A Novel Cyclodextrin Glycosyltransferase from Alkaliphilic Amphibacillus sp. NPST-10: Purification and Properties
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation of CGTase Producing Alkaliphilic Bacteria
2.2. Purification of the CGTase
2.3. Properties of Amphibacillus sp. NPST-10 CGTase
2.3.1. Estimation of Molecular Weight
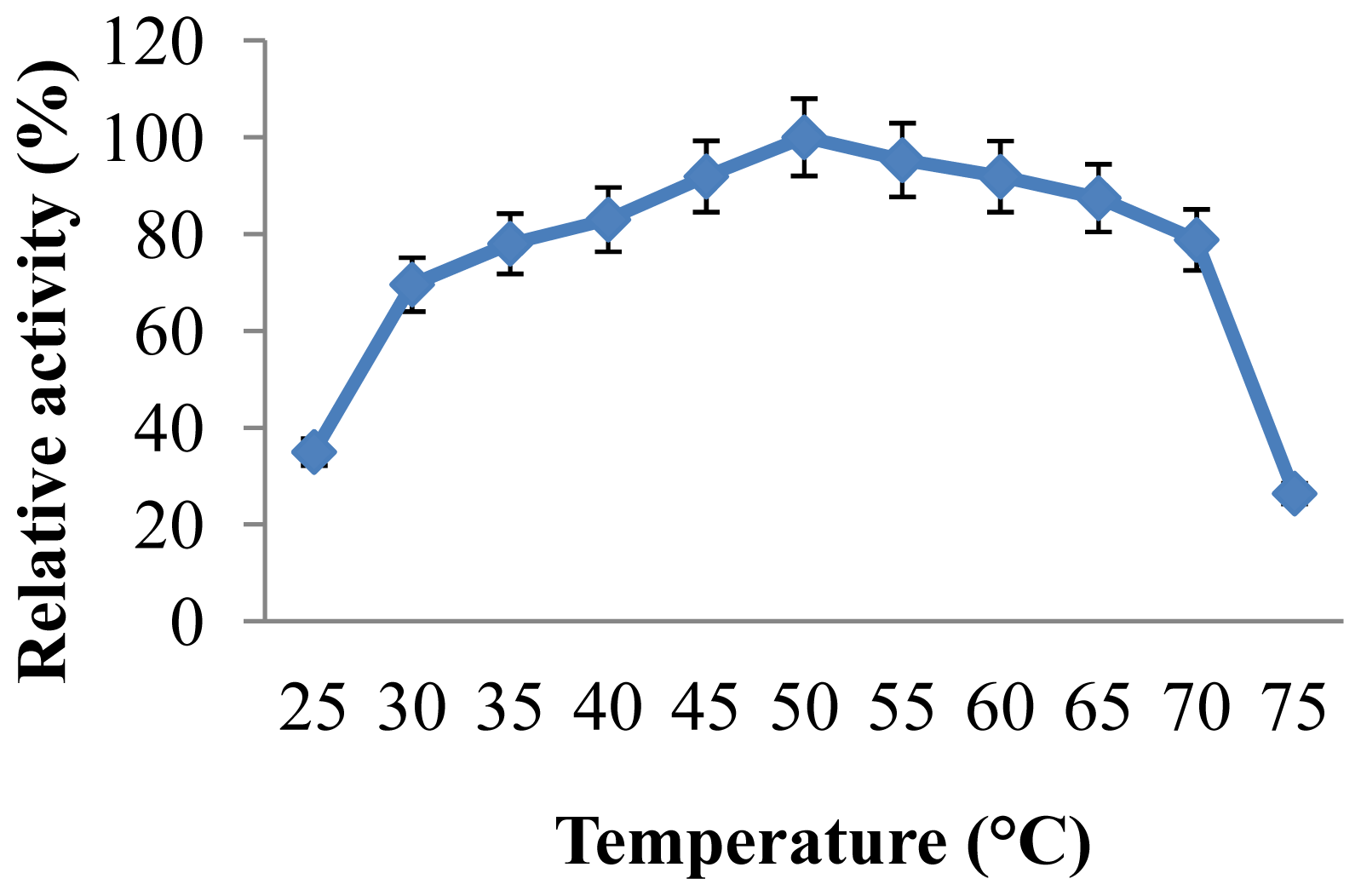
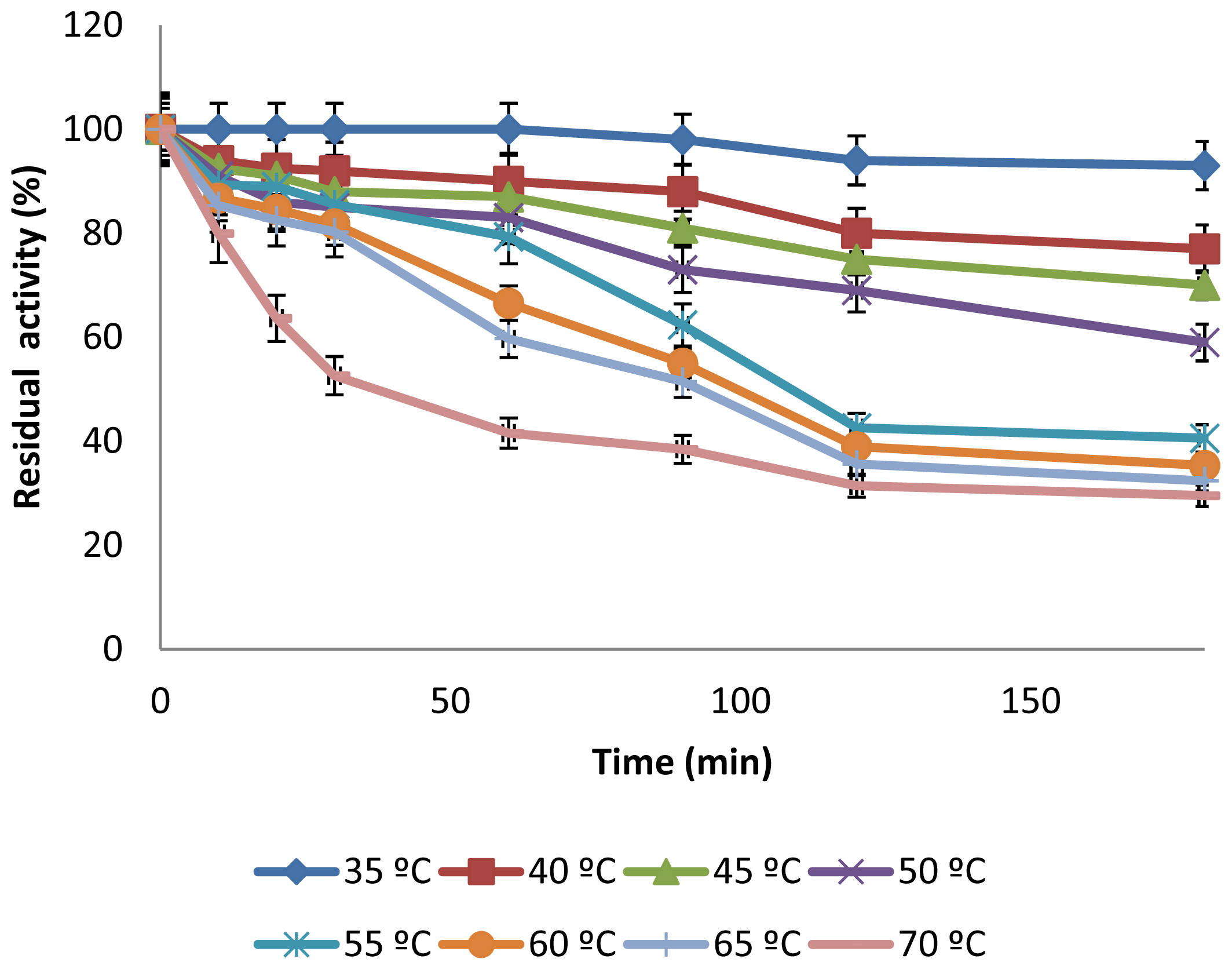
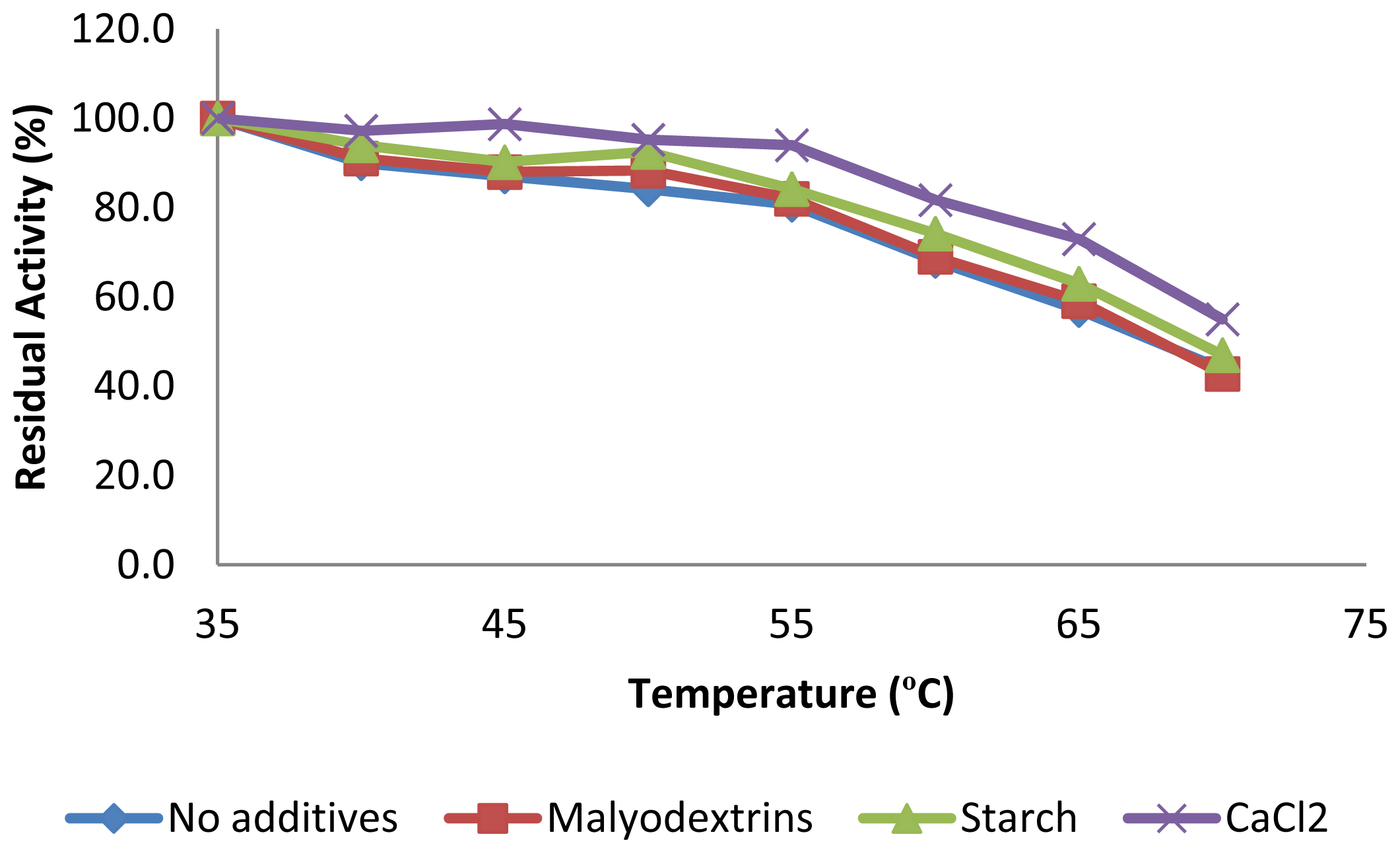
2.3.2. Effect of Temperature on CGTase Activity and Stability
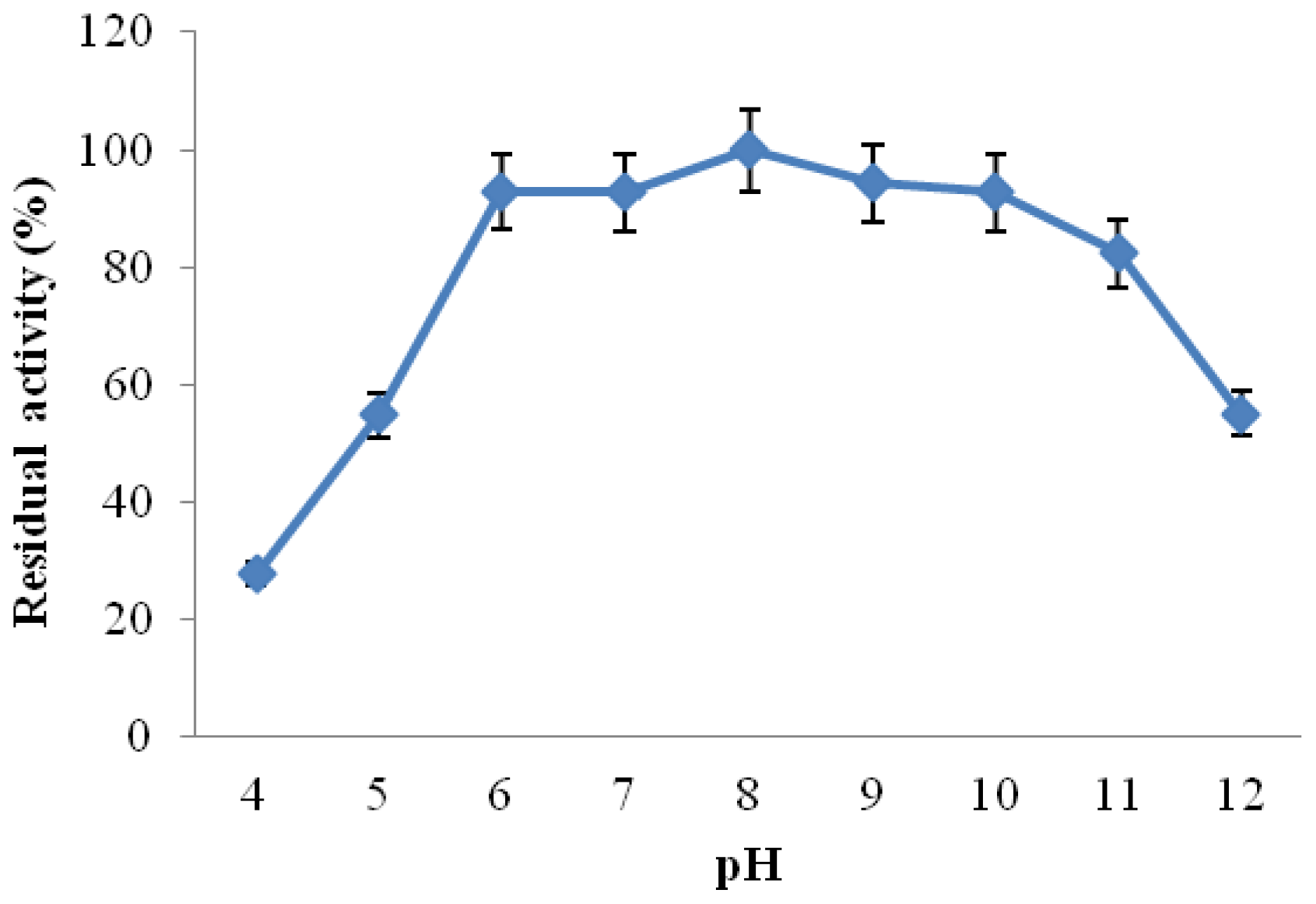
2.3.3. Effect of pH on CGTase Activity and Stability
2.3.4. Effect of Various Reagents and Metal Ions on CGTase Activity
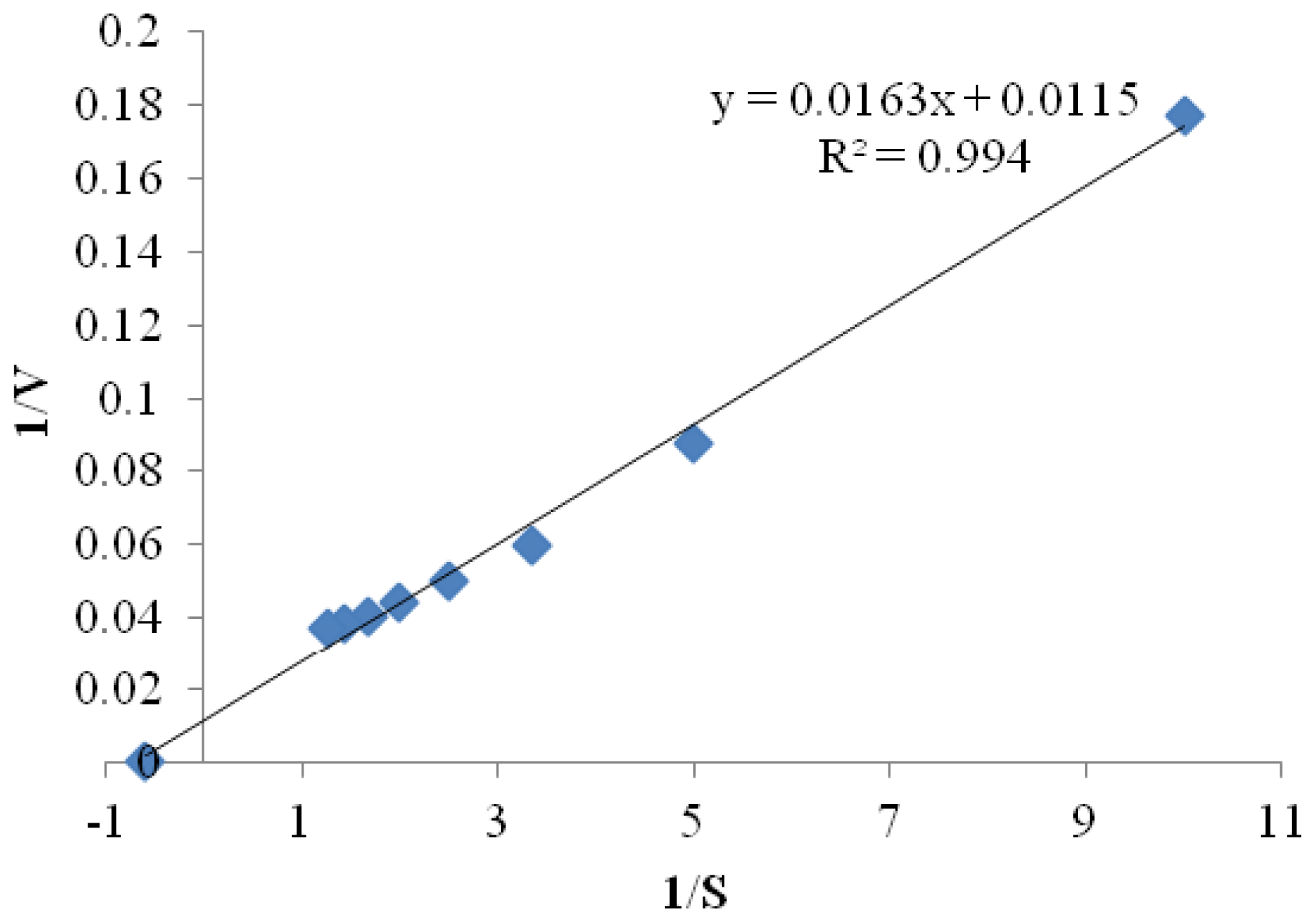
2.3.5. Kinetic Parameters
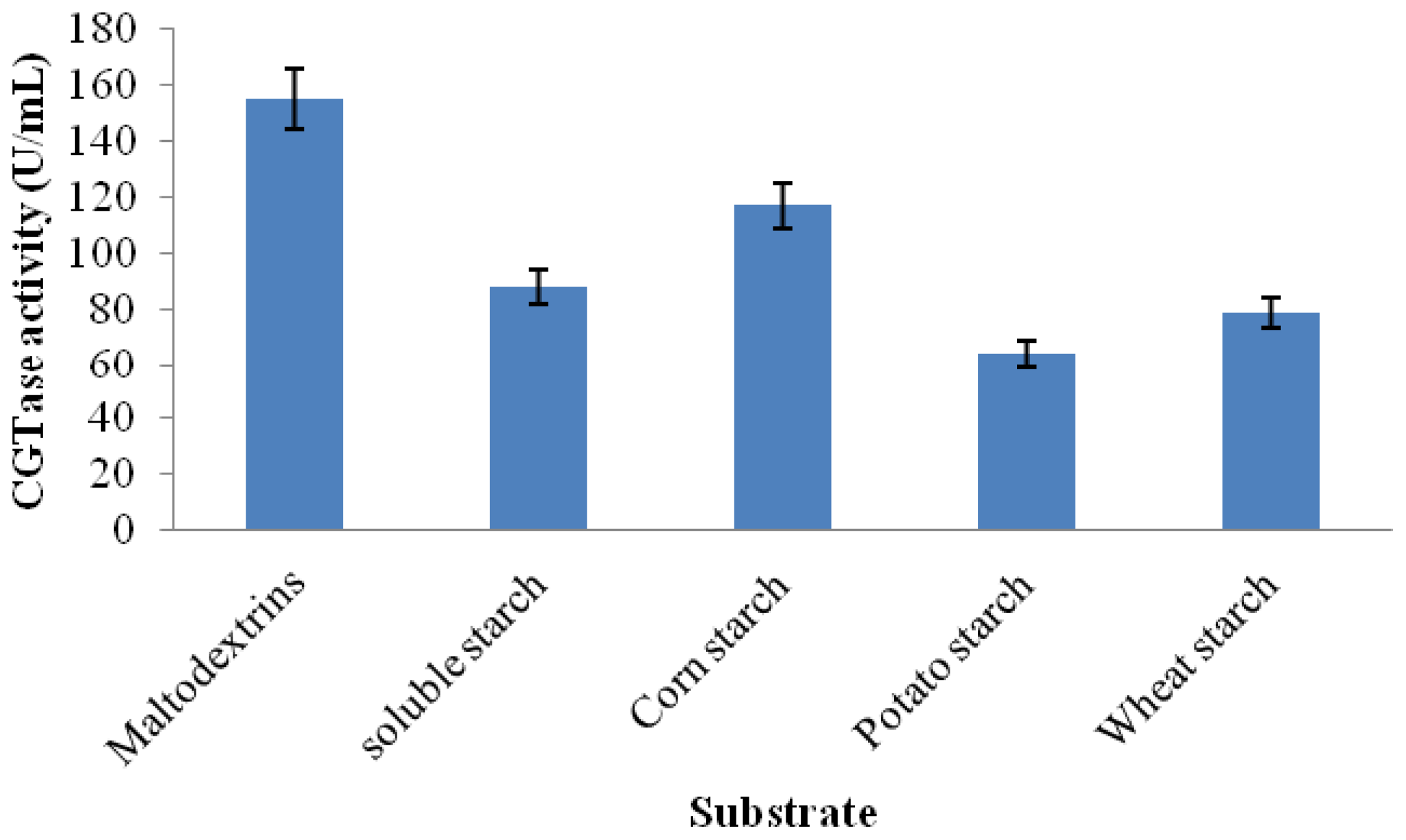
2.3.6. Substrate Specificity
2.3.7. Production of Cyclodextrins
3. Materials and Methods
3.1. Collection of Soil and Water Samples
3.2. Isolation of CGTase Producing Alkaliphilic Bacteria
3.3. Bacterial Identification
3.4. CGTase Production and Purification
3.5. Enzyme Assays
3.5.1. Cyclization Activity
3.5.2. Hydrolytic Activity
3.5.3. Coupling Activity
3.6. Characterization of the Purified CGTase
3.6.1. Estimation of the Molecular Weight of GTase
3.6.2. Zymogram
3.6.3. Effect of Temperature on Activity and Thermostability of CGTase
3.6.4. Effect of pH on Activity Stability of CGTase
3.6.5. Effect of Metal Ions and Inhibitors on CGTase Activity
3.6.6. Kinetic Studies
3.7. Cyclodextrin Production and Product Specificity of CGTase
4. Conclusions
Acknowledgement
Reference
