Tissue Microarray-Based Evaluation of Chromatin Assembly Factor-1 (CAF-1)/p60 as Tumour Prognostic Marker
Abstract
:1. Introduction
2. Results
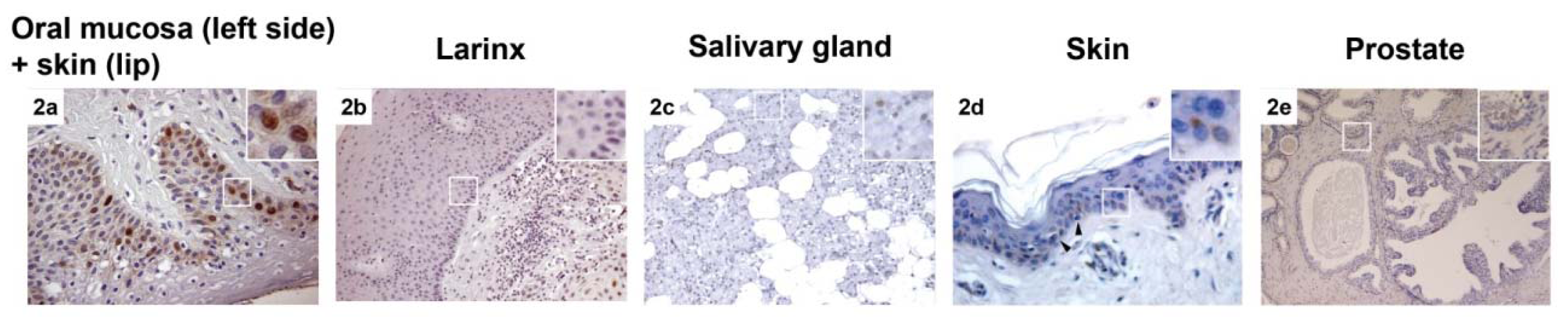
2.1. CAF-1 p60 in Normal Tissue
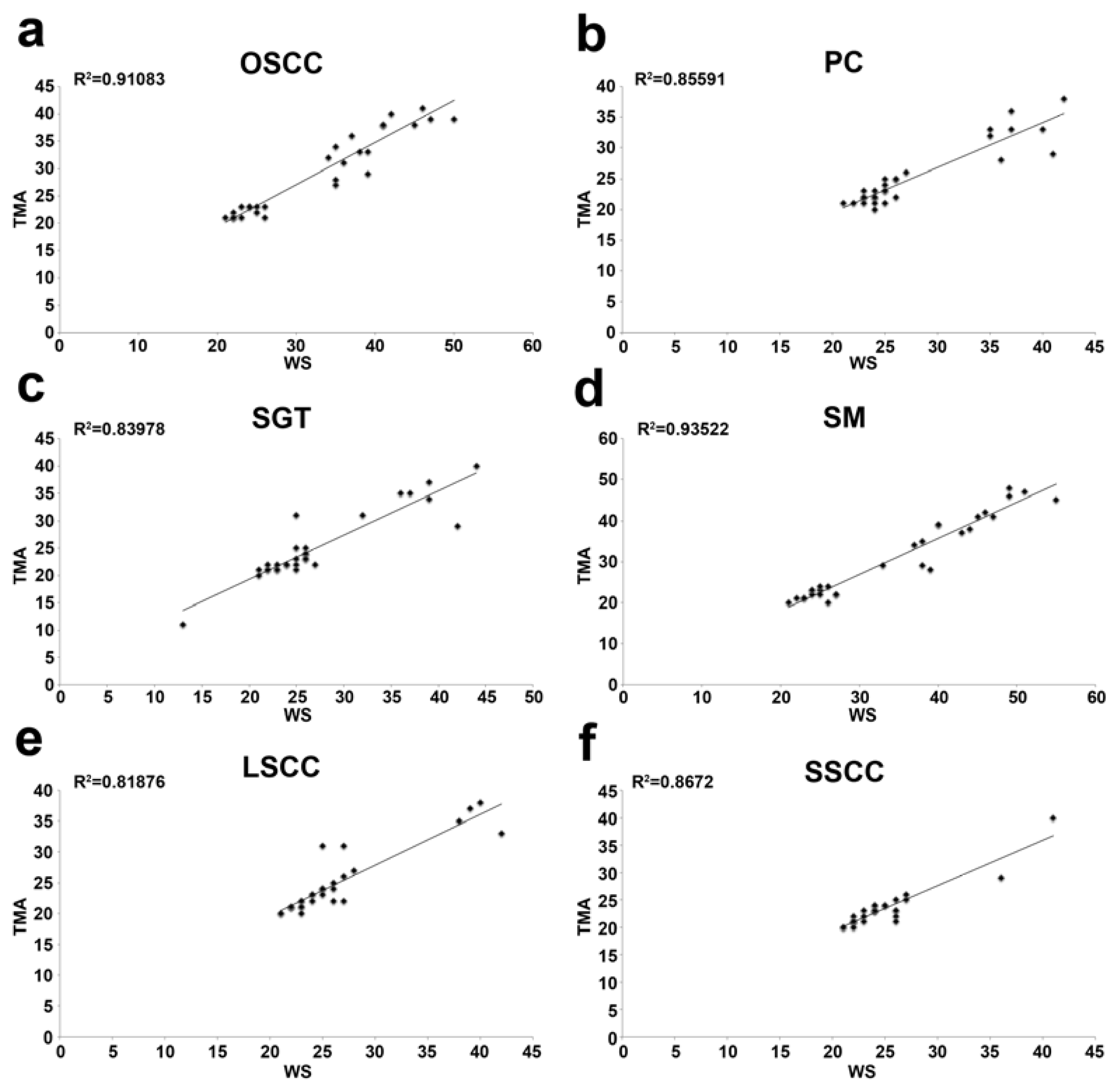
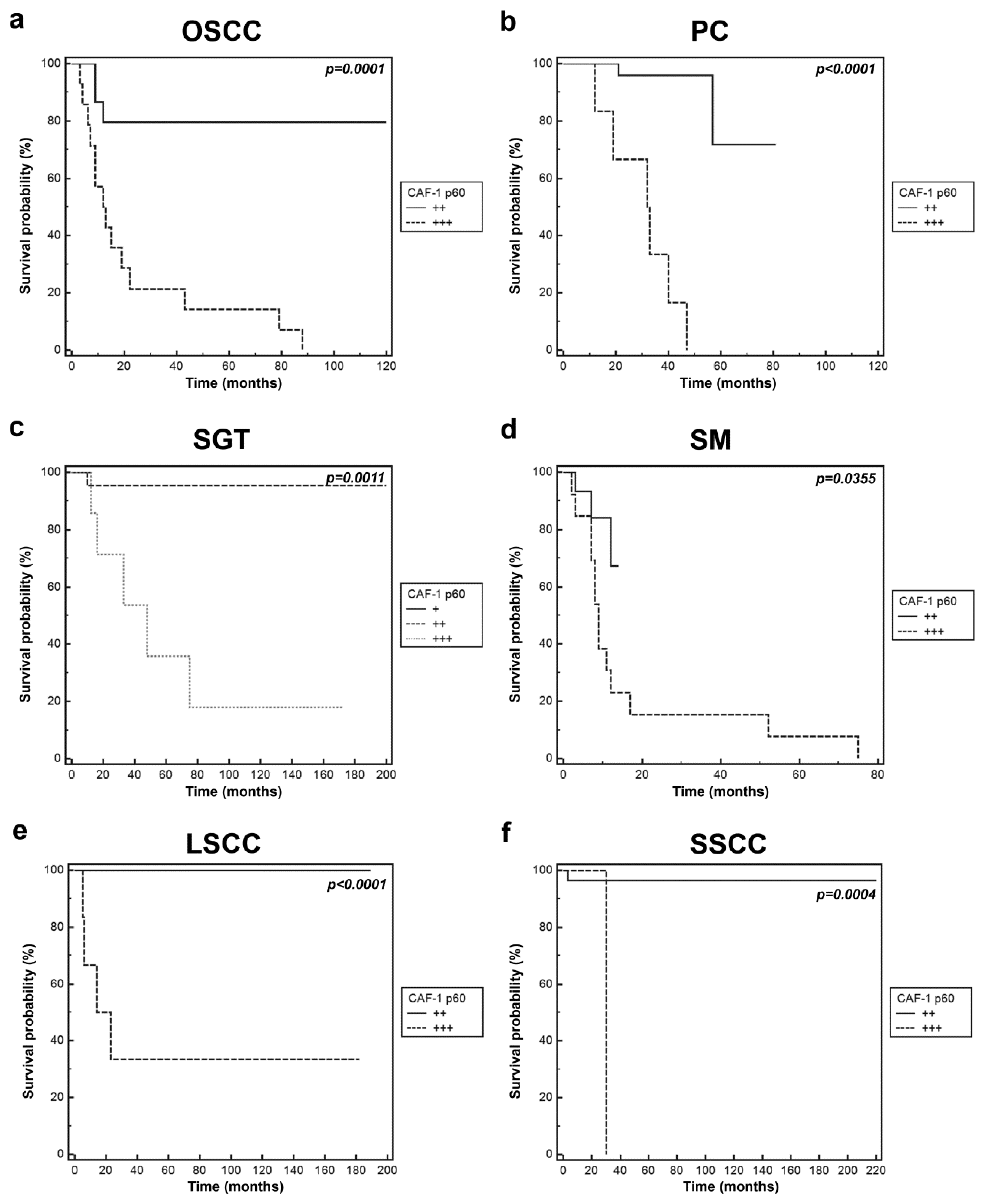
2.2. CAF-1 p60 in Tumours
3. Discussion
4. Experimental Section
4.1. TMAs Construction and Immunohistochemistry
4.2. Statistical Analysis
5. Conclusions
Acknowledgments
- Conflict of InterestThe authors declare no conflict of interest.
References
- Feinberg, A.P.; Ohlsson, R.; Henikoff, S. The epigenetic progenitor origin of human cancer. Nat. Rev. Genet 2006, 7, 21–33. [Google Scholar]
- Feinberg, A.P.; Tycko, B. The history of cancer epigenetics. Nat. Rev. Cancer 2004, 4, 143–153. [Google Scholar]
- Jones, P.A.; Baylin, S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet 2002, 3, 415–428. [Google Scholar]
- Jones, P.A.; Baylin, S.B. The epigenomics of cancer. Cell 2007, 128, 683–692. [Google Scholar]
- Ozanne, S.E.; Constancia, M. Mechanisms of disease: The developmental origins of disease and the role of the epigenotype. Nat. Clin. Pract. Endocrinol. Metab 2007, 3, 539–546. [Google Scholar]
- Schulz, W.A.; Hatina, J. Epigenetics of prostate cancer: Beyond DNA methylation. J. Cell. Mol. Med 2006, 10, 100–125. [Google Scholar]
- Li, L.C. Epigenetics of prostate cancer. Front. Biosci 2007, 12, 3377–3397. [Google Scholar]
- Liu, S.; Ren, S.; Howell, P.; Fodstad, O.; Riker, A.I. Identification of novel epigenetically modified genes in human melanoma via promoter methylation gene profiling. Pigment Cell Melanoma Res 2008, 21, 545–558. [Google Scholar]
- Egger, G.; Liang, G.; Aparicio, A.; Jones, P.A. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004, 429, 457–463. [Google Scholar]
- Heightman, T.D. Therapeutic prospects for epigenetic modulation. Expert Opin. Ther. Targets 2011, 15, 729–740. [Google Scholar]
- Sharma, S.K.; Wu, Y.; Steinbergs, N.; Crowley, M.L.; Hanson, A.S.; Casero, R.A.; Woster, P.M. (Bis)urea and (bis)thiourea inhibitors of lysine-specific demethylase 1 as epigenetic modulators. J. Med. Chem. 2010, 53, 5197–5212. [Google Scholar]
- Rountree, M.R.; Bachman, K.E.; Herman, J.G.; Baylin, S.B. DNA methylation, chromatin inheritance, and cancer. Oncogene 2001, 20, 3156–3165. [Google Scholar]
- Sandoval, J.; Esteller, M. Cancer epigenomics: Beyond genomics. Curr. Opin. Genet. Dev 2012, 22, 50–55. [Google Scholar]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar]
- Ehrenhofer-Murray, A.E. Chromatin dynamics at DNA replication, transcription and repair. Eur. J. Biochem 2004, 271, 2335–2349. [Google Scholar]
- De Koning, L.; Corpet, A.; Haber, J.E.; Almouzni, G. Histone chaperones: An escort network regulating histone traffic. Nat. Struct. Mol. Biol 2007, 14, 997–1007. [Google Scholar]
- Tagami, H.; Ray-Gallet, D.; Almouzni, G.; Nakatani, Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 2004, 116, 51–61. [Google Scholar]
- Krude, T.; Keller, C. Chromatin assembly during S phase: Contributions from histone deposition, DNA replication and the cell division cycle. Cell. Mol. Life Sci 2001, 58, 665–672. [Google Scholar]
