Abstract
Lipopeptide biosurfactants (LPBSs) consist of a hydrophobic fatty acid portion linked to a hydrophilic peptide chain in the molecule. With their complex and diverse structures, LPBSs exhibit various biological activities including surface activity as well as anti-cellular and anti-enzymatic activities. LPBSs are also involved in multi-cellular behaviors such as swarming motility and biofilm formation. Among the bacterial genera, Bacillus (Gram-positive) and Pseudomonas (Gram-negative) have received the most attention because they produce a wide range of effective LPBSs that are potentially useful for agricultural, chemical, food, and pharmaceutical industries. The biosynthetic mechanisms and gene regulation systems of LPBSs have been extensively analyzed over the last decade. LPBSs are generally synthesized in a ribosome-independent manner with megaenzymes called nonribosomal peptide synthetases (NRPSs). Production of active-form NRPSs requires not only transcriptional induction and translation but also post-translational modification and assemblage. The accumulated knowledge reveals the versatility and evolutionary lineage of the NRPSs system. This review provides an overview of the structural and functional diversity of LPBSs and their different biosynthetic mechanisms in Bacillus and Pseudomonas, including both typical and unique systems. Finally, successful genetic engineering of NRPSs for creating novel lipopeptides is also discussed.
1. Introduction
Biosurfactants are biological surface-active compounds largely produced by a wide variety of microorganisms. They have environmentally friendly properties, such as low toxicity and high biodegradability. Biosurfactants are capable of lowering surface and interfacial tensions effectively and thus are potential substitutes for widely used chemically synthesized surfactants. Thus, biosurfactants show promise for use in agricultural, food, pharmaceutical, cosmetic, specialty chemical, and oil production industries as well as in bioremediation technology. The hydrophobic portion of these molecules is commonly made up of fatty acids (saturated, unsaturated, or hydroxylated), whereas the hydrophilic portion is usually composed of peptides or mono-, di-, or polysaccharides [1]. Due to the presence of hydrophobic and hydrophilic moieties within a single molecule, (bio)surfactants tend to migrate toward an interface with different degrees of polarity and hydrogen bonding, such as an air/water or oil/water interface [2].
Production of effective lipopeptide biosurfactants (LPBSs) was first reported from Gram-positive Bacillus subtilis IAM1213 [3]. Since then, various types of LPBSs with significant surface activity and/or anti-microbial activity have been isolated from other Bacillus strains [4–7]. Gram-negative Pseudomonas spp. also produce a variety of LPBSs [8–12]. Gene clusters encoding multi-modular nonribosomal peptide synthetases (NRPSs) for LPBS production have been cloned and characterized from these two genera, and demonstrate their different evolutionary lineages. The systematic modular organization of NRPSs allows structural alteration of lipopeptides by swapping domains or modules of NRPSs to create novel lipopeptides [13,14]. This article aims to summarize current knowledge of the structural diversity of LPBSs and their biosynthetic systems in the genera Bacillus and Pseudomonas. Current possibilities and limits in the modification of their biosynthetic genes for the synthesis of novel compounds are also discussed.
2. Diversity of LPBSs and NRPSs from Bacillus and Pseudomonas
Bacillus and Pseudomonas spp. produce a wide range of LPBSs (Tables 1, 2). These lipopeptides share similar amphipathic structures containing both a hydrophilic peptide portion and a hydrophobic fatty acid portion. Most of the LPBSs have a cyclic structure mediated by the linkage between a C-terminal peptide residue and either a β-hydroxy fatty acid, hydroxyl group of the peptide residue, or β-amino acid. NRPSs are multi-modular enzymes that recognize, activate, modify, and link the amino acid intermediates to the product peptide [15]. They are capable of synthesizing peptides that contain unusual amino acids including d-amino acids, β-amino acids, and hydroxy- or N-methylated amino acids. Biosynthesis of nonribosomal peptides occurs via the function of the catalytic unit, referred to as a module. The order of modules is usually co-linear to the product peptide sequences (Figure 1). Each module is composed of specific domains that are responsible for catalyzing different enzymatic activities.

Table 1.
Primary structure of representative lipopeptides produced by Bacillus strains.

Table 2.
Primary structure of representative lipopeptides produced by Pseudomonas.

Figure 1.