- Biwer, A.; Antranikian, G.; Heinzle, E. Enzymatic production of cyclodextrins. Appl. Microbiol. Biotechnol 2001, 59, 609–617. [Google Scholar]
- Savergave, L.S.; Dhule, S.S.; Jogdand, V.V.; Nene, S.N.; Gadre, R.V. Production and single step purification of cyclodextrin glycosyltransferase from alkalophilic Bacillus firmus by ion exchange chromatography. Biochem. Eng. J 2008, 39, 510–515. [Google Scholar]
- Moriwaki, C.; Ferreira, L.R.; Rodella, J.R.T.; Matioli, G. A novel cyclodextrin glycosyltransferase from Bacillus sphaericus strain 41: Production, characterization and catalytic properties. Biochem. Eng. J 2009, 48, 124–131. [Google Scholar]
- Matte, C.R.; Nunes, M.R.; Benvenutti, E.V.; Schöffer, J.N.; Ayuba, M.A.; Hertz, P.F. Characterization of cyclodextrin glycosyltransferase immobilized on silica microspheres via aminopropyltrimethoxysilane as a “spacer arm”. J. Mol. Catal. B Enzym 2012, 78, 51–56. [Google Scholar]
- Martin del Valle, E.M. Cyclodextrins and their uses: A review. Process Biochem 2009, 39, 1033–1046. [Google Scholar]
- Otero-Espinar, F.J.; Luzardo-Alvarez, A.; Blanco-Mendez, J. Cyclodextrins: More than Pharmaceutical Excipients. Mini-Rev. Med. Chem 2010, 10, 715–725. [Google Scholar]
- Li, Z.; Wang, M.; Wang, F.; Gu, Z.; Du, G.; Wu, J.; Chen, J. Gamma Cyclodextrin: A review on enzymatic production and applications. Appl. Microbiol. Biotechnol 2007, 77, 245–255. [Google Scholar]
- Astray, G.; Gonzalez-Barreiro, C.; Mejuto, J.; Rial-Otero, R.; Simal-Gandara, J. A review on the use of cyclodextrins in foods. Food Hydrocoll 2009, 23, 1631–1641. [Google Scholar]