- Ramirez-Parra, E.; Gutierrez, C. The many faces of chromatin assembly factor 1. Trends Plant Sci 2007, 12, 570–576. [Google Scholar]
- Linger, J.G.; Tyler, J.K. Chromatin disassembly and reassembly during DNA repair. Mutat. Res 2007, 618, 52–64. [Google Scholar]
- Gaillard, P.H.; Martini, E.M.; Kaufman, P.D.; Stillman, B.; Moustacchi, E.; Almouzni, G. Chromatin assembly coupled to DNA repair: A new role for chromatin assembly factor 1. Cell 1996, 86, 887–896. [Google Scholar]
- Verger, A.; Crossley, M. Chromatin modifiers in transcription and DNA repair. Cell. Mol. Life Sci 2004, 61, 2154–2162. [Google Scholar]
- Staibano, S.; Mascolo, M.; Rocco, A.; Lo Muzio, L.; Ilardi, G.; Siano, M.; Pannone, G.; Vecchione, M.L.; Nugnes, L.; Califano, L.; et al. The proliferation marker Chromatin Assembly Factor-1 is of clinical value in predicting the biological behaviour of salivary gland tumours. Oncol. Rep 2011, 25, 13–22. [Google Scholar]
- Mascolo, M.; Vecchione, M.L.; Ilardi, G.; Scalvenzi, M.; Molea, G.; di Benedetto, M.; Nugnes, L.; Siano, M.; de Rosa, G.; Staibano, S. Overexpression of Chromatin Assembly Factor-1/p60 helps to predict the prognosis of melanoma patients. BMC Cancer 2010, 10, 63. [Google Scholar]
- Staibano, S.; Mascolo, M.; Mancini, F.P.; Kisslinger, A.; Salvatore, G.; di Benedetto, M.; Chieffi, P.; Altieri, V.; Prezioso, D.; Ilardi, G.; et al. Overexpression of chromatin assembly factor-1 (CAF-1) p60 is predictive of adverse behaviour of prostatic cancer. Histopathology 2009, 54, 580–589. [Google Scholar]
- Staibano, S.; Mignogna, C.; Lo Muzio, L.; Mascolo, M.; Salvatore, G.; di Benedetto, M.; Califano, L.; Rubini, C.; de Rosa, G. Chromatin assembly factor-1 (CAF-1)-mediated regulation of cell proliferation and DNA repair: A link with the biological behaviour of squamous cell carcinoma of the tongue? Histopathology 2007, 50, 911–919. [Google Scholar]
- Polo, S.E.; Theocharis, S.E.; Klijanienko, J.; Savignoni, A.; Asselain, B.; Vielh, P.; Almouzni, G. Chromatin assembly factor-1, a marker of clinical value to distinguish quiescent from proliferating cells. Cancer Res 2004, 64, 2371–2381. [Google Scholar]
- Polo, S.E.; Theocharis, S.E.; Grandin, L.; Gambotti, L.; Antoni, G.; Savignoni, A.; Asselain, B.; Patsouris, E.; Almouzni, G. Clinical significance and prognostic value of chromatin assembly factor-1 overexpression in human solid tumours. Histopathology 2010, 57, 716–724. [Google Scholar]
- Chen, Y.; Miller, C.; Mosher, R.; Zhao, X.; Deeds, J.; Morrissey, M.; Bryant, B.; Yang, D.; Meyer, R.; Cronin, F.; et al. Identification of cervical cancer markers by cDNA and tissue microarrays. Cancer Res 2003, 63, 1927–1935. [Google Scholar]
- Fons, G.; Burger, M.P.; Ten Kate, F.J.; van der Velden, J. Identification of potential prognostic markers for vulvar cancer using immunohistochemical staining of tissue microarrays. Int. J. Gynecol. Pathol 2007, 26, 188–193. [Google Scholar]
- Gomaa, W.; Ke, Y.; Fujii, H.; Helliwell, T. Tissue microarray of head and neck squamous carcinoma: Validation of the methodology for the study of cutaneous fatty acid-binding protein, vascular endothelial growth factor, involucrin and Ki-67. Virchows Archiv 2005, 447, 701–709. [Google Scholar]