Multidomain organization of the gene clusters encoding NRPSs from Bacillus (A) and Pseudomonas (B).
The adenylation (A) domain is responsible for amino acid recognition and adenylation at the expense of ATP to form an acyl-adenylate intermediate. Then, the adenylated amino acid covalently binds to a phosphopantetheine carrier of the adjacent thiolation (T) or peptidyl carrier protein (PCP) domain. Peptide bond formation of two consecutively bound amino acids is catalyzed by the condensation (C) domain. Modification domains, such as the epimerization (E) domain, catalyze the conversion of l-amino acids to d-isomers, and they are typically associated with the module that incorporates d-amino acids. Lastly, cyclization and release of the product peptide are carried out by C-terminal thioesterase (Te) domain that is associated with a termination module. A number of gene clusters encoding NRPSs for LPBS biosynthesis in both Bacillus and Pseudomonas have been cloned and characterized (Figure 1). These gene clusters show striking similarities in the modular architecture of their repetitive catalytic units and assembly-line mechanism. However, several unique features have also been identified.
2.1. Bacillus
Most LPBSs from Bacillus can be classified into three families: surfactin, fengycin, and iturin [16]. There are several Bacillus strains that have the ability to produce three families of LPBSs simultaneously [17–20]. In addition to these three families, several other lipopeptides have also been identified in Bacillus species (Table 1). Characteristics of these LPBSs families and corresponding NRPSs are as follows.
2.1.1. Surfactin and Lichenysin Synthetases
Surfactin and lichenysin are structurally related LPBSs produced by B. subtilis and B. licheniformis [3,7,21]. Several other forms of surfactin with amino acid variation at position 2, 4, and 7 have been reported [22]. Surfactin carries strong surfactant properties by reducing the surface tension of water from 72 to 27 mN/m at a critical micelle concentration (CMC) of 25–220 mg/L depending on its variants and determined conditions [3,23]. A surfactin-like compound termed lichenysin is at least a 2-fold more efficient biosurfactant than surfactin, probably due to the replacement of Glu1 by Gln1 [6,23]. Surfactin was first identified as an inhibitor of fibrin clot formation. It also exhibits anti-microbial, anti-tumor, anti-viral, and hemolytic properties [24]. Surfactin is required for the biofilm formation of producing cells [25,26], swarming motility [27,28], and fruiting body formation [29]. However, surfactin also inhibits biofilm formation of other bacteria by interfering with attachment of the cells to surfaces [30].
These LPBSs are usually a mixture of compounds with different lengths and types of fatty acid (FA), β-hydroxy FA (FA-β-OH), β-amino FA (FA-β-NH2) or guanidylated-β-OH FA (gFA-β-OH). The β-OH or β-NH2 group of FA forms an ester or peptide bond with the carboxyl group of the C-terminal amino acid. For fengycin, circulocins, fusaricidin, and kurstakin, the carboxyl group of the C-terminal amino acid is lactonised with the hydroxyl group of Tyr3, Thr1, Thr1, and Ser4, respectively.
The biosynthetic gene clusters of surfactin [69] and lichenysin [32], namely srfA and lic, are highly homologous and extend over 25 kb (Figure 1). They contain four open reading frames (ORFs), srfA-A/licA, srfA-B/licB, srfA-C/licC, and srfA-Te/lic-Te. The amino acid sequences of the first three ORFs are homologous to other NRPSs whereas the last ORF encodes a putative type II Te. SrfA-A/B/C and LicA/B/C are composed of three, three, and one module(s), and each ORF can be further subdivided into functional domains. SrfA and Lic bear six typical C-domains that catalyze amide bond formation. An additional C-domain, the N-acyl domain, is located at the N-terminal domain of the first module, suggesting that the first amino acid is initially N-acylated with a β-hydroxy fatty acid in this domain [32,70]. Recently, Kraas and coworkers (2010) have shown that the N-acyl domain in SrfA transfers CoA-activated 3-hydroxy fatty acid to the first T-domain where the N-terminal Glu is bound [71]. SrfA and Lic contain the conventional E-domains essential for the transformation of l-amino acids to d-amino acids and a C-terminal type I Te-domain that releases the final product. In addition to the C-terminal type I Te-domain, SrfA and Lic have an external type II Te protein, SrfA-Te/LicTe. A decreased production of surfactin (84%) is observed in the SrfA-Te mutant [72]. The external type II Te is involved in regenerating misprimed T-domains by removing short acyl chains from the 4′-phosphopantetheine cofactors and thereby regenerates functional NRPSs [73]. Moreover, a study has suggested that the type II Te also hydrolyzes incorrectly loaded amino acids that are not processed by the nonribosomal machinery [74]. SrfA-Te also functions as the thioesterase/acyltransferase that supports and stimulates the formation of β-hydroxymyristoyl-glutamate, an initiation substrate of surfactin synthesis [75,76].
