Nuclei of Tsuga canadensis: Role of Flavanols in Chromatin Organization
Abstract
:1. Introduction
2. Material and Methods
2.1. Collection Sites and Tissue Sampling
2.2. Histochemistry
2.3. Blue Staining of Flavanols Based on the DMACA Reagent
2.4. Incubation of Tissues
2.5. Densitometric Studies
2.6. Analytical Determination of Flavonoids by HPLC
2.7. Peak Identification
3. Results
3.1. Short Cell Lineages
3.2. Long Cell Lineages
3.3. Vacuolate Cells with a Silenced Nuclear Flavanol Pattern
3.4. Daughter Cells Display a Nearly Similar Nuclear Flavanol Pattern
3.5. Variable Nuclear Flavanol Pattern of Fully Differentiated Single Cells
3.6. Nucleoli of Somatic Cells of Tsuga canadensis Are not Recognizable by DMACA Staining
3.7. Nucleoli of Sporogenic Cells Are Detectable by DMACA Staining
3.8. Flavanols and Other Phenolic Compounds during Development of Microspores and Young Needles
3.9. Densitometric Scanning and Expression of Chromatin Activation (Table 1)
4. Discussion
4.1. Highly Mitotic Cells Possess a Mosaic-Like Nuclear Flavanol Pattern
4.2. Different Flavanol Expression of Two Cell Lineages Located Close Together
4.3. Daughter Cells and Beyond: The Nuclear Flavanol Pattern Linked with Progressing Differentiation
4.4. Epigenetic Stress, Loss of Nuclear Flavanols and Recovery by Sucrose and Cytokinin
4.5. Nucleoli of Somatic and Meiotic Cells
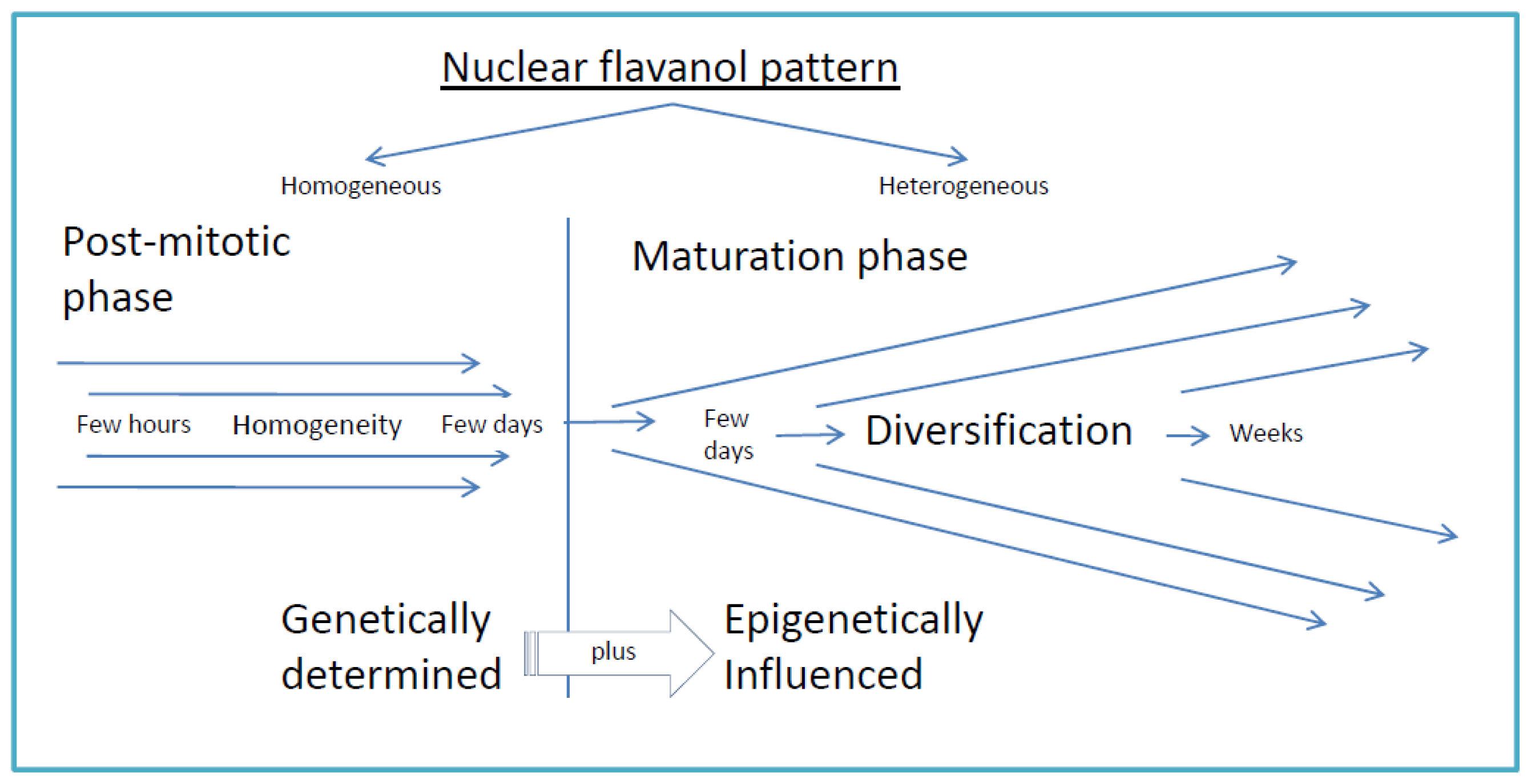
5. Conclusion: Basic Principles of Flavanol Interactions in Nuclear Organization
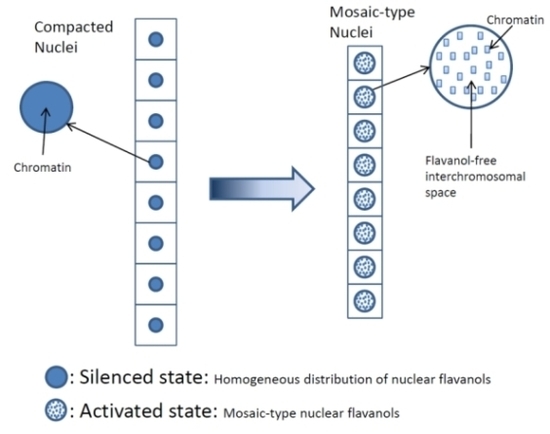
- Low molecular weight flavanols in the nuclei are controlled transcriptionally as shown by the homogeneinity of lineage cells (Figure 2);
- The inherited pattern of nuclear flavanol expression is modified by epigenetic factors (Figure 4) as most dramatically shown during drought periods;