- Su, Y. Immunohistochemical Expressions of Ki-67, Cyclin D1, -Catenin, Cyclooxygenase-2, and epidermal growth factor receptor in human colorectal adenoma: A validation study of tissue microarrays. Cancer Epidemiol. Biomark. Prev 2006, 15, 1719–1726. [Google Scholar]
- Van den Eynden, G.G.; van der Auwera, I.; van Laere, S.; Colpaert, C.G.; van Dam, P.; Merajver, S.; Kleer, C.G.; Harris, A.L.; van Marck, E.A.; Dirix, L.Y.; et al. Validation of a tissue microarray to study differential protein expression in inflammatory and non-inflammatory breast cancer. Breast Cancer Res. Treat 2004, 85, 13–22. [Google Scholar]
- Wan, W.H.; Fortuna, M.B.; Furmanski, P. A rapid and efficient method for testing immunohistochemical reactivity of monoclonal antibodies against multiple tissue samples simultaneously. J. Immunol. Methods 1987, 103, 121–129. [Google Scholar]
- Kononen, J.; Bubendorf, L.; Kallioniemi, A.; Barlund, M.; Schraml, P.; Leighton, S.; Torhorst, J.; Mihatsch, M.J.; Sauter, G.; Kallioniemi, O.P. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med 1998, 4, 844–847. [Google Scholar]
- Monteiro, L.S.; Diniz-Freitas, M.; Garcia-Caballero, T.; Forteza, J.; Fraga, M. EGFR and Ki-67 expression in oral squamous cell carcinoma using tissue microarray technology. J. Oral Pathol. Med 2010, 39, 571–578. [Google Scholar]
- Boone, J.; van Hillegersberg, R.; van Diest, P.J.; Offerhaus, G.J.; Rinkes, I.H.; Kate, F.J. Validation of tissue microarray technology in squamous cell carcinoma of the esophagus. Virchows Arch 2008, 452, 507–514. [Google Scholar]
- Hecht, J.L.; Kotsopoulos, J.; Gates, M.A.; Hankinson, S.E.; Tworoger, S.S. Validation of tissue microarray technology in ovarian cancer: Results from the Nurses’ Health Study. Cancer Epidemiol. Biomark. Prev 2008, 17, 3043–3050. [Google Scholar]
- Alkushi, A. Validation of tissue microarray biomarker expression of breast carcinomas in Saudi women. Hematol. Oncol. Stem Cell Ther 2009, 2, 394–398. [Google Scholar]
- Cunha, K.S.; Caruso, A.C.; Goncalves, A.S.; Bernardo, V.G.; Pires, A.R.; da Fonseca, E.C.; de Faria, P.A.; da Silva, L.E.; Geller, M.; de Moura-Neto, R.S.; et al. Validation of tissue microarray technology in malignant peripheral nerve sheath tumours. J. Clin. Pathol 2009, 62, 629–633. [Google Scholar]
- Camp, R.L.; Neumeister, V.; Rimm, D.L. A decade of tissue microarrays: Progress in the discovery and validation of cancer biomarkers. J. Clin. Oncol 2008, 26, 5630–5637. [Google Scholar]
- Sullivan, C.A.; Chung, G.G. Biomarker validation: In situ analysis of protein expression using semiquantitative immunohistochemistry-based techniques. Clin. Colorectal. Cancer 2008, 7, 172–177. [Google Scholar]
- Giltnane, J.M.; Rimm, D.L. Technology insight: Identification of biomarkers with tissue microarray technology. Nat. Clin. Pract. Oncol 2004, 1, 104–111. [Google Scholar]
- Wang, S.L.; Yang, C.H.; Chen, H.H.; Chai, C.Y. A simple and economical method for the manual construction of well-aligned tissue arrays. Pathol. Res. Pract 2006, 202, 485–486. [Google Scholar]
