Cloning, Characterization and Effect of TmPGRP-LE Gene Silencing on Survival of Tenebrio Molitor against Listeria monocytogenes Infection
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characterization of TmPGRP-LE Full-Length cDNA
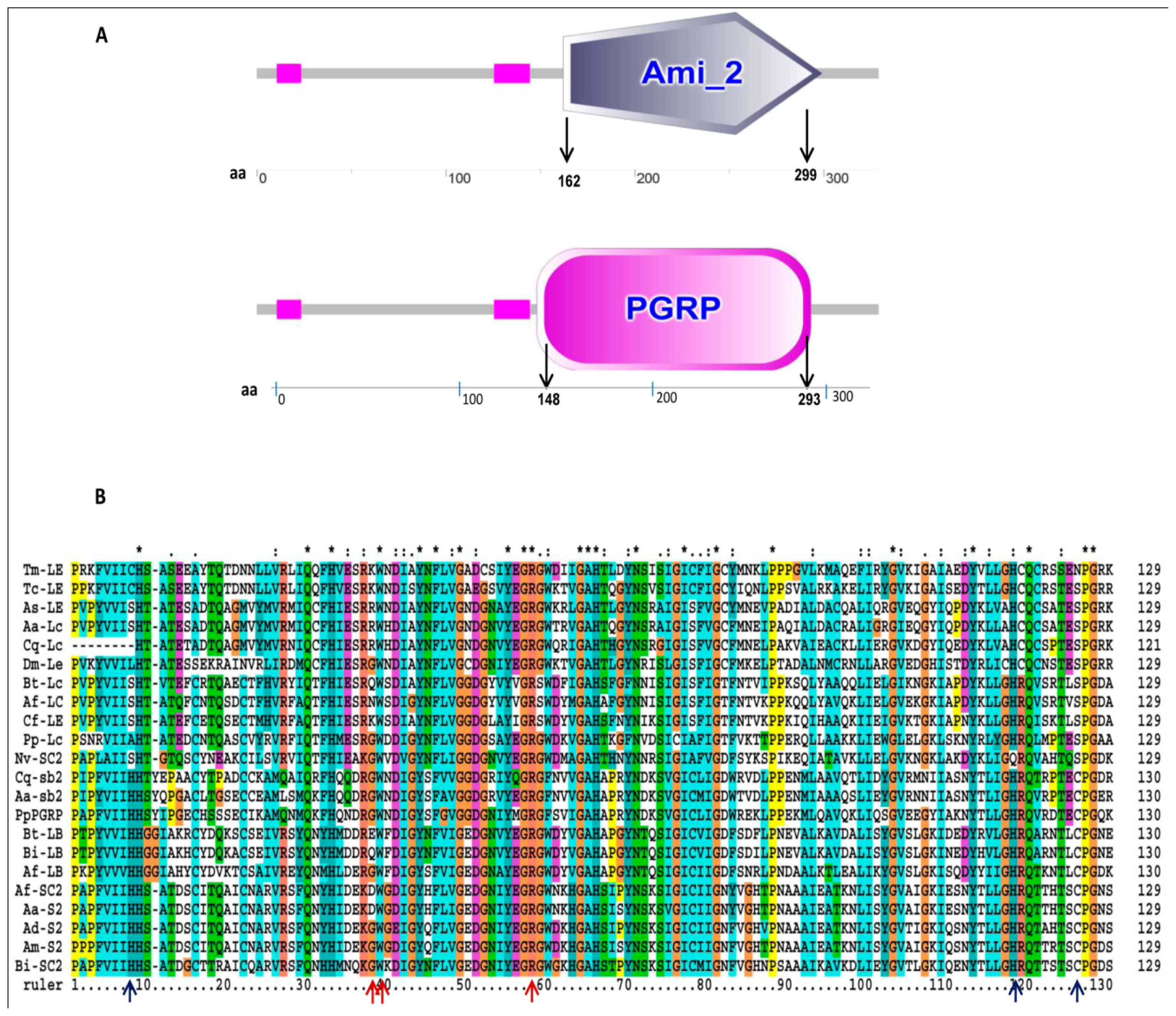
2.2. Phylogenetic Analysis of TmPGRP-LE
2.3. Secondary Structure and Homology Modeling of TmPGRP-LE
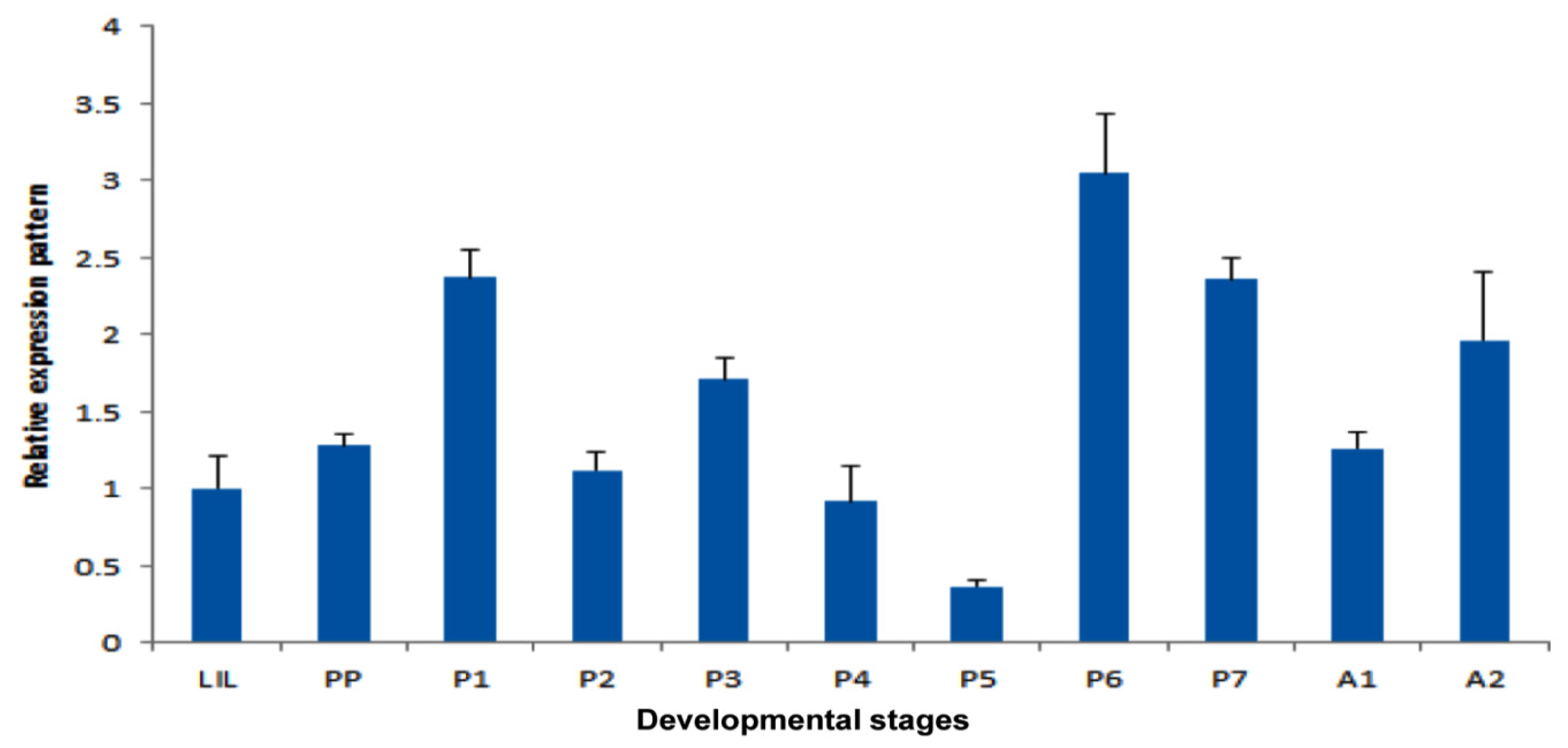
2.4. Temporal Expression Patterns of TmPGRP-LE
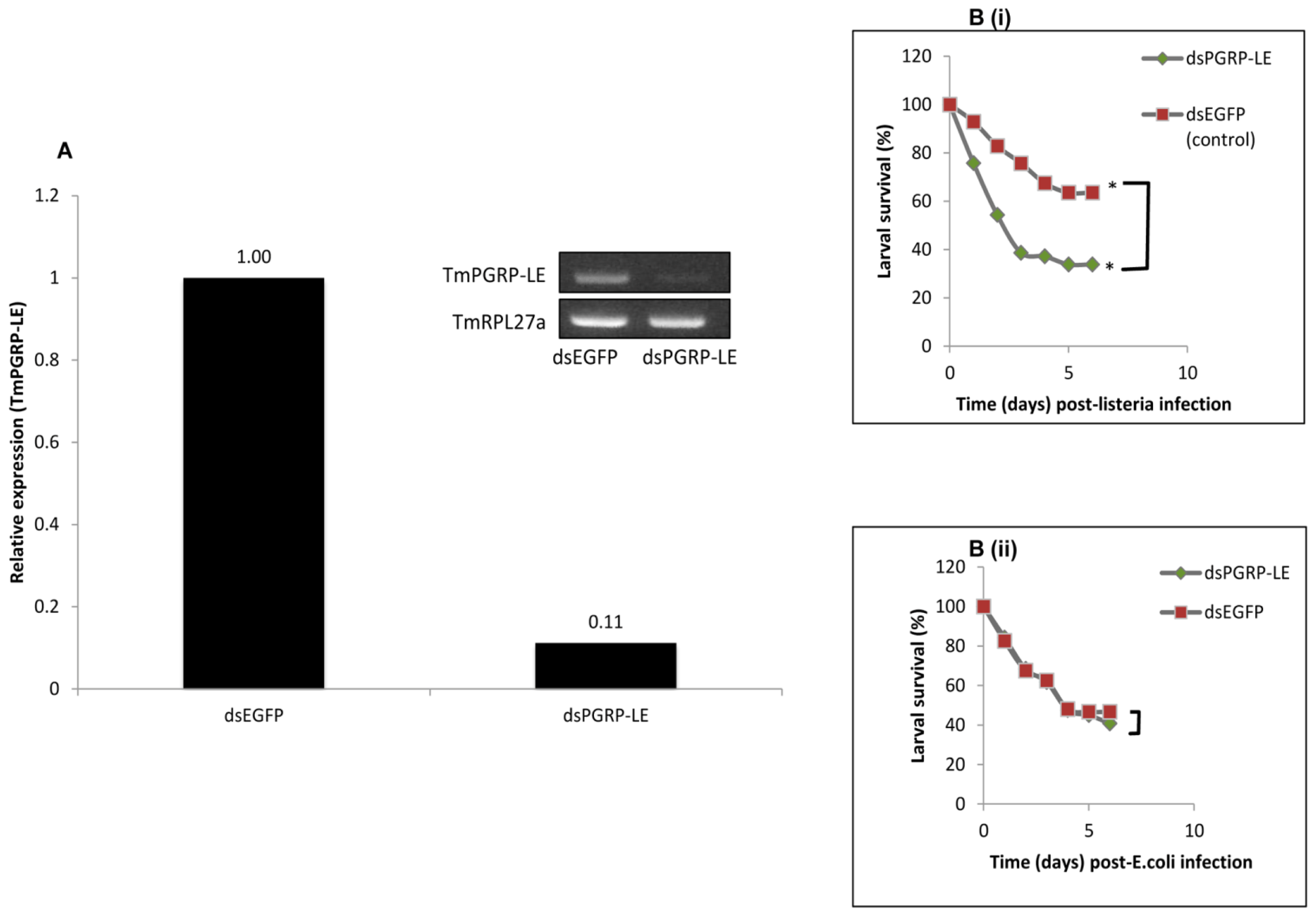
2.5. RNAi-Mediated Knockdown of TmPGRP-LE Leads to Reduced Survival Rate of T. molitor Larvae infected with L. monocytogenes
3. Experimental Section
3.1. Insect Collection and Maintenance
3.2. Chemicals
3.3. Bacterial Strains and Media
3.4. Construction of Full-Length Enriched cDNA Library of T. molitor Larvae
3.5. TmPGRP-LE Sequence Analysis
3.6. Phylogenetic Analysis
3.7. Secondary Structure Prediction and Modeling
3.8. Developmental Expression Pattern of TmPGRP-LE
3.9. RNA Interference of TmPGRP-LE
3.10. Bacterial Injections and Bioassays
3.11. Statistical Analysis
4. Conclusions
Supplementary Information
ijms-14-22462-s001.pdf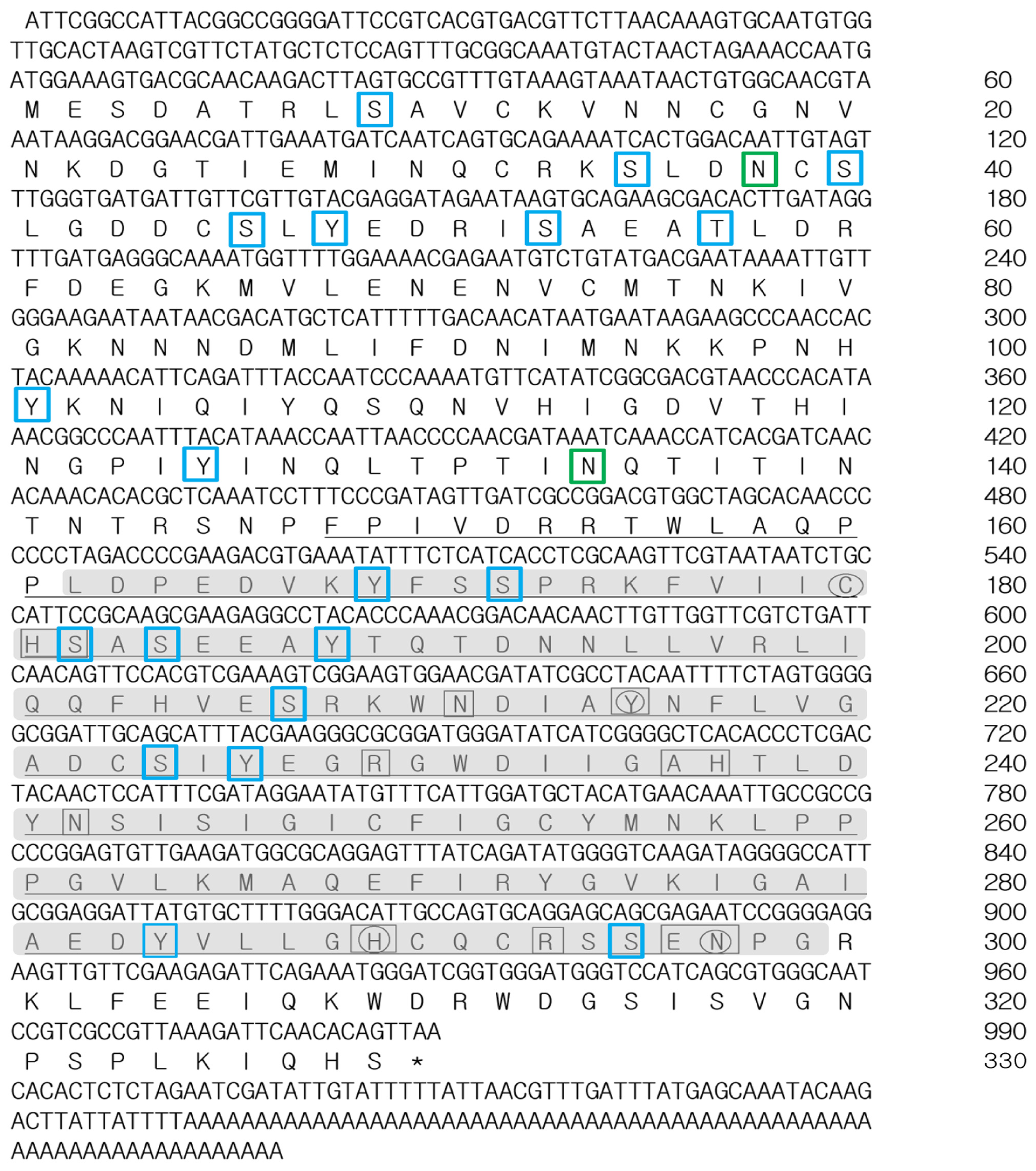

| Primer name | Sequence (5′-3′) |
|---|---|
| Oligo (dT) adaptor | GGCCACGCGTCGACTAGTACT17 |
| M13-F (forward) | GTAAAACGACGGCCAG |
| M13-R (reverse) | CAGGAAACAGCTATGAC |
| RpL27a (forward) | TCATCCTGAAGGCAAAGCTCCAGT |