- Thus, development and final differentiation of a cell is mirrored by changing subnuclear patterns of pale blue euchromatin, dark blue heterochromatin and different amounts of flavanols in the interchromosomal space (Figure 2);
- All in all, the varying spatial densities of anchored or soluble flavanol molecules are expected to modify a series of physico-chemical parameters within the nucleosomal microenvironment (Figure 2).
Abbreviations
| DMACA | p-dimethylaminocinnamaldehyde |
References
- Feucht, W; Treutter, D; Polster, J. Flavanol binding of nuclei from tree species. Plant Cell Rep 2004, 22, 430–436. [Google Scholar]
- Polster, J; Dithmar, H; Burgemeister, R; Friedemann, G; Feucht, W. Flavonoids in plant nuclei: Detection by laser microdissection and pressure catapulting (LMPC), in vivo staining, and uv–visible spectroscopic titration. Physiol. Plant 2006, 128, 163–174. [Google Scholar]
- Polster, J; Dithmar, H; Feucht, W. Are Histones the targets for flavan-3-ols (Catechins) in nuclei? Biol. Chem 2003, 384, 997–1006. [Google Scholar]
- Mueller-Harvey, I; Feucht, W; Polster, J; Burgos, P; Parker, A; Botschway, S. Lasers to probe the nuclear binding of flavanols. In Annual Report for Appletons Laboratory Rutherford; Oxfordshire, UK, 2009. [Google Scholar]
- Feucht, W; Dithmar, H; Polster, J. Nuclei of Taxus baccata: Flavanols linked to chromatin remodeling factors. J Bot 2009. [Google Scholar] [CrossRef]
- Misteli, T; Dernburg, AF. Chromosomes and expression mechanisms - everything old is new again. Curr. Opin. Genet. Dev 2007, 17, 85–87. [Google Scholar]
- Berckmans, B; De Veylder, L. Transcriptional control of the cell cycle. Curr. Opin. Plant Biol 2009, 12, 599–605. [Google Scholar]
- Feucht, W; Dithmar, H; Polster, J. Variation of the nuclear, subnuclear and chromosomal flavanol deposition in hemlock and rye. Int. J. Mol. Sci 2007, 8, 635–650. [Google Scholar]