- Atanasova, N.; Kitayska, T.; Yankova, D.; Safarikova, M.; Tonkova, A. Cyclodextrin glucanotransferase production by cell biocatalysts of alkaliphilic bacilli. Biochem. Eng. J 2009, 46, 278–285. [Google Scholar]
- Antranikian, G.; Vorgias, C.E.; Bertoldo, C. Extreme environments as a resource for microorganisms and novel biocatalysts. Adv. Biochem. Eng. Biotechnol 2005, 96, 219–262. [Google Scholar]
- Horikoshi, K. Alkaliphiles: Some applications of their products for biotechnology. Microbiol. Mol. Biol. Rev 1999, 63, 735–750. [Google Scholar]
- Van den Burg, B. Extremophiles as a source for novel enzymes. Curr. Opin. Microbiol 2003, 6, 213–218. [Google Scholar]
- Grant, W.D.; Jones, B.E. Alkaline Environments. In Encyclopaedia of Microbiology, 2nd ed; Lederberg, J., Ed.; Academic Press: New York, NY, USA, 2000; pp. 126–133. [Google Scholar]
- Park, C.S.; Park, K.H.; Kim, S.H. A rapid screening method for alkaline β cyclodextrin-methyl orange containing solid medium. Agric. Biol. Chem 1989, 53, 1167–1169. [Google Scholar]
- Niimura, Y.; Koh, E.; Yanagida, F.; Suzuki, K.; Komagata, K.; Kozaki, M. Amphibacillus xylanus gen. nov., sp. nov., a facultatively anaerobic sporeforming xylan digesting bacterium which lacks cytochrome, quinone, and catalase. Int. J. Syst. Bacteriol 1990, 40, 297–301. [Google Scholar]
- Zhilina, T.N.; Garnova, E.S.; Tourova, T.P.; Kostrikina, N.A.; Zavarzin, G.A. Amphibacillus fermentum sp. nov. and Amphibacillus tropicus sp. nov., new alkaliphilic, facultatively anaerobic, saccharolytic bacilli from Lake Magadi. Microbiology 2001, 70, 711–722. [Google Scholar]
- An, S.Y.; Shu Ishikawa, S.; Kasai, H.; Goto, K.; Yokota, A. Amphibacillus sediminis sp. nov., an endosporeforming bacterium isolated from lake sediment in Japan. Int. J. Syst. Bacteriol 2007, 57, 2489–2492. [Google Scholar]