- Eguiluz, C.; Viguera, E.; Millan, L.; Perez, J. Multitissue array review: A chronological description of tissue array techniques, applications and procedures. Pathol. Res. Pract 2006, 202, 561–568. [Google Scholar]
- Edge, S.B.; Byrd, D.R.; Compton, C.C.; Fritz, A.G.; Greene, F.L.; Trotti, A. AJCC Cancer Staging Manual, 7th ed; Springer: New York, NY, USA, 2010; p. 130. [Google Scholar]
- Hoos, A.; Urist, M.J.; Stojadinovic, A.; Mastorides, S.; Dudas, M.E.; Leung, D.H.; Kuo, D.; Brennan, M.F.; Lewis, J.J.; Cordon-Cardo, C. Validation of tissue microarrays for immunohistochemical profiling of cancer specimens using the example of human fibroblastic tumors. Am. J. Pathol 2001, 158, 1245–1251. [Google Scholar]
- Kundel, H.L.; Polansky, M. Measurement of observer agreement. Radiology 2003, 228, 303–308. [Google Scholar]

| Histotype | N° | m | f | Age Range (average) | F-up Range (average) | Grade | Outcome |
|---|---|---|---|---|---|---|---|
| OSCC | 30 | 18 | 12 | 48–95 years (66.7) | 4–120 months (49.23) | 10 G1 (33%) | 3 NED, 2 M, 1 D, 1 M,D, 3 R,M,D |
| 15 G2 (50%) | 8 NED, 3 M,D, 2 R,D, 2 R,M,D | ||||||
| 5 G3 (17%) | 2 NED, 1 M,D, 1 R,D, 1 R,M,D | ||||||
| PC | 30 | 30 | 0 | 55–80 years (66.7) | 30–64 months (40) | 3 GS ≤ 6 (10%) | 1 NED, 1 M, 1 M,D |
| 18 GS 7 (60%) | 15 NED, 2 M, 1 M,D | ||||||
| 9 GS > 8 (30%) | 6 NED, 2 M, 1 M,D | ||||||
| SM | 30 | 15 | 15 | 21–81 years (46.2) | 1–22 years (8.67) | 6 BT < 1 (20%) | 4 NED, 1 R, 1 N |
| 8BT 1.01–2 (27%) | 3 NED, 3 N, 1 R,N, 1 N,M,D | ||||||
| 10 BT 2.01–4 (33%) | 4 NED, 1 M, 2 N, 1 N,M, 2 N,M,D | ||||||
| 6 BT > (20%) | 3 NED, 2 N, 1 R,M,D | ||||||
| SGT | 30 | 12 | 18 | 18–80 years (48.77) | 12–200 months (67) | 1 PLGC (3%) | 1 NED |
| 4 ACC (13%) | 3 NED, 1 M,R | ||||||
| 16 MEC (54%) | 12 NED, 3 R, 1 M | ||||||
| 3 AC (10%) | 3 NED | ||||||
| 5 CXPA (17%) | 4 NED; 1 R,N | ||||||
| 1 PA (3%) | 1 NED | ||||||
| LSCC | 30 | 30 | 0 | 38–76 years (63.3) | 38–189 months (130.33) | 7 G1 (23%) | 6 NED, 1 M |
| 7 G2 (23%) | 6 NED, 1 M | ||||||
| 16 G3 (54%) | 14 NED, 2 M | ||||||
| SSCC | 30 | 17 | 13 | 32–95 years (68.04) | 12–223 months (138.83) | 8 G1 (26%) | 8 NED |
| 11 G2 (37%) | 10 NED, 1 R,N | ||||||
| 11 G3 (37%) | 10 NED, 1 R | ||||||
| Sex | Age | Histotype | Grading | TNM Stage | WS CAF-1 p60 | WS CAF-1 p60 (%) | TMA CAF-1 p60 | TMA CAF-1 p60 (%) | F-up |
|---|---|---|---|---|---|---|---|---|---|
| m | 95 | SCC | 1 | I | ++ | 25 | ++ | 22 | 17 NED |
| m | 64 | SCC | 1 | I | ++ | 22 | ++ | 21 | 21 NED |
| f | 81 | SCC | 1 | I | ++ | 23 | ++ | 21 | 4 NED |
| m | 49 | SCC | 1 | I | +++ | 34 | +++ | 32 | 43 D |
| m | 66 | SCC | 1 | III | +++ | 39 | +++ | 33 | 120 M |
| m | 49 | SCC | 1 | III | +++ | 41 | +++ | 38 | 79 M |
| m | 86 | SCC | 1 | III | +++ | 42 | +++ | 40 | 22 M,D |
| m | 75 | SCC | 1 | IV | +++ | 45 | +++ | 38 | 11 R,M,D |
| m | 56 | SCC | 1 | IV | +++ | 47 | +++ | 39 | 77 R,M,D |
| f | 68 | SCC | 1 | III | +++ | 46 | +++ | 41 | 88 R,M,D |
| m | 66 | SCC | 2 | II | ++ | 23 | ++ | 23 | 120 NED |
| m | 50 | SCC | 2 | I | ++ | 22 | ++ | 21 | 109 NED |
| m | 78 | SCC | 2 | III | ++ | 25 | ++ | 23 | 11 NED |
