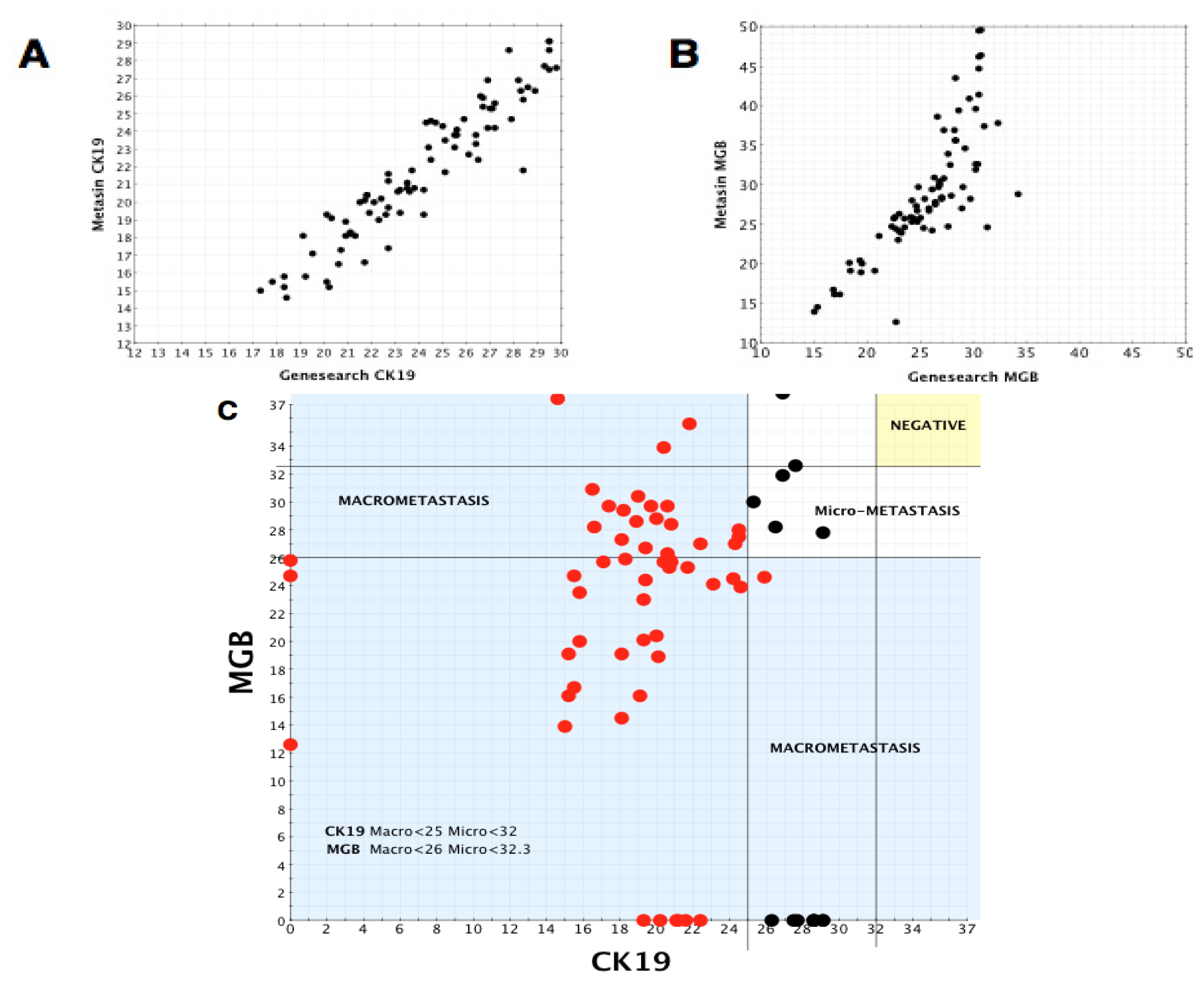
2.3. Cut-Off Values for Positive and Negative Nodes and Macro- and Micro-Metastases
2.4. Comparison of Metasin with Histology
2.5. Comparison of Metasin with GeneSearch
2.6. Statistical Analysis: Comparison of Cq Values of Metasin with GeneSearch
2.7. Independent Verification of Positive/Negative Macro/Micro Cut-Offs for Metasin
2.8. Statistical Analysis of JBI Samples
2.9. CK19 and MGB IHC
Limits of the Detection of Metasin: Copy Number and Cell Count Number Determination
3. Discussion
- amplification products run on agarose gels showed only a single band of the correct size (128 bp).