- Feucht, W; Treutter, D; Dithmar, H; Polster, J. Microspore development of three coniferous species: Affinity of nuclei for flavonoids. Tree Physiol 2008, 28, 1783–1791. [Google Scholar]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar]
- Haslam, E. Practical Polyphenolics: From Structure to Molecular Recognition and Physiological Action; Cambridge University Press: Cambridge, UK, 1998. [Google Scholar]
- Coleman, AW; Maguire, MJ; Coleman, JR. Mithramycin- and 4′-6-diamidino-2-phenylindole (DAPI)-DNA staining for fluorescence microspectrophotometric measurement of DNA in nuclei, plastids, and virus particles. J. Histochem. Cytochem 1981, 29, 959–968. [Google Scholar]
- Shorack, GR; Wellner, JA. Empirical Processes with Applications to Statistics; Wiley: New York, NY, USA, 1986. [Google Scholar]
- Treutter, D; Santos-Buelga, C; Gutmann, M; Kolodziej, H. Identification of flavan-3-ols and procyanidins by high-performance liquid chromatography and chemical reaction detection. J. Chromatogr. A 1994, 667, 290–297. [Google Scholar]
- Weigel, D; Jurgens, G. Stem cells that make stems. Nature 2002, 415, 751–754. [Google Scholar]
- Mazumdar, M; Misteli, T. Chromokinesins: Multitalented players in mitosis. Trends Cell Biol 2005, 15, 349–355. [Google Scholar]
- Haywood, V; Kragler, F; Lucas, WJ. Plasmodesmata: Pathways for protein and ribonucleoprotein signaling. Plant Cell 2002, 14, S303–S325. [Google Scholar]
- Grewal, SIS; Moazed, D. heterochromatin and epigenetic control of gene expression. Science 2003, 301, 798–802. [Google Scholar]
- Misteli, T. Protein Dynamics: Implications for nuclear architecture and gene expression. Science 2001, 291, 843–847. [Google Scholar]
- Gunjan, A; Paik, J; Verreault, A. Regulation of histone synthesis and nucleosome assembly. Biochimie 2005, 87, 625–635. [Google Scholar]