| RpL27a (reverse) | AGGTTGGTTAGGCAGGCACCTTTA |
| TmPGRP-LE Real-time PCR (Forward) | CTTCGCTTGCGGAATGGCAGATTA |
| TmPGRP-LE Real-time PCR (Reverse) | AACACACGCTCAAATCCTTTCCCG |
| TmPGRP-LE dsRNA (forward) | TAATACGACTCACTATAGGGAGAGGCAACGTAAATAAGGACGG |
| TmPGRP-LE dsRNA (Reverse) | TAATACGACTCACTATAGGGAGAGTAGGCGATATCGTTCCACTTC |
Acknowledgments
Conflicts of Interest
References
- Yano, T.; Kurata, S. Intracellular recognition of pathogens and autophagy as an innate immune host defence. J. Biochem 2011, 150, 143–149. [Google Scholar]
- Schleifer, K.H.; Stackerbrandt, E. Molecular systematics of prokaryotes. Annu. Rev. Microbiol 1983, 37, 143–187. [Google Scholar]
- Dziarski, R.; Gupta, D. The peptidoglycan recognition proteins (PGRPs). Genome Biol 2006. [Google Scholar] [CrossRef]
- Kang, D.; Liu, G.; Lundstrom, A.; Gelius, E.; Steiner, H. A peptidoglycan recognition protein in innate immunity conserved from insects to humans. Proc. Natl. Acad. Sci. USA 1998, 95, 10078–10082. [Google Scholar]
- Yoshida, H.; Kinoshita, K.; Ashida, M. Purification of a peptidoglycan recognition protein from hemolymph of the silkwormBombyx mori. J. Biol. Chem 1996, 271, 13854–13860. [Google Scholar]
- Dziarski, R.; Gupta, D. Mammalian PGRPs: Novel antibacterial proteins. Cell. Microbiol 2006, 8, 1059–1069. [Google Scholar]