- Rahman, K.; Illias, R.M.; Hassan, O.; Mahmood, N.A.; Rashid, N.A. Molecular cloning of a cyclodextrin glucanotransferase gene from alkalophilic Bacillus sp. TS1-1 and characterization of the recombinant enzyme. Enzym. Microbial. Technol 2006, 39, 74–78. [Google Scholar]
- Charoensakdi, R.; Murakami, S.; Aoki, K.; Rimphanitchayakit, V.; Limpaseni, T. Cloning and expression of cyclodextrin glycosyltransferase gene from Paenibacillus sp. T16 isolated from hot spring soil in Northern Thailand. J. Biochem. Mol. Biol 2007, 40, 333–340. [Google Scholar]
- Alves-Prado, H.F.; Carneiro, A.J.; Pavezzi, F.C.; Gomes, E.; Boscolo, M.; Franco, C.L. Production of cyclodextrins by CGTase from Bacillus clausii using different starches as substrates. Appl. Biochem. Biotechnol 2008, 146, 3–13. [Google Scholar]
- Li, Z.; Li, B.; Gu, Z.; Du, G.; Wu, J.; Chen, J. Extracellular expression and biochemical characterization of α-cyclodextrin glycosyltransferase from Paenibacillus macerans. Carbohyd. Res 2010, 345, 886–892. [Google Scholar]
- Doukyu, N.; Kuwahara, H.; Ano, R. Isolation of Paenibacillus illiniosensis that produces cyclodextrin glucanotransferase resistant to organic solvents. Biosci. Biotechnol. Biochem 2003, 67, 334–340. [Google Scholar]
- Cao, X.; Jin, Z.; Wang, X.; Chen, F. A novel cyclodextrin glycosyl transferase from an alkalophilic Bacillus species: Purification and characterization. Food Res. Int 2005, 38, 309–314. [Google Scholar]