2.1.2. Fengycin Synthetase
Fengycin, also referred to as plipastatin when Tyr3 and Tyr9 is present as the l-and d-form, repectively. It is an anti-fungal antibiotic that inhibits filamentous fungi but is ineffective against yeast and bacteria. It is also capable of inhibiting phospholipase A2 and biofilm formation of several bacteria [4,37,77,78]. These types of lipodecapeptides are produced by various strains of Bacillus spp. and exhibit moderate surfactant activities [4,17,79]. Fengycin is expected to form a lactone between the hydroxyl group of l-Tyr3 and the C-terminal carboxyl group of l-Ile. Fengycin synthetase (Fen) contains five NRPS subunits: FenC (287 kDa), FenD (290 kDa), FenE (286 kDa), FenA (406 kDa), and FenB (146 kDa). Like SrfA and Lic, Fen is also composed of an N-acyl domain at the N-terminus of FenC, conventional E-domains, and a typical type I Te-domain. Fen assembles to form a co-linear chain ordered as FenC-Fend-FenE-FenA-FenB [80].
2.1.3. Bacillomycin, Iturin, and Mycosubtilin Synthetases
The iturin family comprises bacillomycin, iturin, and mycosubtilin, which are cyclic lipoheptapeptides linked by a β-amino acid residue. Members of this family have strong antibiotic activity, moderate surfactant activity, and enhanced swarming motility [17,38,81]. The NRPS gene cluster of bacillomycin D (bam/bmy), mycosubtilin (myc), and iturin A (itu) is composed of four large ORFs [18,40,82,83]. bam and bmy are identical gene clusters found in B. subtilis AU195 and B. amyloliquefaciens FZB42, respectively. The gene encodes multifunctional hybrid enzymes of a fatty acid synthase, an aminotransferase, and peptide synthetases. The first ORF-bmyD, ituD, and fenF-encodes malonyl-CoA transacylase. The second ORF-bmyA, ituA, and mycA-encodes acyl-CoA ligase, acyl carrier protein (ACP), β-ketoacyl synthetase and aminotransferase domains before a conventional module of NRPS. The MycA loading module activates free fatty acids through an acyl-adenylate intermediate and loads on the adjacent ACP1 domain [84]. Meanwhile, FenF reveals broad acyl-substrate specificity and loads malonyl-CoA onto ACP2 in MycA [85]. The aminotransferase domain catalyzes the transfer of an amino group to the β-position of the growing acyl chain [86]. The resulting β-amino fatty thioester is then presumably passed on to the third and fourth ORFs that encode four and two functional modules of a typical NRPS, respectively. Mycosubtilin and iturin A have almost the same structure except that d-Ser6 and l-Asn7 residues in mycosubtilin are inverted to d-Asn6 and l-Ser7 in iturin A. Amino acid sequence homology between the two A-domains for Ser and Asn in the two synthetases is high, suggesting that an intragenic domain change occurred in either Myc or Itu synthetase to evolve a counterpart gene [82].
2.1.4. Fusaricidin Synthetase
Fusaricidin is a unique hexapeptide linked to guanidylated β-hydroxyl fatty acid that possesses a potent anti-fungal activity produced by Paenibacillus polymyxa PKB1 (formerly called Bacillus polymyxa). It is a candidate for a biocontrol medicine used to treat blackleg disease [44]. Fusaricidin synthetase (FusA) comprises six NRPS modules that are encoded by a single ORF. The second, fourth, and fifth, modules of FusA incorporate d-amino acids and carry E-domains. However, no E-domain was detected in the sixth module that would incorporate d-Ala [44].