- amplification of 100 DNA samples with the Metasin assay did not result in any detectable fluorescence signals and gel electrophoresis of amplified material yielded a series of smears.
- We draw on our observations relating to the low discordant rates of the Metasin assay having analysed over 1700 cases maintaining an overall discordant rate under 4 percent.
4. Experimental Section
4.1. Ethical Approval/PAH Samples
4.2. Histopathological Processing and Assessment of Sentinel Lymph Nodes
4.3. Node Processing for GeneSearch/Metasin
4.4. Ethical Approval/JBI RNA
4.5. Quantitation and Identification of Template (RNA and DNA)
4.6. Primer and Probe Sequences
4.7. RT-PCR Assay
4.8. Positive and Negative Controls
4.9. PCR Efficiency and Copy Number Calculations
4.10. Cut-Off Cq Value for CK19 and MGB for Positive and Negative Nodes
4.11. Statistical Analysis
4.12. CK19 and MGB IHC on Breast Cores
5. Conclusions
Supplementary Information
CK19 Pseudogene Alignments. CK19 Pseudogene alignments [18] (A) Sequence alignment of members of the CK19 gene family. CK19. Primer alignment (B & D) shows several mismatches with both the forward and reverse primer binding sequences. The detection probe sequence (C) also has a number of mismatches with pseudogene target sequences.
| Case No | Metasin | GeneSearch BLNA | Histopathology | Node-based Concordance or Discordance | Case-based- Concordance/discordance | Notes | |
|---|---|---|---|---|---|---|---|
| Initial Analysis | Deeper Levels | ||||||
| 1 16837 | Negative | Macro-metastasis | Macro-metastasis | NA | Macro-metastasis Discordant | Macro-metastasis Discordant | 1 2008 case-earliest batch of cases (? RNA degradation) Macro detected in one of 2 slices given for histology in 2 levels only. Not present in third slice or in deeper level |
| 2 7587B | Negative | Negative | Macro-metastasis | NA | Macro-metastasis Discordant | Concordant | 2 Another lymph node from this case showed both histological and molecular evidence of metastatic disease-hence not a true discordant case. 3 Macro-metastasis 3 mm tumour in 1 slice |
| 3 12395 | Negative | Negative | 3Macro-metastasis 3 mm in 1 slice | NA | Macro-metastasis Discordant | Macro-metastasis Discordant | 3 Macro-metastasis 3 mm tumour in 1 slice |
| 4 12082 | Positive Micromet | Negative | Negative | Negative | Micro-metastasis Discordant | Micro-metastasis Discordant | 4 Micro met MGB 31.5, Metasin Cq for CK19 is 29.9 (Veridex cut off is 30) |
| 6 9041B | 6 Positive Micromet | 6 Positive Micromet | Negative | Negative | Micro-metastasis Discordant | Micro-metastasis Discordant | 6 9041B Cp MGB is 30.4, Metasin 29.8. Positive independent of Metasin on GeneSearch BLN Assay |
| 7 8489 | Positive Micromet | Positive Micromet | Negative | 7 ITC | Micro-metastasis Discordant | 7ITC’s present in axillary clearance | 7 ITCs seen in deeper levels and in axillary clearance specimen. Positive independent of Metasin on GeneSearch BLN Assay |
| 8 10931B | Positive Macro | Positive Micromet | Negative | Negative | Micro/Macro metastasis Discordant | Micro/Macro metastasis Discordant | 8 Positive independent of Metasin on GeneSearch BLN Assay |
Acknowledgments
Conflict of Interest
References
- Veronesi, U.; Paganelli, G.; Galimberti, V.; Viale, G.; Zurrida, S.; Bedoni, M.; Costa, A.; de Cicco, C.; Geraghty, J.G.; Luini, A.; et al. Sentinel node biopsy to avoid axillary dissection in breast cancer with clinically negative lymph nodes. Lancet 1997, 349, 1864–1867. [Google Scholar]