| m | 48 | SCC | 2 | I | ++ | 21 | ++ | 21 | 14 NED |
| m | 48 | SCC | 2 | I | ++ | 24 | ++ | 23 | 85 NED |
| f | 74 | SCC | 2 | I | ++ | 26 | ++ | 21 | 19 NED |
| f | 67 | SCC | 2 | I | ++ | 22 | ++ | 22 | 97 NED |
| f | 59 | SCC | 2 | II | ++ | 24 | ++ | 23 | 98 NED |
| m | 69 | SCC | 2 | III | +++ | 35 | +++ | 34 | 12 R,D |
| m | 58 | SCC | 2 | IV | +++ | 36 | +++ | 31 | 8 R,D |
| f | 77 | SCC | 2 | I | +++ | 38 | +++ | 33 | 114 M,D |
| f | 75 | SCC | 2 | IV | +++ | 37 | +++ | 36 | 27 M,D |
| f | 73 | SCC | 2 | III | +++ | 41 | +++ | 38 | 10 M,D |
| m | 65 | SCC | 2 | III | +++ | 35 | ++ | 27 | 20 R,M,D |
| m | 60 | SCC | 2 | IV | +++ | 50 | +++ | 39 | 24 R,M,D |
| f | 60 | SCC | 3 | I | ++ | 26 | ++ | 23 | 106 NED |
| m | 66 | SCC | 3 | I | ++ | 22 | ++ | 21 | 97 NED |
| f | 80 | SCC | 3 | II | +++ | 35 | ++ | 28 | 23 R,D |
| m | 63 | SCC | 3 | III | +++ | 39 | ++ | 29 | 17 M,D |
| m | 65 | SCC | 3 | II | +++ | 41 | +++ | 38 | 15 R,M,D |
| Sex | Age | Histotype | Gleason | TNM Stage | WS CAF-1 p60 | WS CAF-1 p60 (%) | TMA CAF-1 p60 | TMA CAF-1 p60 (%) | F-up |
|---|---|---|---|---|---|---|---|---|---|
| m | 70 | AC | 5 (3 + 2) | pT2bN0 | +++ | 36 | ++ | 28 | 57 M |
| m | 51 | AC | 5 (3 + 2) | pT2bN0 | +++ | 40 | +++ | 33 | 50 M,D |
| m | 72 | AC | 6 (3 + 3) | pT2bN0 | ++ | 23 | ++ | 22 | 81 NED |
| m | 68 | AC | 7 (3 + 4) | pT3aN0 | ++ | 25 | ++ | 25 | 49 NED |
| m | 67 | AC | 7 (4 + 3) | pT3aNx | ++ | 27 | ++ | 26 | 33 NED |
| m | 58 | AC | 7 (3 + 4) | pT2bN0 | ++ | 24 | ++ | 21 | 32 NED |
| m | 45 | AC | 7 (3 + 4) | pT2bN0 | ++ | 23 | ++ | 23 | 30 NED |
| m | 53 | AC | 7 (3 + 4) | pT2bN0 | ++ | 24 | ++ | 22 | 75 NED |
| m | 65 | AC | 7 (4 + 3) | pT4N1 | +++ | 37 | +++ | 36 | 49 M |
| m | 63 | AC | 7 (4 + 3) | pT3aN0 | ++ | 24 | ++ | 23 | 32 NED |
| m | 68 | AC | 7 (4 + 3) | pT3bN0 | ++ | 26 | ++ | 25 | 33 NED |
| m | 73 | AC | 7 (3 + 4) | pT3aN1 | ++ | 23 | ++ | 22 | 56 NED |
| m | 71 | AC | 7 (4 + 3) | pT3bN1 | ++ | 26 | ++ | 25 | 30 NED |
| m | 80 | AC | 7 (4 + 3) | pT3bN1 | ++ | 24 | ++ | 20 | 40 NED |
| m | 57 | AC | 7 (3 + 4) | pT2bN0 | ++ | 21 | ++ | 21 | 33 NED |
| m | 70 | AC | 7 (3 + 4) | pT2aN0 | ++ | 25 | ++ | 23 | 32 NED |
| m | 56 | AC | 7 (3 + 4) | pT2bN0 | ++ | 23 | ++ | 22 | 33 NED |
| m | 61 | AC | 7 (3 + 4) | pT2aN0 | ++ | 25 | ++ | 24 | 32 NED |
| m | 65 | AC | 7 (4 + 3) | pT3bN1 | +++ | 41 | ++ | 29 | 43 M,D |
| m | 60 | AC | 7 (3 + 4) | pT2aN0 | ++ | 24 | ++ | 22 | 33 NED |
| m | 62 | AC | 7 (4 + 3) | pT3aNo | +++ | 35 | +++ | 33 | 64 M |
| m | 64 | AC | 8 (4 + 4) | pT2bN0 | ++ | 23 | ++ | 21 | 59 NED |
| m | 66 | AC | 8 (4 + 4) | pT3bN1 | ++ | 26 | ++ | 22 | 33 NED |
| m | 71 | AC | 8 (4 + 4) | pT3bN1 | +++ | 42 | +++ | 38 | 57 M,D |
| m | 60 | AC | 8 (4 + 4) | PT3aN0 | ++ | 25 | ++ | 21 | 33 NED |
| m | 70 | AC | 8 (4 + 4) | pT3bN0 | ++ | 22 | ++ | 21 | 32 NED |
| m | 74 | AC | 8 (4 + 4) | pT3aN0 | ++ | 25 | ++ | 23 | 32 NED |
| m | 64 | AC | 8 (4 + 4) | pT3bN0 | +++ | 37 | +++ | 33 | 48 M |
| m | 57 | AC | 8 (4 + 4) | pT3aN0 | ++ | 24 | ++ | 22 | 30 NED |
| m | 73 | AC | 8 (4 + 4) | pT2bN1 | +++ | 35 | +++ | 32 | 34 M |