- Ong, R.M.; Goh, K.M.; Mahadi, N.M.; Hassan, O.; Rahman, R.Z.; Illias, R.M. Cloning, extracellular expression and characterization of a predominant β CGTase from Bacillus sp. G1 in E. coli. J. Ind. Microbiol. Biotechnol 2008, 35, 1705–1714. [Google Scholar]
- Martins, R.F.; Hatti-Kaul, R. Bacillus agaradhaerens LS-3C cyclodextrin glycosyltransferase: Activity and stability features. Enzym. Microb. Technol 2003, 33, 819–827. [Google Scholar]
- Van der Veen, B.A.; van Alebeek, G.W.M.; Uitdehaag, J.C.M.; Dijkstra, B.W.; Dijkhuizen, L. The three transglycosylation reactions catalyzed by cyclodextrin glycosyltransferase from Bacillus circulans (strain 251) proceed via different kinetic mechanisms. Eur. J. Biochem 2000, 267, 658–665. [Google Scholar]
- Hirano, K.; Ishihara, T.; Ogasawara, S.; Maeda, H.; Abe, K.; Nakajima, T. Molecular cloning and characterization of a novel γ-CGTase from alkalophilic Bacillus sp. Appl. Microbiol. Biotechnol 2006, 70, 193–201. [Google Scholar]
- Avci, A.; Dönmez, S. A novel thermophilic anaerobic bacteria producing cyclodextrin Glycosyltransferase. Process Biochem 2009, 44, 36–42. [Google Scholar]
- Atanasova, N.; Kitayska, D.; Bojadjieva, I.; Yankov, D.; Tonkova, A. A novel cyclodextrin glucanotransferase from alkaliphilic Bacillus pseudalcaliphilus 20RF: Purification and properties. Process Biochem 2011, 46, 116–122. [Google Scholar]
- Sian, H.K.; Said, M.; Hassan, O.; Kamaruddin, K.; Ismail, A.F.; Rahman, R.A. Purification and characterization of cyclodextrin glucanotransferase f sphaericus rom alkalophilic Bacillus sp. G1. Process Biochem 2005, 40, 1101–1111. [Google Scholar]