To date, three different mechanisms have been reported for incorporation of d-amino acids in the LPBS products. In the typical NRPS system in Gram-positive bacilli, an E-domain responsible for epimerization of l-amino acids to the d-forms is located downstream of d-amino acid-incorporating modules. A second mechanism is the direct incorporation of d-amino acids by the respective A-domains. This system is found in eukaryotic fungal NRPS systems such as cyclosporine and HC-toxin [87,88]. A third system for incorporating d-amino acid is a novel type of C-domain with dual epimerization and condensation activities (C/E domain), which has recently been identified in several NRPSs in both actinomycete and Gram-negative pseudomonads [89].
Regarding FusA synthetase, two different mechanisms for incorporation of d-amino acids are employed. Conversion from l- to d-amino acid in the three modules of FurA synthetase is mediated by conventional E-domains, whereas direct incorporation of d-Ala is found in the last module that does not contain the E-domain or C/E domain [44]. Thus, horizontal gene transfer has potentially occurred between Gram-positive bacterial and fungal NRPS genes.
2.2. Pseudomonas
Pseudomonas also produces a variety of cyclic lipopeptides. Recently, lipopeptides of pseudomonads have been classified into six groups: viscosin, syringomycin, amphisin, putisolvin, tolaasin, and syringopeptin [90]. In addition to these six main LPBS groups, other lipopeptides have been also identified in Pseudomonas species, but no cyclic lipopeptides linked by a β-amino acid residue have been reported (Table 2).
2.2.1. Syringomycin Synthetase
Syringomycin is a phytotoxin and a key determinant of Pseudomonas syringae B301D virulence. It has moderate surfactant activity with a CMC of 1250 mg/L and minimum surface tension of 33 mN/m [91]. Syringomycin is synthesized by two NRPSs (SyrB1, SyrE) and three modifying protein systems (SyrB2, SyrC, SyrP). SyrB1 and SyrE do not follow the co-linearity rule and also lack E-domains. Eight modules for the first eight amino acids in SyrE are arranged in a line, but the ninth module (SyrB1), which is necessary for incorporation of the last amino acid (l-Thr9), is located in the upstream region [51]. This observation suggests that absolute co-linearity is not essential for NRPS synthesis, which is similar to SrfA data [92]. l-Thr9 is activated and loaded by SyrB1 and is then chlorinated to 4-Cl-l-Thr by the non-heme Fe(II) halogenase SyrB2 [93]. This intermediate is transferred from the T-domain of SyrB1 to SyrE by aminoacyltransferase SyrC to form the final product [94]. Hydroxylation of Asp at module 8 is catalyzed by SyrP, whose gene is located upstream of syrB1 [95]. Although three amino acid residues in syringomycin are in the d-form, the E-domain is not associated with the modules incorporating the respective d-amino acids. However, Balibar and coworkers (2005) demonstrated that SyrE contains unique dual C/E domains, which contribute to the conversion of l-amino acids to the d-form [89].
2.2.2. Syringopeptin Syntheatase
P. syringae B301D also produces another class of lipodepsipeptide phytotoxins called syringopeptin. Syringopeptin contains a larger peptide moiety than syringomycin, with 22 or 25 amino acid residues, and is one of the largest LPBS ever reported. Syringopeptin has a CMC of 820 mg/L and reduces surface tension to 40.2 mN/m [91]. Three NRPSs, SypA, SypB, SypC, are involved in the biosynthesis of syringopeptin. The order and number of the modules are co-linear to the amino acid sequence of syringopeptin SP22. SypA/B/C represents the largest NRPSs among those reported for prokaryotes. Similar to Syr synthetase, no E-domain is present in Syp synthetase, despite the presence of several d-amino acids. In contrast to the Syr synthetase, SypC contains two unique C-terminal Te-domains predicted to catalyze the release and cyclization of syringopeptin [96].