- Samaniego, R; Jeong, SY; de la Torre, C; Meier, I; Díaz de la Espina, SM. CK2 phosphorylation weakens 90 kDa MFP1 association to the nuclear matrix in Allium cepa. J. Exp. Bot 2006, 57, 113–124. [Google Scholar]
- Scheres, B. Plant Cell Identity. The role of position and lineage. Plant Physiol 2001, 125, 112–114. [Google Scholar]
- Wu, X; Dinneny, JR; Crawford, KM; Rhee, Y; Citovsky, V; Zambryski, PC; Weigel, D. Modes of intercellular transcription factor movement in the Arabidopsis apex. Development 2003, 130, 3735–3745. [Google Scholar]
- Kurata, T; Okada, K; Wada, T. Intercellular movement of transcription factors. Curr. Opin. Plant Biol 2005, 8, 600–605. [Google Scholar]
- Scholten, S; Lörz, H; Kranz, E. Paternal mRNA and protein synthesis coincides with male chromatin decondensation in maize zygotes. Plant J 2002, 32, 221–231. [Google Scholar]
- Jasencakova, Z; Soppe, WJJ; Meister, A; Gernand, D; Turner, BM; Schubert, I. Histone modifications in Arabidopsis–high methylation of H3 lysine 9 is dispensable for constitutive heterochromatin. Plant J 2003, 33, 471–480. [Google Scholar]
- Krogan, NT; Long, JA. Why so repressed? Turning off transcription during plant growth and development. Curr. Opin. Plant Biol 2009, 12, 628–636. [Google Scholar]
- Richards, EJ; Elgin, SCR. Epigenetic codes for heterochromatin formation and silencing: Rounding up the usual suspects. Cell 2002, 108, 489–500. [Google Scholar]
- Soria-Guerra, R; Rosales-Mendoza, S; Chang, S; Haudenshield, J; Padmanaban, A; Rodriguez-Zas, S; Hartman, G; Ghabrial, S; Korban, S. Transcriptome analysis of resistant and susceptible genotypes of Glycine tomentella during Phakopsora pachyrhizi infection reveals novel rust resistance genes. Theor. Appl. Genet 2010, 120, 1315–1333. [Google Scholar]