- Thiemann, V.; Donges, C.; Prowe, S.G.; Sterner, R.; Antranikian, G. Characterization of a thermoalkali-stable cyclodextrin glycosyltransferase from the anaerobic thermoalkaliphilic bacterium Anaerobranca gottschalkii. Arch. Microbiol 2004, 182, 226–235. [Google Scholar]
- Higuti, I.H.; Grande, S.W.; Sacco, R.; Nascimento, A.J. Isolation of alkalophilic CGTase producing bacteria and characterization of cyclodextrin glycosyltransferase. Braz. Arch. Biol. Technol 2003, 46, 183–186. [Google Scholar]
- Martins, R.F.; Hatti-Kaul, R. A new cyclodextrin glycosyltransferse from alkaliphilic Bacillus agaradhaerens isolate: Purification and characterisation. Enzym. Microb. Technol 2002, 30, 116–124. [Google Scholar]
- Alves-Prado, H.F.; Gomes, E.; da Silva, R. Purification and characterization of a cyclomaltodextrin glucanotransferase from Paenibacillus campinasensis H69-3. Appl. Biochem. Biotechnol 2007, (136–140), 41–55. [Google Scholar]
- Zhekova, B.Y.; Pishtiyski, I.G.; Stanchev, V.S. Investigation on cyclodextrin production with cyclodextrin glucanotransferase from Bacillus megaterium. Food Technol. Biotechnol 2008, 46, 328–334. [Google Scholar]
- Gastón, J.R.; Szerman, N.; Costa, H.; Krymkiewicz, N.; Ferrarotti, S.A. Cyclodextrin glycosyltransferase from Bacillus circulans DF 9R: Activity and kinetic studies. Enzym. Microbial. Technol 2009, 45, 36–41. [Google Scholar]
- Goh, K.M.; Mahadi, N.M.; Hassan, O.; Abdul Rahman, R.N.; Illias, R.M. The effects of reaction conditions on the production of γ-cyclodextrin from tapioca starch by using a novel recombinant engineered CGTase. J. Mol. Catal. B Enzym 2007, 49, 118–126. [Google Scholar]