2.2.3. Arthrofactin Synthetase
Arthrofactin is a cyclic lipoundecapeptide produced by Pseudomonas sp. MIS38, which was initially misidentified as Arthrobacter sp. [56], and belongs to the amphisin group. The molecule is cyclized through the formation of an ester bond between the carboxyl group of the C-terminal Asp and the β-hydroxyl group of d-allo-Thr [97]. Arthrofactin is one of the most effective cyclic LPBSs; it reduces the surface tension of water from 72 to 24 mN/m with a CMC of 13.5 mg/L [56]. Arthrofactin appears to be essential for swarming activity and inhibits initial attachment of the planktonic cells in biofilm formation [98]. Several arthrofactin-like compounds with remarkable biosurfactant and antifungal properties or an enzyme inhibitor have been reported from Pseudomonas spp. [9–12,57]. Like arthrofactin, amphisin is involved in swarming motility of Pseudomonas sp. DSS73 [99].
Biosynthesis of arthrofactin is catalyzed by the arthrofactin synthetase (Arf), which consists of three NRPS protein subunits, ArfA (234 kDa), ArfB (474 kDa), and ArfC (648 kDa), which contain two, four, and five functional modules, respectively (Figure 2) [98]. An additional C-domain was identified in the first module of ArfA, suggesting that the first amino acid could be initially acylated with a fatty acid. Site-directed mutagenesis changing the histidine residue of conserved core motif (HHXXXDG) to alanine impairs arthrofactin production [100]. This result suggested that the first C-domain is essential for biosynthesis of lipopeptide. Indeed, the β-hydroxydecanoyl thioester may be coupled to the activated leucine by the action of this C-domain to yield β-hydroxydecanoyl-l-Leu as the initial intermediate. A phylogenetic tree showed that the first C-domain of Arf belongs to N-acyl groups that use fatty acyl-CoA as their starter unit [70]. Although seven of the 11 amino acid residues in arthrofactin are in the d-form, Arf contains no E-domains, as found in syringomycin and syringopeptin [98]. The A-domain of d-Leu1 specifically recognizes only l-Leu in vitro. Based on these observations, we initially hypothesized that an external racemase may be responsible for incorporation of the d-amino acids in arthrofactin. Different amino acid sequences downstream of a conserved core motif [FFELGGHSLLA(V/M)] in the T-domains were expected to reflect the recognition by external racemase. However, Balibar and coworkers later demonstrated that Arf contains unique dual C/E domains, which contribute to the conversion of l-amino acids to the d-form [89]. This novel C/E domain is cryptically embedded with the C-domain located downstream of the d-amino acid–incorporating modules. Dual C/E domains can be recognized by an elongated His motif (HHI/LXXXXGD). This feature was also identified in the Syr and Syp synthetases. Another unique characteristic of Arf is the presence of C-terminal tandem Te-domains like syringopeptin. By site-directed mutagenesis, the first Te-domain (ArfC-Te1) was shown to be essential for the completion of macrocyclization and the release of the final product. The second Te-domain (ArfC-Te2) was suggested to be involved in the evolution of Arf to improve the macrocyclization efficiency [101]. Moreover, we found that the gene encoding putative ArfA/B/C exists in the genome sequence of Pseudomonas fluorescens Pf0-1 (YP_347943/YP_347944/YP_347945) [102]. Arf represents a novel NRPS architecture that features tandem Te-domains and dual C/E domains. Interestingly, another type of NRPS involved in biosynthesis of a siderophore, pyoverdine, was also identified in arthrofactin-producing Pseudomonas sp. MIS38. A gene encoding NRPS for the chromophore part of pyoverdine contains a conventional E-domain [102]. This observation suggests that different NRPS systems with dual C/E domains and a conventional E-domain are both functional in Pseudomonas spp.