- Carmo-Fonseca, M. The contribution of nuclear compartmentalization to gene regulation. Cell 2002, 108, 513–521. [Google Scholar]
- Liu, Z; Karmarkar, V. Groucho/Tup1 family co-repressors in plant development. Trends Plant Sci 2008, 13, 137–144. [Google Scholar]
- Misteli, T. Beyond the sequence: Cellular organization of genome function. Cell 2007, 128, 787–800. [Google Scholar]
- Zhao, Y. The role of local biosynthesis of auxin and cytokinin in plant development. Curr. Opin. Plant Biol 2008, 11, 16–22. [Google Scholar]
- Vanneste, S; Friml, J. Auxin: A trigger for change in plant development. Cell 2009, 136, 1005–1016. [Google Scholar]
- Bernard, P; Maure, JF; Partridge, JF; Genier, S; Javerzat, JP; Allshire, RC. Requirement of heterochromatin for cohesion at centromeres. Science 2001, 294, 2539–2542. [Google Scholar]
- Li, B; Carey, M; Workman, JL. The role of chromatin during transcription. Cell 2007, 128, 707–719. [Google Scholar]
- Ries, G; Heller, W; Puchta, H; Sandermann, H; Seidlitz, HK; Hohn, B. Elevated UV-B radiation reduces genome stability in plants. Nature 2000, 406, 98–101. [Google Scholar]
- Treutter, D. Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biol 2005, 7, 581–591. [Google Scholar]
- Kuhnle, JA; Fuller, G; Corse, J; Mackey, BE. Antisenescent activity of natural cytokinins. Physiol. Plant 1977, 41, 14–21. [Google Scholar]
- Treutter, D; Santos-Buelga, C; Gutmann, M; Kolodziej, K. Taxonomically relevant flavonoid glycosides of Prunus phloem and their response according to various physiological conditions. Groupe Polyphenols Bull. Liaison 1986, 13, 45–49. [Google Scholar]
- Ferreira, FJ; Kieber, JJ. Cytokinin signaling. Curr. Opin. Plant Biol 2005, 8, 518–525. [Google Scholar]
- Hutchison, CE; Kieber, JJ. Cytokinin signaling in Arabidopsis. Plant Cell Online 2002, 14, S47–S59. [Google Scholar]