- Ni, D.; Song, L.; Wu, L.; Chang, Y.; Yu, Y.; Qiu, L.; Wang, L. Molecular cloning and mRNA expression of peptidoglycan recognition protein (PGRP) gene in bay scallop (Argopecten irradians, Lamarck 1819). Dev. Comp. Immunol 2007, 31, 548–558. [Google Scholar]
- Lemaitre, B.; Hoffmann, J.A. The host defense of Drosophila melanogaster. Annu. Rev. Immunol 2007, 25, 697–743. [Google Scholar]
- Yano, T.; Mita, S.; Ohmori, H.; Oshima, Y.; Fujimoto, Y.; Ueda, R.; Takada, H.; Goldman, W.E.; Fukase, K.; Silverman, N.; et al. Autophagic control of Listeria through intracellular innate immune recognition in Drosophila. Nat. Immunol 2008, 9, 908–916. [Google Scholar]
- Kim, M.S.; Byun, M.; Oh, B.H. Crystal structure of peptidoglycan recognition protein LB from Drosophila melanogaster. Nat. Immunol 2003, 4, 787–793. [Google Scholar]
- Mellroth, P.; Karlsson, J.; Steiner, H. Scavenger function for a Drosophila peptidoglycan recognition protein. J. Biol. Chem 2003, 278, 7059–7064. [Google Scholar]
- Takehana, A.; Katsuyama, T.; Yano, T.; Oshima, Y.; Takada, H.; Aigaki, T.; Kurata, S. Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc. Natl. Acad. Sci. USA 2002, 99, 13705–13710. [Google Scholar]
- Takehana, A.; Yano, T.; Mita, S.; Kotani, A.; Oshima, Y.; Kurata, S. Peptidoglycan recognition protein (PGRP)-LE and PGRP-LC act synergistically in Drosophila immunity. EMBO J 2004, 23, 4690–4700. [Google Scholar]