- Veronesi, U.; Paganelli, G.; Viale, G.; Galimberti, V.; Luini, A.; Zurrida, S.; Robertson, C.; Sacchini, V.; Veronesi, P.; Orvieto, E.; et al. Sentinel lymph node biopsy and axillary dissection in breast cancer: Results in a large series. J. Nat. Cancer Inst 1999, 91, 368–373. [Google Scholar]
- Lyman, G.H.; Giuliano, A.E.; Somerfield, M.R.; Benson, A.B., III; Bodurka, D.C.; Burstein, H.J.; Cochran, A.J.; Cody, H.S., III; Edge, S.B.; Galper, S.; et al. American Society of Clinical Oncology guideline recommendations for sentinel lymph node biopsy in early-stage breast cancer. J. Clin. Oncol. Off. J. 2005, 23, 7703–7720. [Google Scholar]
- MacNeill, F.A. New start: The UK SLNB training programme–A progress report. Ann. R. Coll Surg. Engl 2007, 89, 60–61. [Google Scholar]
- Mansel, R.E.; Goyal, A.; Douglas-Jones, A.; Woods, V.; Goyal, S.; Monypenny, I.; Sweetland, H.; Newcombe, R.G.; Jasani, B. Detection of breast cancer metastasis in sentinel lymph nodes using intra-operative real time GeneSearch BLN Assay in the operating room: Results of the Cardiff study. Breast Cancer Res. Treat 2009, 115, 595–600. [Google Scholar]
- Krishnamurthy, S. A prospective study comparing touch imprint cytology, frozen section analysis and rapid cytokeratin immunostain for intraoperative evaluation of axillary sentinel lymph nodes in breast cancer. Cancer 2009, 115, 1555–1562. [Google Scholar]
- Liu, L.-C. Intraoperative frozen section analysis of sentinel lymph nodes in breast cancer patients, a meta-analysis and single-institution experience. Cancer 2011, 117, 250–258. [Google Scholar]
- Lumachi, F.; Marino, F.; Zanella, S.; Chiara, G.B.; Basso, S.M. Touch imprint cytology and frozen-section analysis for intraoperative evaluation of sentinel nodes in early breast cancer. Anticancer Res 2012, 32, 3523–3526. [Google Scholar]
- Blumencranz, P.; Whitworth, P.W.; Deck, K.; Rosenberg, A.; Reintgen, D.; Beitsch, P.; Chagpar, A.; Julian, T.; Saha, S.; Mamounas, E.; et al. Scientific impact recognition award. sentinel node staging for breast cancer: Intraoperative molecular pathology overcomes conventional histologic sampling errors. Am. J. Surg 2007, 194, 426–432. [Google Scholar]
- Martin Martinez, M.D.; Veys, I.; Majjaj, S.; Lespagnard, L.; Schobbens, J.C.; Rouas, G.; Filippov, V.; Noterman, D.; Hertens, D.; Feoli, F.; et al. Clinical validation of a molecular assay for intra-operative detection of metastases in breast sentinel lymph nodes. Eur. J. Surg. Oncol. J. Eur. Soc. Surg. Oncol. Br. Assoc. Surg. Oncol 2009, 35, 387–392. [Google Scholar]
- Viale, G.; Dell’Orto, P.; Biasi, M.O.; Stufano, V.; De Brito Lima, L.N.; Paganelli, G.; Maisonneuve, P.; Vargo, J.M.; Green, G.; Cao, W.; et al. Comparative evaluation of an extensive histopathologic examination and a real-time reverse-transcription-polymerase chain reaction assay for mammaglobin and cytokeratin 19 on axillary sentinel lymph nodes of breast carcinoma patients. Ann. Surg 2008, 247, 136–142. [Google Scholar]
- Healthcare Industry & Policy Community. Available online: http://www.wellsphere.com/healthcare-industry-policy-article/johnson-and-johnson-pulls-genesearch-breast-cancer-diagnosis-test-8211-trials-ended/944105 (on accessed 17 May 2013).