- Gaudino, RJ; Pikaard, CS. Cytokinin induction of RNA polymerase I transcription in Arabidopsis thaliana. J. Biol. Chem 1997, 272, 6799–6804. [Google Scholar]
- Creasy, LL. The increase in phenylalanine ammonia-lyase activity in strawberry leaf disks and its correlation with flavonoid synthesis. Phytochemistry 1968, 7, 441–446. [Google Scholar]
- Gibson, SI. Control of plant development and gene expression by sugar signaling. Curr. Opin. Plant Biol 2005, 8, 93–102. [Google Scholar]
- van Attikum, H; Gasser, SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol 2009, 19, 207–217. [Google Scholar]
- Nitsch, JP. Experimental androgenesis in Nicotiana. Phytomorphology 1969, 19, 389–404. [Google Scholar]
- Burger, EC; Müller, M. Multiplied nucleolus organizer regions (NORs) in polyploid nuclei Vicia faba revealed by ammoniacal silver staining. Cell. Mol. Life Sci 1987, 43, 421–423. [Google Scholar]
- Rose, RJ. Changes in nucleolar activity during the growth and development of the wheat coleoptile. Protoplasma 1974, 79, 127–143. [Google Scholar]
- Hubbell, HR. Silver staining as an indicator of active ribosomal genes. Biotech. Histochem 1985, 60, 285–294. [Google Scholar]
- Heslop-Harrison, J. Comparative genome organisation in plants: From sequence and markers to chromatin and chromosomes. Plant Cell 2000, 14, 617–635. [Google Scholar]
- Jasencakova, Z; Meister, A; Walter, J; Turner, BM; Schubert, I. Histone H4 acetylation of euchromatin and heterochromatin is cell cycle dependent and correlated with replication rather than with transcription. Plant Cell 2000, 12, 2087–2100. [Google Scholar]
- Kaszas, E; Cande, WZ. Phosphorylation of histone H3 is correlated with changes in the maintenance of sister chromatid cohesion during meiosis in maize, rather than the condensation of the chromatin. J. Cell Sci 2000, 113, 3217–3226. [Google Scholar]