- Doukyu, N.; Kuwahara, H.; Ano, R. Isolation of Paenibacillus illiniosensis that produces cyclodextrin glucanotransferase resistant to organic solvents. Biosci. Biotechnol. Biochem 2003, 67, 334–340. [Google Scholar]
- Taher, A.G. Inland saline lakes of Wadi El Natrun depression. Egypt Int. J Salt Lake Res 1999, 8, 149–169. [Google Scholar]
- National Center for Biotechnology Information. Available online: http://www.ncbi.nlm.nih.gov Accessed on 6 May 2012.
- Bradford, M.M. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 1976, 72, 248–254. [Google Scholar]
- Miller, G.L. Use of dinitrosalycilic acid reagent for determination of reducing sugar. Anal. Chem 1959, 31, 426–428. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar]
- Bluum, H.; Beier, H.; Gross, H.J. Improved silver staining method of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 1987, 8, 93–99. [Google Scholar]
- Krisman, C.R. A method for the colorimetric estimation of glycogen with iodine. Anal. Biochem 1962, 4, 17–23. [Google Scholar]
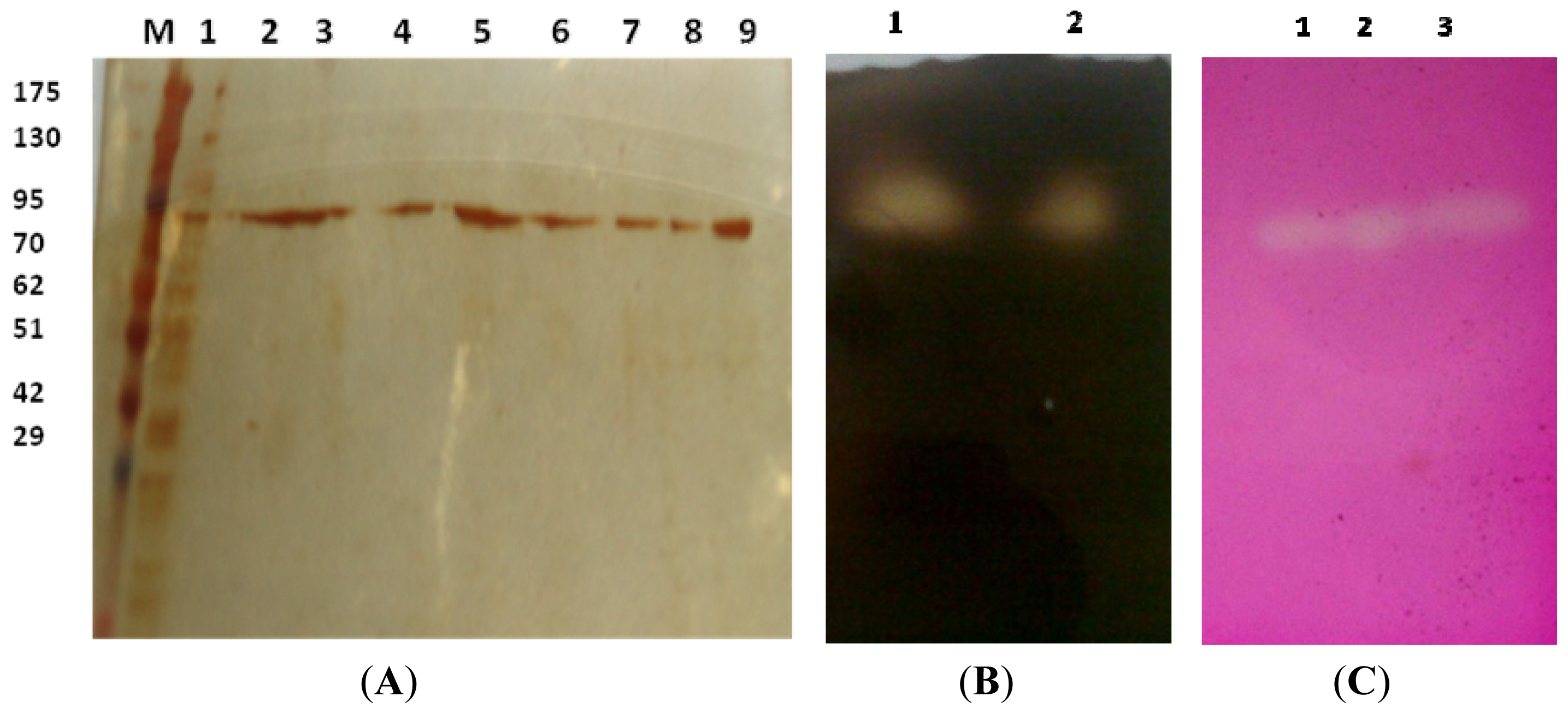

| Purification step | Volume(mL) | Activity(U mL−1) | Protein(mg mL−1) | Specific activity (U mg−1) | Yield(%) | Purification (Fold) |
|---|---|---|---|---|---|---|
| Crude | 1000 | 379.0 | 94.5 | 4.0 | 100 | 1.0 |
| Starch adsorption eluate | 285 | 177.5 | 2.0 | 88.8 | 44.7 | 22.2 |
| DEAE-Cellulose fractions | 8 | 144.2 | 1.6 | 89.0 | 36.3 | 23.1 |
| Salts/Reagent | Residual Activity (%) | |
|---|---|---|
| 1 mM | 10 mM | |
| CuCl2 | 99.2 ± 1.0 | 32.8 ± 1.5 |
| CaCl2 | 101.0 ± 2.0 | 108.7 ± 1.9 |
| ZnCl2 | 89.1 ± 1.9 | 40.3± 2.5 |
| MnSO4 | 93.1 ± 0.9 | 88.0 ± 0.8 |
| MgCl2 | 90.9 ± 0.85 | 96.6 ± 0.7 |
| CdCl2 | 82.6 ± 1.1 | 29.3 ± 1.0 |