- Park, J.W.; Kim, C.H.; Kim, J.H.; Je, B.R.; Roh, K.B.; Kim, S.J.; Lee, H.H.; Ryu, J.H.; Lim, J.H.; Oh, B.H.; et al. Clustering of peptidoglycan recognition protein-SA is required for sensing lysine-type peptidoglycan in insects. Proc. Natl. Acad. Sci. USA 2007, 104, 6602–6607. [Google Scholar]
- Yu, Y.; Park, J.W.; Kwon, H.M.; Hwang, H.O.; Jang, I.H.; Masuda, A.; Kurokawa, K.; Nakayama, H.; Lee, W.J.; Dohmae, N.; et al. Diversity of innate immune recognition mechanism for bacterial polymeric meso-diaminopimelic acid-type peptidoglycan in insects. J. Biol. Chem 2010, 285, 32937–32945. [Google Scholar]
- Kaneko, T.; Yano, T.; Aggarwal, K.; Lim, J.H.; Ueda, K.; Oshima, Y.; Peach, C.; Erturk-Hasdemir, D.; Goldman, W.E.; Oh, B.H.; et al. PGRP-LC and PGRP-LE have essential yet distinct functions in the drosophila immune response to monomeric DAP-type peptidoglycan. Nat. Immunol 2006, 7, 715–723. [Google Scholar]
- Ghosh, J.; Lun, C.M.; Majeske, A.J.; Sacchi, S.; Schrankel, C.S.; Smith, L.C. Invertebrate immune diversity. Dev. Comp. Immunol 2011, 35, 959–974. [Google Scholar]
- Bao, Y.Y.; Qu, L.Y.; Zhao, D.; Chen, L.; Jin, H.Y.; Xu, L.M.; Cheng, J.A.; Zhang, C.X. The genome- and transcriptome-wide analysis of in innate immunity in the brown planthopper Nilaparvata lugens. BMC Genomics 2013, 14, 160. [Google Scholar]
- Blom, N.; Sicheritz-Ponten, T.; Gupta, R.; Gammeltoft, S.; Brunak, S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 2004, 4, 1633–1649. [Google Scholar]