| Sex | Age | Histotype | Grading | TNM Stage | WS CAF-1 p60 | WS CAF-1 p60 (%) | TMA CAF-1 p60 | TMA CAF-1 p60 (%) | F-up |
|---|---|---|---|---|---|---|---|---|---|
| m | 49 | PLGC | - | pT1N0M0 | ++ | 24 | ++ | 22 | 26 NED |
| f | 33 | AC | - | pT3N0M0 | ++ | 25 | ++ | 21 | 67 NED |
| m | 45 | AC | - | pT2NxM0 | ++ | 26 | ++ | 24 | 70 NED |
| f | 78 | AC | - | pT3NxM0 | ++ | 21 | ++ | 20 | 44 NED |
| f | 39 | ACC | - | pT2N0M0 | ++ | 25 | +++ | 31 | 29 NED |
| f | 41 | ACC | - | pT3N0M0 | ++ | 26 | ++ | 24 | 54 NED |
| m | 48 | ACC | - | pT4aNxM0 | +++ | 39 | +++ | 37 | 71 M,R |
| f | 63 | ACC | - | pT2NxM0 | ++ | 25 | ++ | 22 | 44 NED |
| f | 20 | LG-MEC | low | pT1N0M0 | ++ | 22 | ++ | 21 | 49 NED |
| f | 57 | LG-MEC | low | pT3N0M0 | +++ | 32 | +++ | 31 | 173 NED |
| f | 18 | LG-MEC | low | pT2N0M0 | ++ | 26 | ++ | 23 | 92 NED |
| f | 40 | LG-MEC | low | pT4aN0M0 | ++ | 27 | ++ | 22 | 69 NED |
| f | 62 | LG-MEC | low | pT3NxM0 | ++ | 23 | ++ | 22 | 103 NED |
| m | 39 | IG-MEC | intermediate | pT2N0M0 | +++ | 37 | +++ | 35 | 140 R |
| f | 51 | IG-MEC | intermediate | pT2N2bM0 | +++ | 39 | +++ | 34 | 132 M |
| m | 57 | IG-MEC | intermediate | pT1NxM0 | +++ | 44 | +++ | 40 | 94 R |
| m | 72 | IG-MEC | intermediate | pT1NxM0 | ++ | 25 | ++ | 23 | 31 NED |
| f | 74 | IG-MEC | intermediate | pT1NxM0 | ++ | 21 | ++ | 21 | 95 NED |
| f | 80 | IG-MEC | intermediate | pT3NxM0 | ++ | 26 | ++ | 24 | 97 NED |
| m | 56 | HG-MEC | high | pT2N0M0 | +++ | 36 | +++ | 35 | 131 R |
| f | 32 | HG-MEC | high | pT4NxM0 | ++ | 25 | ++ | 25 | 200 NED |
| f | 33 | HG-MEC | high | pT1NxM0 | ++ | 22 | ++ | 21 | 23 NED |
| m | 51 | HG-MEC | high | pT2N0M0 | ++ | 24 | ++ | 22 | 72 NED |
| m | 60 | HG-MEC | high | pT3N0M0 | ++ | 26 | ++ | 23 | 34 NED |
| f | 56 | CXPA | - | pT2NxM0 | ++ | 23 | ++ | 21 | 23 NED |
| f | 24 | CXPA | - | pT2N0M0 | ++ | 26 | ++ | 25 | 97 NED |
| m | 42 | CXPA | - | pT1N0M0 | +++ | 42 | ++ | 29 | 55 R,N |
| m | 43 | CXPA | - | pT3N0M0 | ++ | 23 | ++ | 21 | 63 NED |
| m | 72 | CXPA | - | pT2NxM0 | ++ | 22 | ++ | 22 | 166 NED |
| f | 28 | PA | - | - | + | 13 | + | 11 | 12 NED |
| Sex | Age | Histotype | Breslow | TNM Stage | WS CAF-1 p60 | WS CAF-1 p60 (%) | TMA CAF-1 p60 | TMA CAF-1 p60 (%) | F-up |
|---|---|---|---|---|---|---|---|---|---|
| m | 39 | MM | ≤/=1.00 | IA | +++ | 37 | +++ | 34 | 6 R |
| f | 36 | MM | ≤/=1.00 | IB | +++ | 33 | ++ | 29 | 4 N |
| m | 56 | MM | ≤/=1.00 | IB | ++ | 24 | ++ | 23 | 2 NED |
| m | 35 | MM | ≤/=1.00 | IB | ++ | 23 | ++ | 21 | 7 NED |
| m | 41 | MM | ≤/=1.00 | IB | ++ | 25 | ++ | 23 | 7 NED |
| f | 32 | MM | ≤/=1.00 | IA | ++ | 27 | ++ | 22 | 6 NED |
| f | 66 | MM | 1.01–2.00 | IB | +++ | 38 | +++ | 35 | 12 N |
| f | 47 | MM | 1.01–2.00 | IIA | +++ | 55 | +++ | 45 | 12 N,M,D |
| f | 39 | MM | 1.01–2.00 | IB | +++ | 46 | +++ | 42 | 9 N |
| f | 37 | MM | 1.01–2.00 | IIA | +++ | 51 | +++ | 47 | 7 R,N |
| f | 43 | MM | 1.01–2.00 | IB | ++ | 23 | ++ | 21 | 9 NED |
| m | 22 | MM | 1.01–2.00 | IA | ++ | 24 | ++ | 22 | 4 NED |
| m | 37 | MM | 1.01–2.00 | IA | ++ | 26 | ++ | 20 | 3 NED |
| m | 43 | MM | 1.01–2.00 | IB | +++ | 40 | +++ | 39 | 7 N |
| m | 45 | MM | 2.01–4.00 | IIIC | +++ | 49 | +++ | 46 | 11 N,M,D |