- Singletary, S.E.; Greene, F.L. Revision of breast cancer staging: The 6th edition of the TNM Classification. Semin. Surg. Oncol 2003, 21, 53–59. [Google Scholar]
- Singletary, S.E.; Greene, F.L.; Sobin, L.H. Classification of isolated tumor cells: Clarification of the 6th edition of the American Joint Committee on Cancer Staging Manual. Cancer 2003, 98, 2740–2741. [Google Scholar]
- Sobin, L.; Gospodarowicz, M.; Wittekind, C.H. UICC TNM Classification of Malignant Tumours; John Wiley & Sons: New York, NY, USA, 2009. [Google Scholar]
- Alvarenga, C.A.; Paravidino, P.I.; Alvarenga, M.; Dufloth, R.; Gomes, M.; Zeferino, L.C.; Schmitt, F. Expression of CK19 in invasive breast carcinomas of special histological types: Implications for the use of one-step nucleic acid amplification. J. Clin. Pathol 2011, 64, 493–497. [Google Scholar]
- Vilardell, F.; Novell, A.; Martin, J.; Santacana, M.; Velasco, A.; Diez-Castro, M.J.; Cuevas, D.; Panades, M.J.; Gonzalez, S.; Llombart, A.; et al. Importance of assessing CK19 immunostaining in core biopsies in patients subjected to sentinel node study by OSNA. Virchows Archiv. An Int. J. Pathol 2012, 460, 569–575. [Google Scholar]
- Zhang, Z.; Harrison, P.M.; Liu, Y.; Gerstein, M. Millions of years of evolution preserved: A comprehensive catalog of the processed pseudogenes in the human genome. Genome Res 2003, 13, 2541–2558. [Google Scholar]
- Cserni, G.; Bianchi, S.; Vezzosi, V.; Peterse, H.; Sapino, A.; Arisio, R.; Reiner-Concin, A.; Regitnig, P.; Bellocq, J.P.; Marin, C.; et al. The value of cytokeratin immunohistochemistry in the evaluation of axillary sentinel lymph nodes in patients with lobular breast carcinoma. J. Clin. Pathol 2006, 59, 518–522. [Google Scholar]
- Pathology Reporting of Breast Disease. Available online: http://www.cancerscreening.nhs.uk/breastscreen/publications/nhsbsp58-low-resolution.pdf (on accessed 21 May 2013).
- Cserni, G. Metastases in axillary sentinel lymph nodes in breast cancer as detected by intensive histopathological work up. J. Clin. Pathol 1999, 52, 922–924. [Google Scholar]
- Cserni, G. Intraoperative analysis of sentinel lymph nodes in breast cancer by one-step nucleic acid amplification. J. Clin. Pathol 2012, 65, 193–199. [Google Scholar]
- Maiorano, E.; Mazzarol, G.M.; Pruneri, G.; Mastropasqua, M.G.; Zurrida, S.; Orvieto, E.; Viale, G. Ectopic breast tissue as a possible cause of false-positive axillary sentinel lymph node biopsies. Am. J. Surg. Pathol 2003, 27, 513–518. [Google Scholar]
- Turner, D.R.; Millis, R.R. Breast tissue inclusions in axillary lymph nodes. Histopathology 1980, 4, 631–636. [Google Scholar]
- McCluggage, W.G.; el-Agnaff, M.; O’Hara, M.D. Cytokeratin positive T cell malignant lymphoma. J. Clin. Pathol 1998, 51, 404–406. [Google Scholar]
- McCluggage, W.G.; Walsh, M.Y.; Bharucha, H. Anaplastic large cell malignant lymphoma with extensive eosinophilic or neutrophilic infiltration. Histopathology 1998, 32, 110–115. [Google Scholar]
- Cserni, G.; Bianchi, S.; Vezzosi, V.; van Diest, P.; van Deurzen, C.; Sejben, I.; Regitnig, P.; Asslaber, M.; Foschini, M.P.; Sapino, A.; et al. Variations in sentinel node isolated tumour cells/micrometastasis and non-sentinel node involvement rates according to different interpretations of the TNM definitions. Eur. J. Cancer 2008, 44, 2185–2191. [Google Scholar]
- Edge, S.B.; Byrd, D.R.; Compton, C.C. Cancer Staging Handbook: From the AJCC Cancer Staging Manual; Springer: New York, NY, USA, 2009. [Google Scholar]
- Cody, H.S., 3rd; Fey, J.; Akhurst, T.; Fazzari, M.; Mazumdar, M.; Yeung, H.; Yeh, S.D.; Borgen, P.I. Complementarity of blue dye and isotope in sentinel node localization for breast cancer: Univariate and multivariate analysis of 966 procedures. Ann. Surg. Oncol. 2001, 8, 13–19. [Google Scholar]
- Prieto, V.G.; McNutt, N.S. Immunohistochemical detection of keratin with the monoclonal antibody MNF116 is useful in the diagnosis of epidermolysis bullosa simplex. J. Cutan. Pathol 1994, 21, 118–122. [Google Scholar]
- GeneSearch Breast Lymph Node (BLN) Test Kit. Instructions for use. Available online: http://www.fda.gov/ohrms/dockets/ac/06/briefing/2006-4249b1_02.pdf (on accessed 17 May 2013).