- Nishikawa, K.; Ooi, T. Radial locations of amino acid residues in a globular protein: Correlation with the sequence. J. Biochem 1986, 100, 1043–1047. [Google Scholar]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol 1982, 157, 105–132. [Google Scholar]
- Werner, T.; Liu, G.; Kang, D.; Ekengren, S.; Steiner, H.; Hultmark, D. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2000, 97, 13772–13777. [Google Scholar]
- Liu, C.; Xu, Z.; Gupta, D.; Dziarski, R. Peptidoglycan recognition proteins: A novel family of four human innate immunity pattern recognition molecules. J. Biol. Chem 2001, 276, 34686–34694. [Google Scholar]
- Gelius, E.; Persson, C.; Karlsson, J.; Steiner, H. A mammalian peptidoglycan recognition protein with N-acetylmuramoyl-l-alanine amidase activity. Biochem. Biophys. Res. Commun 2003, 306, 988–994. [Google Scholar]
- Wang, Z.M.; Li, X.; Cocklin, R.R.; Wang, M.; Wang, M.; Fukase, K.; Inamura, S.; Kusumoto, S.; Gupta, D.; Dziarski, R. Human peptidoglycan recognition protein-l is an N-Acetylmuramoyl-l-alanine amidase. J. Biol. Chem 2003, 278, 49044–49052. [Google Scholar]
- Mellroth, P.; Steiner, H. PGRP-SB1: An N-acetylmuramoyl l-alanine amidase with antibacterial activity. Biochem. Biophys. Res. Commun 2006, 350, 994–999. [Google Scholar]
- Zou, Z.; Evans, D.J.; Lu, Z.; Zhao, Z.; Williams, M.; Sumathipala, N.; Hetru, C.; Hultmark, D.; Jiang, H. Comparative genomic analysis of the Tribolium immune system. Genome Biol 2007, 8, R177. [Google Scholar]