- Mizushina, Y; Saito, A; Tanaka, A; Nakajima, N; Kuriyama, I; Takemura, M; Takeuchi, T; Sugawara, F; Yoshida, H. Structural analysis of catechin derivatives as mammalian DNA polymerase inhibitors. Biochem. Biophys. Res. Commun 2005, 333, 101–109. [Google Scholar]
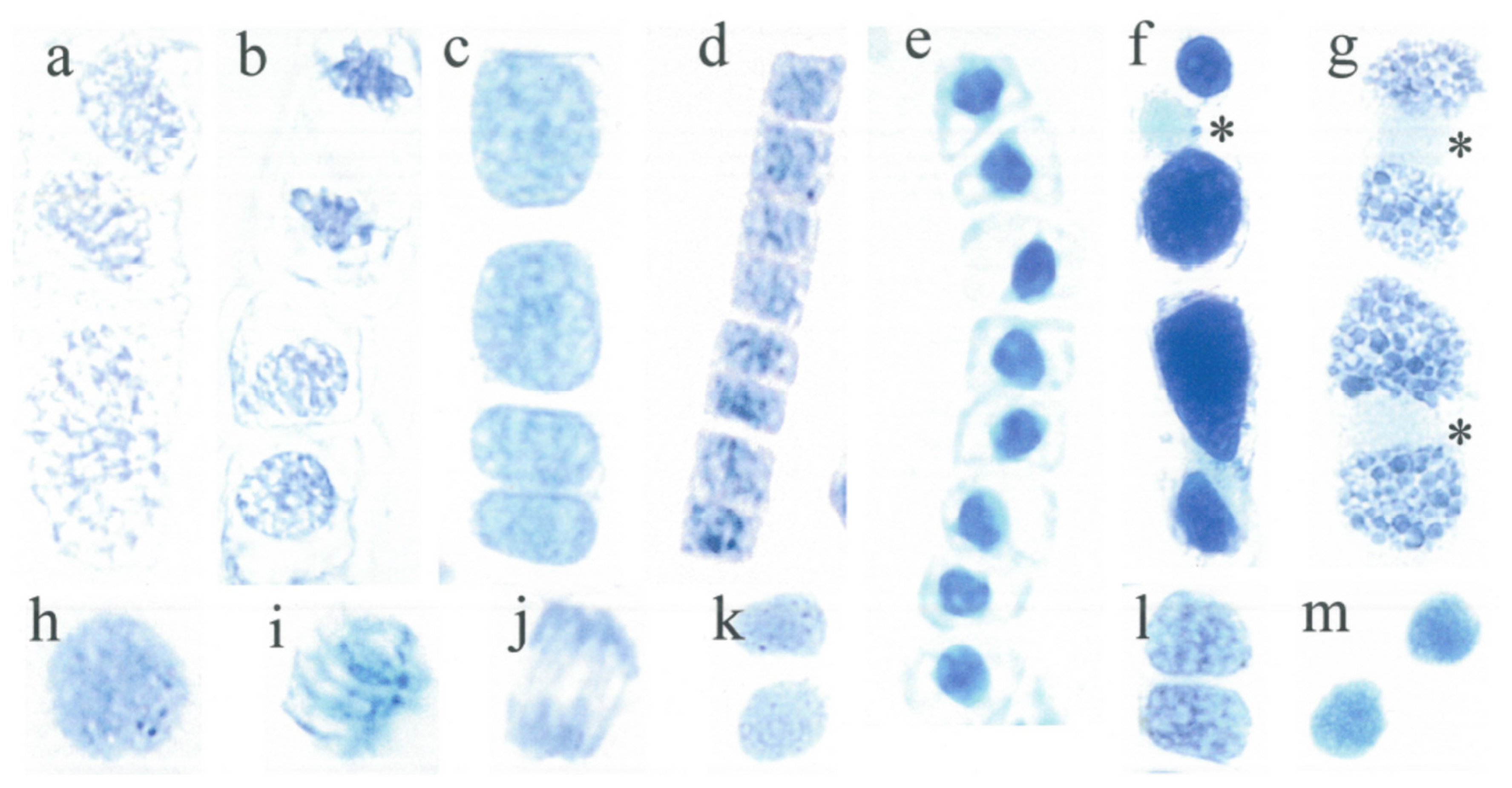
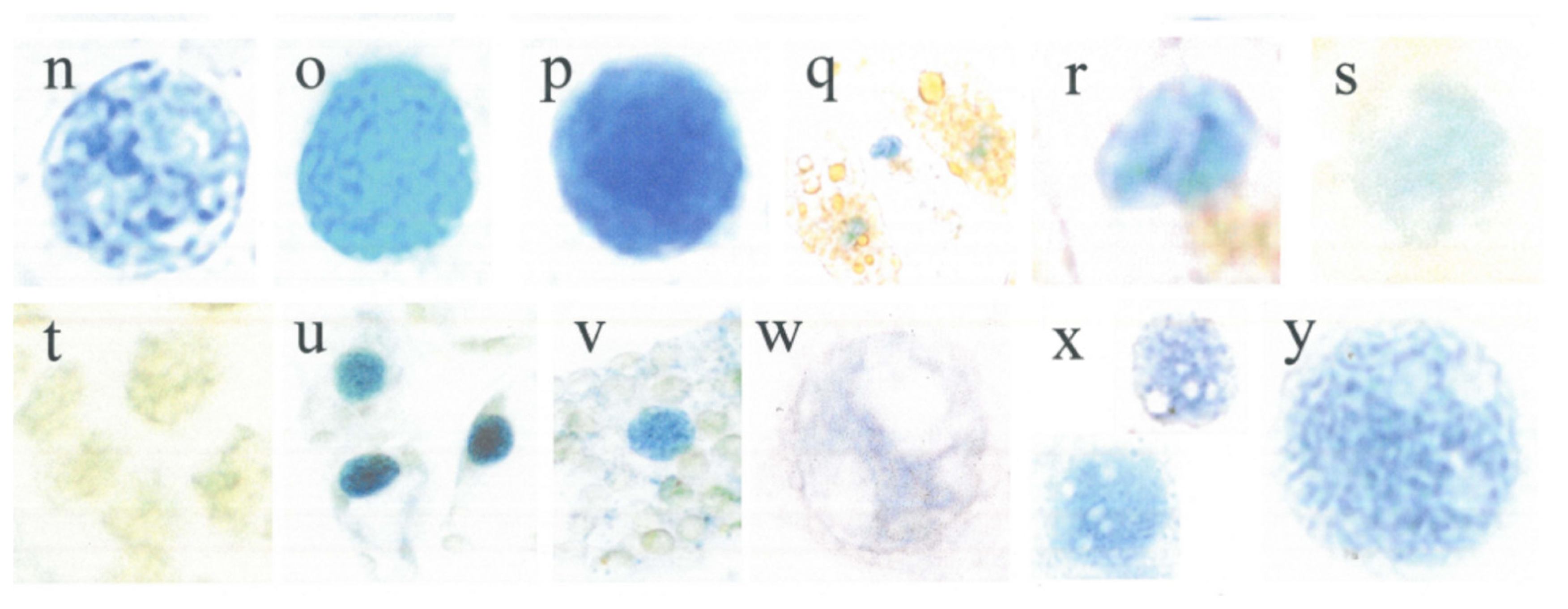

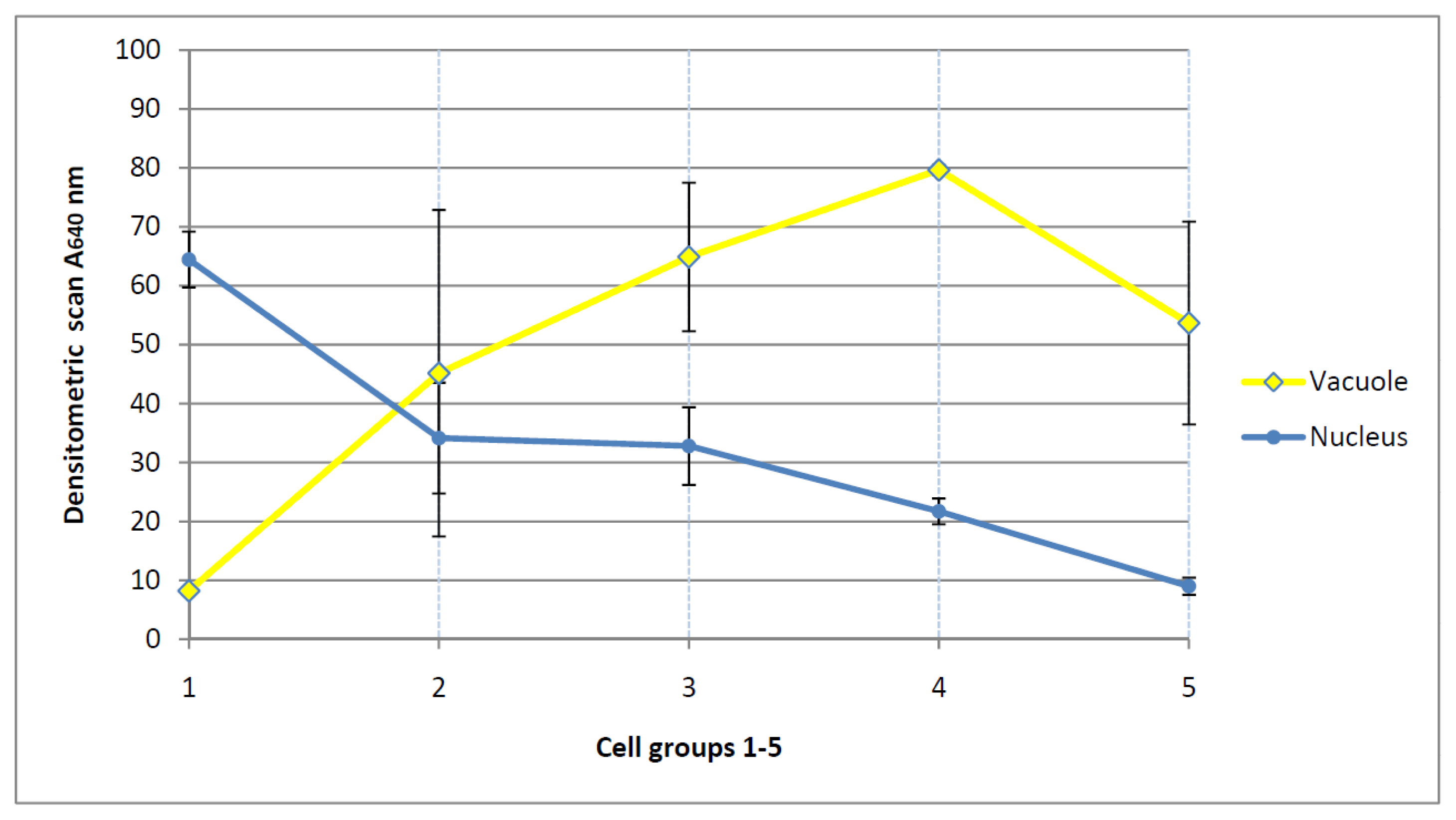
| Figures | Absorbance (640 nm) | Standard deviation SD | State | Relative SD | Relative SD |
|---|---|---|---|---|---|
| a1 | 21.6 | 4.6 | active | 21 | 16 |
| a2 | 17.8 | 1.4 | active | 8 | |
| b1 | 49.9 | 8.8 | active | 18 | |
| b2 | 30.3 | 2.9 | active | 10 | |
| c1 | 37.5 | 5.9 | active | 16 | |
| c2 | 36.9 | 7.0 | active | 19 | |
| d1 | 52.0 | 20.8 | active | 40 | |
| n1 | 35.0 | 8.5 | active | 24 | |
| w1 | 24.1 | 2.2 | active | 9 | |
| x1 | 34.8 | 4.5 | active | 13 | |
| x2 | 20.4 | 1.8 | active | 9 | |
| y1 | 47.8 | 4.7 | active | 10 | |
| h1 | 49.4 | 0.1 | intermediate | 0 | 11 |
| i1 | 35.5 | 8.6 | intermediate | 24 | |
| j1 | 82.7 | 7.9 | intermediate | 10 | |
| k1 | 35.8 | 5.9 | intermediate | 16 | |
| l1 | 53.7 | 6.6 | intermediate | 12 | |
| m1 | 72.4 | 3.7 | intermediate | 5 | |
| e1 | 96.0 | 8.6 | silent | 9 | 4 |
| f1 | 31.3 | 1.0 | silent | 3 | |
| g1 | 19.6 | 0.2 | silent | 1 | |
| o1 | 49.4 | 4.0 | silent | 8 | |
| p1 | 100.0 | 0.0 | silent | 0 | |
| q1 | magnification see r1 | - | silent | - | |
| r1 | 45.7 | 3.3 | silent | 7 | |
| s1 | 21.5 | 2.2 | silent | 10 | |
| t1 | 0 --> heat | - | silent | - | |
| u1 | 88.4 | 0.5 | silent | 1 | |
| v1 | 44.3 | 0.4 | silent | 1 |
| Lineage | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 1–8 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Silenced | A | 91 | 106 | 101 | 93 | 104 | 93 | 79 | 97 | 96 |