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem 2009, 55, 611–622. [Google Scholar]
- Soule, H.D.; Vazguez, J.; Long, A.; Albert, S.; Brennan, M. A human cell line from a pleural effusion derived from a breast carcinoma. J. Natl. Cancer Inst 1973, 51, 1409–1416. [Google Scholar]


| Nodal status | All patients | Total | Type of tumour | Nodal status | Total | ||
|---|---|---|---|---|---|---|---|
| Patients Enrolled age, years | 156 | IDC | positive SN | 26 | 103 | ||
| negative SN | 74 | ||||||
| mean age at diagnosis | positive SN | 60.2 | 62.2 | discordant SN | 3 | ||
| negative SN | 63.2 | ILC | positive SN | 6 | 16 | ||
| discordant SN | 60.2 | negative SN | 9 | ||||
| median age at diagnosis | positive SN | 59 | discordant SN | 1 | |||
| negative SN | 63 | 63 | Others | positive SN | 6 | 32 | |
| discordant SN | 62 | negative SN | 24 | ||||
| age range | positive SN | 39–92 | discordant SN | 2 | |||
| negative SN | 34–87 | 34–92 | DCIS | positive SN | 0 | 11 | |
| discordant SN | 43–68 | negative SN | 10 | ||||
| Surgery Performed | discordant SN | 1 | |||||
| Mastectomy | positive SN | 18 | positive SN | 0 | |||
| negative SN | 18 | 42 | LCIS | negative SN | 1 | 1 | |
| discordant SN | 6 | discordant SN | 0 | ||||
| Wide local excision | positive SN | 18 | 100 | Hormone receptor status | |||
| discordant SN | 0 | negative SN | 17 | ||||
| Tumour Size (TNM) (15) | discordant SN | 0 | |||||
| pT1 (<2 cm) | positive SN | 18 | positive SN | 7 | |||
| negative SN | 61 | 79 | Mean ER score | negative SN | 6 | 7 | |
| discordant SN | 0 | discordant SN | 7 | ||||
| pT2 (>2 cm, <5 cm) | positive SN | 15 | positive SN | 27 | |||
| negative SN | 28 | 46 | PR positive | negative SN | 71 | 103 | |
| discordant SN | 3 | discordant SN | 5 | ||||
| pT3 (>5 cm) | positive SN | 2 | positive SN | 10 | |||
| negative SN | 2 | 4 | PR negative | negative SN | 34 | 45 | |
| discordant SN | 0 | discordant SN | 1 | ||||
| Tumour Grade | Mean PR score | positive SN | 6 | 5 | |||
| Grade 1 | positive SN | 5 | negative SN | 5 | |||
| negative SN | 15 | 20 | Her2 positive | discordant SN | 5 | ||
| discordant SN | 0 | positive SN | 4 | 13 | |||
| Grade 2 | positive SN | 20 | 78 | negative SN | 8 | ||
| negative SN | 55 | Her2 negative | discordant SN | 1 | |||
| discordant SN | 3 | positive SN | 33 | 127 | |||
| Grade 3 | positive SN | 12 | 29 | negative SN | 90 | ||
| negative SN | 16 | Triple negative | discordant SN | 4 | |||
| discordant SN | 1 | positive SN | 1 | 14 | |||
| Ungradeable | positive SN | 0 | 10 | negative SN | 13 | ||
| negative SN | 9 | discordant SN | 0 | ||||
| discordant SN | 1 | ||||||
| Concordant cases | Discordant cases | Total | Discordant cases | |||
|---|---|---|---|---|---|---|
| H+ M+ | H− M− | H+ M− | H− M+ | |||
| Initial analysis | 33 (21.4%) | 111 (7.2%) | 2 (1.3%) | 8 (5.2%) | 154 | 10 (6.49%) |
| Deeper levels | 37 (24%) | 111 (77%) | 2 (1.3%) | 4 (2.6%) | 154 | 6 (3.89%) |
| Metasin vs. GeneSearch A | Metasin vs. Histology | ||
|---|---|---|---|
| Node Based-Analysis | Node Based-Analysis | ||
| Positive nodes | 64 (18.2%) | Positive nodes | 61 (17.4) |
| Negative nodes | 280 (80%) | Negative nodes | 280 (80%) |
| Discordant nodes | 6 (1.7%) | Discordant nodes | 9 (2.5%) |