- Zhang, Y.; Yu, Z. The first evidence of positive selection in peptidoglycan recognition protein (PGRP) genes of Crassostrea gigas. Fish Shellfish Immunol 2013, 34, 1352–1355. [Google Scholar]
- Li, M.F.; Zhang, M.; Wang, C.L.; Sun, L. A peptidoglycan recognition protein from Sciaenops ocellatus is a zinc amidase and a bactericide with a substrate range limited to Gram-positive bacteria. Fish Shellfish Immunol 2012, 32, 322–330. [Google Scholar]
- Swaminathan, C.P.; Brown, P.H.; Roychowdhury, A.; Wang, Q.; Guan, R.; Silverman, N.; Goldman, W.E.; Boons, G.J.; Mariuzza, R.A. Dual strategies for peptidoglycan discrimination by peptidoglycan recognition proteins (PGRPs). Proc. Natl. Acad. Sci. USA 2006, 10, 684–689. [Google Scholar]
- Mendes, C.; Felix, R.; Sousa, A.M.; Lamego, J.; Charlwood, D.; do Rosário, V.E.; Pinto, J.; Silveira, H. Molecular evolution of the three short PGRPs of the malaria vectors Anopheles gambiae and Anopheles arabiensis in East Africa. BMC Evol. Biol 2010. [Google Scholar] [CrossRef]
- Lim, J.H.; Kim, M.S.; Kim, H.E.; Yano, T.; Oshima, Y.; Aggarwal, K.; Goldman, W.E.; Silverman, N.; Kurata, S.; Oh, B.H. Structural basis for recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan recognition proteins. J. Biol. Chem 2006, 281, 8286–8295. [Google Scholar]
- Kopp, J.; Schwede, T. The SWISS-MODEL Repository: New features and functionalities. Nucl. Acids Res 2006, 34, D315–D318. [Google Scholar]