| SD | 7.0 | 2.8 | 2.8 | 0.4 | 1.3 | 3.3 | 4.8 | 8.5 | 8.6 | |
| Activated | A | 27 | 49 | 29 | 48 | 48 | 85 | 54 | 79 | 52 |
| SD | 2.6 | 13.9 | 5.2 | 11.8 | 17.6 | 7.3 | 10.8 | 18.4 | 20.8 |
| Lineage | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 1–8 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Silenced | C | 51 | 49 | 70 | 54 | 53 | 66 | 45 | 60 | 56 |
| N | 16 | 17 | 17 | 17 | 15 | 19 | 15 | 19 | 17 | |
| R | 3.2 | 2.9 | 4.1 | 3.2 | 3.5 | 3.5 | 3.0 | 3.2 | 3.3 | |
| Activated | C | 40 | 50 | 35 | 37 | 41 | 39 | 48 | 45 | 42 |
| N | 31 | 37 | 27 | 31 | 36 | 26 | 40 | 29 | 32 | |
| R | 1.3 | 1.4 | 1.3 | 1.2 | 1.1 | 1.5 | 1.2 | 1.6 | 1.3 |
| Catechin | Epigallocatechin | Epicatechin | Oligomers | Hydroxycinnamic acids | Flavonols | |
|---|---|---|---|---|---|---|
| Microspores | 8.6 | 0.6 | 7.1 | 3.3 | 2.9 | 0.06 |
| Young needles | 1.6 | 0.2 | 0.7 | 0.9 | 161.0 | 0.3 |
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Feucht, W.; Schmid, M.; Treutter, D. Nuclei of Tsuga canadensis: Role of Flavanols in Chromatin Organization. Int. J. Mol. Sci. 2011, 12, 6834-6855. https://doi.org/10.3390/ijms12106834
Feucht W, Schmid M, Treutter D. Nuclei of Tsuga canadensis: Role of Flavanols in Chromatin Organization. International Journal of Molecular Sciences. 2011; 12(10):6834-6855. https://doi.org/10.3390/ijms12106834
Chicago/Turabian StyleFeucht, Walter, Markus Schmid, and Dieter Treutter. 2011. "Nuclei of Tsuga canadensis: Role of Flavanols in Chromatin Organization" International Journal of Molecular Sciences 12, no. 10: 6834-6855. https://doi.org/10.3390/ijms12106834
APA StyleFeucht, W., Schmid, M., & Treutter, D. (2011). Nuclei of Tsuga canadensis: Role of Flavanols in Chromatin Organization. International Journal of Molecular Sciences, 12(10), 6834-6855. https://doi.org/10.3390/ijms12106834