| m | 47 | MM | 2.01–4.00 | IIB | +++ | 39 | ++ | 28 | 10 N |
| f | 42 | MM | 2.01–4.00 | IIA | +++ | 43 | +++ | 37 | 9 N,M,D |
| f | 46 | MM | 2.01–4.00 | IIB | +++ | 40 | +++ | 39 | 3 N |
| f | 81 | MM | 2.01–4.00 | IIA | +++ | 45 | +++ | 41 | 2 M |
| m | 32 | MM | 2.01–4.00 | IIB | +++ | 47 | +++ | 41 | 3 N,M |
| f | 50 | MM | 2.01–4.00 | IIA | ++ | 25 | ++ | 22 | 11 NED |
| m | 38 | MM | 2.01–4.00 | IIA | ++ | 25 | ++ | 24 | 3 NED |
| f | 56 | MM | 2.01–4.00 | IIA | ++ | 24 | ++ | 23 | 13 NED |
| m | 21 | MM | 2.01–4.00 | IIB | ++ | 27 | ++ | 22 | 14 NED |
| f | 55 | MM | >4.00 | IIC | ++ | 22 | ++ | 21 | 14 NED |
| m | 54 | MM | >4.00 | IIB | ++ | 21 | ++ | 20 | 12 NED |
| m | 35 | MM | >4.00 | IIC | ++ | 26 | ++ | 24 | 2 NED |
| f | 38 | MM | >4.00 | IIC | +++ | 44 | +++ | 38 | 1 N |
| f | 52 | MM | >4.00 | IIC | +++ | 38 | ++ | 29 | 2 R,M,D |
| m | 44 | MM | >4.00 | IIC | +++ | 49 | +++ | 48 | 12 N |
| Sex | Age | Histotype | Grading | TNM Stage | WS CAF-1 p60 | WS CAF-1 p60 (%) | TMA CAF-1 p60 | TMA CAF-1 p60 (%) | F-up |
|---|---|---|---|---|---|---|---|---|---|
| m | 38 | SCC | G1 | II | ++ | 22 | ++ | 21 | 187 NED |
| m | 66 | SCC | G1 | IVA | +++ | 42 | +++ | 33 | 184 M |
| m | 53 | SCC | G1 | IVA | ++ | 27 | ++ | 22 | 182 NED |
| m | 68 | SCC | G1 | III | ++ | 23 | ++ | 20 | 181 NED |
| m | 70 | SCC | G1 | II | ++ | 22 | ++ | 21 | 180 NED |
| m | 70 | SCC | G1 | I | ++ | 24 | ++ | 23 | 82 NED |
| m | 53 | SCC | G1 | I | ++ | 25 | ++ | 24 | 189 NED |
| m | 76 | SCC | G2 | II | ++ | 26 | ++ | 22 | 188 NED |
| m | 65 | SCC | G2 | I | ++ | 21 | ++ | 20 | 186 NED |
| m | 75 | SCC | G2 | IVA | ++ | 27 | +++ | 31 | 182 NED |
| m | 72 | SCC | G2 | IVA | ++ | 23 | ++ | 21 | 58 NED |
| m | 52 | SCC | G2 | I | +++ | 40 | +++ | 38 | 45 M |
| m | 61 | SCC | G2 | II | ++ | 22 | ++ | 21 | 39 NED |
| m | 65 | SCC | G2 | IVA | ++ | 25 | +++ | 31 | 38 NED |
| m | 57 | SCC | G3 | IVA | ++ | 26 | ++ | 25 | 69 NED |
| m | 73 | SCC | G3 | III | ++ | 25 | ++ | 24 | 66 NED |
| m | 62 | SCC | G3 | III | ++ | 23 | ++ | 22 | 45 NED |
| m | 63 | SCC | G3 | IVA | ++ | 23 | ++ | 22 | 186 NED |
| m | 58 | SCC | G3 | IVA | ++ | 25 | ++ | 24 | 185 NED |
| m | 60 | SCC | G3 | I | ++ | 24 | ++ | 23 | 153 NED |
| m | 75 | SCC | G3 | II | ++ | 22 | ++ | 21 | 65 NED |
| m | 52 | SCC | G3 | IVB | +++ | 39 | +++ | 37 | 187 M |
| m | 66 | SCC | G3 | IVA | ++ | 23 | ++ | 21 | 183 NED |
| m | 59 | SCC | G3 | III | ++ | 21 | ++ | 20 | 181 NED |
| m | 75 | SCC | G3 | IVA | ++ | 25 | ++ | 23 | 124 NED |
| m | 58 | SCC | G3 | IVA | ++ | 27 | ++ | 26 | 69 NED |
| m | 71 | SCC | G3 | IVA | ++ | 24 | ++ | 22 | 53 NED |
| m | 50 | SCC | G3 | IVA | ++ | 26 | ++ | 24 | 119 NED |
| m | 72 | SCC | G3 | III | +++ | 38 | +++ | 35 | 180 M |
| m | 65 | SCC | G3 | IVA | ++ | 28 | ++ | 27 | 124 NED |
| Sex | Age | Histotype | Grading | TNM Stage | WS CAF-1/p60 | WS CAF-1/p60 (%) | TMA CAF-1 p60 | TMA CAF-1 p60 (%) | F-up |
|---|---|---|---|---|---|---|---|---|---|
| m | 76 | SCC | G1 | I | ++ | 23 | ++ | 21 | 223 NED |
| m | 73 | SCC | G1 | I | ++ | 24 | ++ | 23 | 222 NED |
| f | 86 | SCC | G1 | I | ++ | 22 | ++ | 20 | 221 NED |