- Neyen, C.; Poidevin, M.; Roussel, A.; Lemaitre, B. Tissue- and ligand-specific sensing of gram-negative infection in Drosophila by PGRP-LC isoforms and PGRP-LE. J. Immunol 2012, 189, 1886–1897. [Google Scholar]
- Wang, S.; Beerntsen, B.T. Insights into the different functions of multiple peptidoglycan recognition proteins in the immune response against bacteria in the mosquito Armigeres subalbatus. Insect Biochem. Mol. Biol 2013, 43, 533–543. [Google Scholar]
- Ewing, B.; Green, P. Base calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 1998, 8, 186–194. [Google Scholar]
- Untergrasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucl. Acids Res 2012. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. Available online: http://www.ncbi.nlm.nih.gov/gorf/gorf.html/ (accessed on 23 April 2013).
- Expert Protein Analysis System (ExPASy) Bioinformatics Resource Portal. Available online: http://www.expasy.org/ (accessed on 23 April 2013).
- Petersen, T.N.; Brunak, S.; Heijne, G.; Nielsen, H. Signal 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar]
- ProtParam Tool. Available online: http://web.expasy.org/protscale/ (accessed on 23 April 2013).
- PeptideCutter Tool. Available online: http://web.expasy.org/peptide_cutter/ (accessed on 5 May 2013).
- ProtScale Tool. Available online: http://web.expasy.org/protscale/ (accessed on 5 May 2013).