| Total | 350 | Total | 350 |
| Case Based-Analysis | Case Based-Analysis | ||
| Positive patients | 37 (24%) | Positive patients | 37 (24%) |
| Negative patients | 111 (72%) | Negative patients | 111 72% |
| Discordant patients | 6 (3.89%) | Discordant patients | 6 (3.89%) |
| Total | 154 | Total | 154 |
| Metasin | Total | ||
|---|---|---|---|
| Histology | Macro | Micro | |
| Macro | 50 (81.9%) | 2 (3.2%) | 52 (85.2) |
| Micro | 1 (1.6%) | 8 (13%) | 9 (14.7) |
| Total | 51 (84%) | 10 (16.4%) | 61 (100%) |
| Node based discordance | |||||
|---|---|---|---|---|---|
| Concordant | Discordant | Total | Discordant nodes | ||
| 1 G+ 2 M+ | G− M− | G+ M− | G− M+ | ||
| 64 (18.2%) | 280 (80%) | 2 (0.6%) | 4 (1.14%) | 350 | 6 (1.17%) |
| Case based discordance | |||||
| Concordant | Discordant | Total | Discordant cases | ||
| G+ M+ | G− M− | G+ M− | G− M+ | ||
| 37 (24%) | 111 (72%) | 2 (1.2%) | 4 (2.6%) | 154 | 6 (3.89%) |
| Genesearch analysed nodes from JBI 1 | Total | |||
|---|---|---|---|---|
| Macro-Metastasis 1 | Micro-metastasis | Negative Nodes | ||
| Metasin Positive Nodes 2 | 56 (29%) | 18 (9.3%) | – | 74 (38.3%) |
| Metasin Negative Nodes | – | 15 (7.7%) | 104 (54%) | 119 (61.7%) |
| Total | 56 (29%) | 33 (17.1%) | 104 (53.9%) | 193 (100%) |
| Marker | Hydrolysis probe | Amplicon length | Forward primer 5′ to 3′ | Reverse primer 5′ to 3′ |
|---|---|---|---|---|
| PBGD 1 | ctcctgaactccagatgcggga | 92 | tgtggtgggaaccagctc | tgttgaggtttccccgaat |
| CK19 2 | cagccagacgggcattgtcg | 128 | cagccactactacacgaccatc | caaacttggttcggaagtcatc |
| MGB 3 | ctctggctgccccttattggag | 69 | ctcccagcactgctacgc | ggattgattgtcttggaaatcaca |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Al-Ramadhani, S.; Sai-Giridhar, P.; George, D.; Gopinath, P.; Arkoumani, E.; Jader, S.; Sundaresan, M.; Salgado, R.; Larsimont, D.; Bustin, S.A.; et al. Metasin—An Intra-Operative RT-qPCR Assay to Detect Metastatic Breast Cancer in Sentinel Lymph Nodes. Int. J. Mol. Sci. 2013, 14, 12931-12952. https://doi.org/10.3390/ijms140712931
Al-Ramadhani S, Sai-Giridhar P, George D, Gopinath P, Arkoumani E, Jader S, Sundaresan M, Salgado R, Larsimont D, Bustin SA, et al. Metasin—An Intra-Operative RT-qPCR Assay to Detect Metastatic Breast Cancer in Sentinel Lymph Nodes. International Journal of Molecular Sciences. 2013; 14(7):12931-12952. https://doi.org/10.3390/ijms140712931
Chicago/Turabian StyleAl-Ramadhani, Salma, Priya Sai-Giridhar, Dilushana George, Preethi Gopinath, Evdokia Arkoumani, Samar Jader, Maryse Sundaresan, Roberto Salgado, Dennis Larsimont, Stephen A. Bustin, and et al. 2013. "Metasin—An Intra-Operative RT-qPCR Assay to Detect Metastatic Breast Cancer in Sentinel Lymph Nodes" International Journal of Molecular Sciences 14, no. 7: 12931-12952. https://doi.org/10.3390/ijms140712931
APA StyleAl-Ramadhani, S., Sai-Giridhar, P., George, D., Gopinath, P., Arkoumani, E., Jader, S., Sundaresan, M., Salgado, R., Larsimont, D., Bustin, S. A., & Sundaresan, V. (2013). Metasin—An Intra-Operative RT-qPCR Assay to Detect Metastatic Breast Cancer in Sentinel Lymph Nodes. International Journal of Molecular Sciences, 14(7), 12931-12952. https://doi.org/10.3390/ijms140712931