| m | 72 | SCC | G1 | II | ++ | 26 | ++ | 22 | 218 NED |
| m | 36 | SCC | G1 | II | ++ | 24 | ++ | 23 | 209 NED |
| f | 95 | SCC | G1 | II | ++ | 22 | ++ | 21 | 122 NED |
| m | 80 | SCC | G1 | II | ++ | 26 | ++ | 23 | 69 NED |
| m | 55 | SCC | G1 | I | ++ | 24 | ++ | 23 | 69 NED |
| f | 67 | SCC | G2 | II | ++ | 22 | ++ | 22 | 210 NED |
| m | 49 | SCC | G2 | II | +++ | 36 | ++ | 29 | 207 R,M |
| f | 78 | SCC | G2 | IV | ++ | 25 | ++ | 24 | 147 NED |
| f | 83 | SCC | G2 | I | ++ | 26 | ++ | 23 | 127 NED |
| m | 80 | SCC | G2 | I | ++ | 24 | ++ | 24 | 126 NED |
| f | 68 | SCC | G2 | II | ++ | 23 | ++ | 23 | 125 NED |
| m | 79 | SCC | G2 | II | ++ | 22 | ++ | 21 | 90 NED |
| f | 51 | SCC | G2 | I | ++ | 26 | ++ | 21 | 24 NED |
| m | 63 | SCC | G2 | I | ++ | 24 | ++ | 23 | 69 NED |
| m | 59 | SCC | G2 | II | ++ | 26 | ++ | 25 | 70 NED |
| m | 85 | SCC | G2 | I | ++ | 21 | ++ | 20 | 66 NED |
| m | 32 | SCC | G3 | IV | ++ | 22 | ++ | 21 | 213 NED |
| m | 67 | SCC | G3 | II | +++ | 41 | +++ | 40 | 212 R |
| f | 71 | SCC | G3 | I | ++ | 22 | ++ | 21 | 209 NED |
| f | 76 | SCC | G3 | I | ++ | 27 | ++ | 25 | 201 NED |
| f | 72 | SCC | G3 | IV | ++ | 23 | ++ | 22 | 189 NED |
| m | 67 | SCC | G3 | II | ++ | 24 | ++ | 23 | 155 NED |
| f | 73 | SCC | G3 | II | ++ | 22 | ++ | 21 | 12 NED |
| f | 42 | SCC | G3 | III | ++ | 21 | ++ | 20 | 12 NED |
| m | 74 | SCC | G3 | II | ++ | 27 | ++ | 26 | 68 NED |
| f | 77 | SCC | G3 | II | ++ | 24 | ++ | 24 | 64 NED |
| m | 66 | SCC | G3 | II | ++ | 25 | ++ | 24 | 216 NED |
| OSCC | PC | SM | SGT | LSCC | SSCC | |
|---|---|---|---|---|---|---|
| K-coefficient | 0.8018 | 0.8148 | 0.8018 | 0.8529 | 0.8696 | 0.8076 |
| S.E. | 0.07361111 | 0.08611111 | 0.07361111 | 0.07638889 | 0.08819444 | 0.14513889 |
| 95% CI | 0.593–1 | 0.571–1 | 0.593–1 | 0.627–1 | 0.620–1 | 0.374–1 |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Mascolo, M.; Ilardi, G.; Merolla, F.; Russo, D.; Vecchione, M.L.; De Rosa, G.; Staibano, S. Tissue Microarray-Based Evaluation of Chromatin Assembly Factor-1 (CAF-1)/p60 as Tumour Prognostic Marker. Int. J. Mol. Sci. 2012, 13, 11044-11062. https://doi.org/10.3390/ijms130911044
Mascolo M, Ilardi G, Merolla F, Russo D, Vecchione ML, De Rosa G, Staibano S. Tissue Microarray-Based Evaluation of Chromatin Assembly Factor-1 (CAF-1)/p60 as Tumour Prognostic Marker. International Journal of Molecular Sciences. 2012; 13(9):11044-11062. https://doi.org/10.3390/ijms130911044
Chicago/Turabian StyleMascolo, Massimo, Gennaro Ilardi, Francesco Merolla, Daniela Russo, Maria Luisa Vecchione, Gaetano De Rosa, and Stefania Staibano. 2012. "Tissue Microarray-Based Evaluation of Chromatin Assembly Factor-1 (CAF-1)/p60 as Tumour Prognostic Marker" International Journal of Molecular Sciences 13, no. 9: 11044-11062. https://doi.org/10.3390/ijms130911044
APA StyleMascolo, M., Ilardi, G., Merolla, F., Russo, D., Vecchione, M. L., De Rosa, G., & Staibano, S. (2012). Tissue Microarray-Based Evaluation of Chromatin Assembly Factor-1 (CAF-1)/p60 as Tumour Prognostic Marker. International Journal of Molecular Sciences, 13(9), 11044-11062. https://doi.org/10.3390/ijms130911044