- Solovyev, V.; Kosarev, P.; Seledsov, I.; Vorobyev, D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol 2006, 7, 1–10. [Google Scholar]
- Tamura, K.; Petersen, D.; Petersen, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol. Biol. Evol 2011. [Google Scholar] [CrossRef]
- Sneath, P.H.A.; Sokal, R.R. Numerical Taxonomy; W.H. Freeman: San Franscisco, CA, USA, 1973; p. 115. [Google Scholar]
- Buchan, D.W.A.; Minneci, F.; Nugent, T.C.O.; Bryson, K.; Jones, D.T. Scalable web services for the PSIPRED protein analysis workbench. Nucl. Acids Res 2013, 41, 340–348. [Google Scholar]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL Workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2000, 22, 195–201. [Google Scholar]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem 2011, 55, 611–622. [Google Scholar]
- Mouillet, J.F.; Bousquet, F.; Sedano, N.; Alabouvette, J.; Nicolai, M.; Zelus, D.; Laudet, V.; Delachambre, J. Cloning and characterization of new orphan nuclear receptors and their developmental profiles during Tenebrio metamorphosis. Eur. J. Biochem 1999, 265, 972–981. [Google Scholar]
- Dobson, A.J.; Johnston, P.R.; Vilcinskas, A.; Rolff, J. Identification of immunological expressed sequence tags in the mealworm beetle Tenebrio molitor. J. Insect Physiol 2012, 58, 1556–1561. [Google Scholar]
- Roh, K.B.; Kim, C.H.; Lee, H.; Kwon, H.M.; Park, J.W.; Ryu, J.H.; Kurokawa, K.; Ha, N.C.; Lee, W.J.; Lemaitre, B.; et al. Proteolytic cascade for the activation of the insect toll pathway induced by the fungal cell wall component. J. Biol. Chem 2009, 284, 19474–19481. [Google Scholar]
- Taliaferro, J.M.; Aspden, J.L.; Bradley, T.; Marwha, D.; Blanchette, M.; Rio, D.C. Two new and distinct roles for Drosophila Argonaute-2 in the nucleus: Alternative pre-mRNA splicing and transcriptional repression. Genes Dev 2013, 27, 378–389. [Google Scholar]
- Khan, A.M.; Ashfaq, M.; Kiss, Z.; Khan, A.B.; Mansoor, S.; Bryce, W.; Falk, B.W. Use of recombinant Tobacco Mosaic virus to achieve RNA interference in plants against the Citrus Mealybug, Planococcus citri (Hemiptera: Pseudococcidae). PLoS One 2013, 8, e73657. [Google Scholar]
- Fabrick, J.; Oppert, C.; Lorenzen, M.D.; Morris, K.; Oppert, B.; Jurat-Fuentes, J.L. A novel Tenebrio molitor cadherin is a functional receptor for Bacillus thuringiensis Cry3Aa toxin. J. Biol. Chem 2009, 284, 18401–18410. [Google Scholar]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc 2008, 3, 1101–1108. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Tindwa, H.; Patnaik, B.B.; Kim, D.H.; Mun, S.; Jo, Y.H.; Lee, B.L.; Lee, Y.S.; Kim, N.J.; Han, Y.S. Cloning, Characterization and Effect of TmPGRP-LE Gene Silencing on Survival of Tenebrio Molitor against Listeria monocytogenes Infection. Int. J. Mol. Sci. 2013, 14, 22462-22482. https://doi.org/10.3390/ijms141122462
Tindwa H, Patnaik BB, Kim DH, Mun S, Jo YH, Lee BL, Lee YS, Kim NJ, Han YS. Cloning, Characterization and Effect of TmPGRP-LE Gene Silencing on Survival of Tenebrio Molitor against Listeria monocytogenes Infection. International Journal of Molecular Sciences. 2013; 14(11):22462-22482. https://doi.org/10.3390/ijms141122462
Chicago/Turabian StyleTindwa, Hamisi, Bharat Bhusan Patnaik, Dong Hyun Kim, Seulgi Mun, Yong Hun Jo, Bok Luel Lee, Yong Seok Lee, Nam Jung Kim, and Yeon Soo Han. 2013. "Cloning, Characterization and Effect of TmPGRP-LE Gene Silencing on Survival of Tenebrio Molitor against Listeria monocytogenes Infection" International Journal of Molecular Sciences 14, no. 11: 22462-22482. https://doi.org/10.3390/ijms141122462
APA StyleTindwa, H., Patnaik, B. B., Kim, D. H., Mun, S., Jo, Y. H., Lee, B. L., Lee, Y. S., Kim, N. J., & Han, Y. S. (2013). Cloning, Characterization and Effect of TmPGRP-LE Gene Silencing on Survival of Tenebrio Molitor against Listeria monocytogenes Infection. International Journal of Molecular Sciences, 14(11), 22462-22482. https://doi.org/10.3390/ijms141122462