| NaSo4 | 96.0 ± 0.9 | 94.7 ± 0.7 |
| BaCl2 | 85.1 ± 2.1 | 65.7 ± 1.9 |
| HgBr2 | 85.1 ± 2.0 | 0.0 |
| NiCl2 | 98.1 ± 0.8 | 101.2 ± 0.5 |
| Co(NO3)2 | 100.9 ± 1.0 | 82.1 ± 1.2 |
| EDTA | 100.8 ± 0.7 | 99.5 ± 0.6 |
| 2-ME | 98.8 ± 0.6 | 76.7 ± 0.7 |
| α-CD | 68.7 ± 2.3 | N/A |
| β-CD | 45.7 ± 2.2 | N/A |
| γ-CD | 54.2 ± 1.8 | N/A |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ibrahim, A.S.S.; Al-Salamah, A.A.; El-Tayeb, M.A.; El-Badawi, Y.B.; Antranikian, G. A Novel Cyclodextrin Glycosyltransferase from Alkaliphilic Amphibacillus sp. NPST-10: Purification and Properties. Int. J. Mol. Sci. 2012, 13, 10505-10522. https://doi.org/10.3390/ijms130810505
Ibrahim ASS, Al-Salamah AA, El-Tayeb MA, El-Badawi YB, Antranikian G. A Novel Cyclodextrin Glycosyltransferase from Alkaliphilic Amphibacillus sp. NPST-10: Purification and Properties. International Journal of Molecular Sciences. 2012; 13(8):10505-10522. https://doi.org/10.3390/ijms130810505
Chicago/Turabian StyleIbrahim, Abdelnasser S. S., Ali A. Al-Salamah, Mohamed A. El-Tayeb, Yahya B. El-Badawi, and Garabed Antranikian. 2012. "A Novel Cyclodextrin Glycosyltransferase from Alkaliphilic Amphibacillus sp. NPST-10: Purification and Properties" International Journal of Molecular Sciences 13, no. 8: 10505-10522. https://doi.org/10.3390/ijms130810505
APA StyleIbrahim, A. S. S., Al-Salamah, A. A., El-Tayeb, M. A., El-Badawi, Y. B., & Antranikian, G. (2012). A Novel Cyclodextrin Glycosyltransferase from Alkaliphilic Amphibacillus sp. NPST-10: Purification and Properties. International Journal of Molecular Sciences, 13(8), 10505-10522. https://doi.org/10.3390/ijms130810505