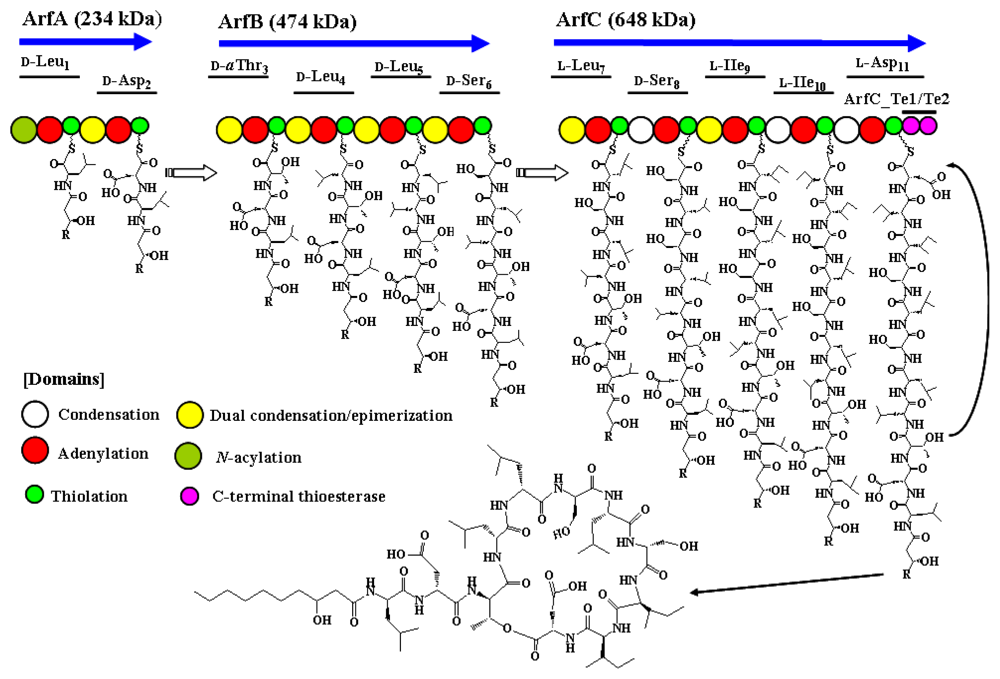
Figure 2.
The arthrofactin biosynthesis assembly line.
2.2.4. Viscosin and Massetolide Synthetases
Viscosin and massetolide are structurally related lipononapeptides produced by P. fluorescens SBW25 and P. fluorescens SS101, respectively [46,48]. Viscosin has significant surfactant activity by reducing surface tension of water to 28 mN/m with a CMC of 10–15 mg/L and forms stable emulsions [46,103]. It also inhibits migration of a metastatic prostate cancer cell line without visible toxicity [103]. In addition, viscosin and massetolide are required for biofilm formation and swarming motility of Pseudomonas cells [46,48]. Viscosin/massetolide is synthesized by NRPS systems that are encoded by three large ORFs, termed viscA/massA, viscB/massB, and viscC/massC. The viscA/massA gene is not clustered with the latter genes, but is located at a different locus of the Pseudomonas genome. The distance between viscA/massA and the latter genes is more than 1.5 MB. Analysis of the amino acid sequences revealed two modules in ViscA/MassA, four modules in ViscB/MassB, and three modules in ViscC/MassC. Each module bears A-, T-, and C-domains like other NRPSs. However, none of the five d-amino acid-incorporating modules possesses a cognate E-domain, but contains a C/E domain similar to Arf. Tandem Te-domains were also identified in the last ViscC/MassC module and are likely to be functional for the biosynthesis of both lipopeptides as was shown for the two Te-domains in Arf [101]. Similar to other NRPSs involved in lipopeptide biosynthesis, the N-terminal C-domain in the first module is highly similar to the N-acyl domain and is presumably involved in N-acylation of the first amino acid.
2.2.5. Orfamide Synthetase
Orfamide and its biosynthetic genes (ofaA/B/C) were discovered from the P. fluorescens Pf-5 genome using a genome isotope approach that employs a combination of genome sequence analysis and isotope-guided fractionation to identify the corresponding compounds [66]. Orfamide is a lipodecapeptide consisting of a β-hydroxy fatty acid linked to a 10-amino acid cyclic peptide in which five amino acids are in the d-form. Orfamide is essential for swarming activity and exhibits strong zoosporicidal activity, but it is not involved in biofilm formation [66]. Although orfamide is composed of 10 amino acids, its structure is most similar to the lipononapeptide viscosin, suggesting a common evolutionary lineage. It is also interesting that the gene structure, ofaABC, is rather similar to arfABC (Figure 1). Structural analysis of OfaA/B/C identified ten modules. The first NRPS, OfaA, consists of two modules with an N-acyl domain at its N-terminus. Four modules were identified in the second NRPS, OfaB, and the last NRPS, OfaC. Similar to other Pseudomonas NRPSs, no cognate E-domains are found in Ofa modules. Although five amino acid residues in orfamide are in the d-form, Ofa seems to contain a total of six dual C/E domains. This inconsistency suggests that the NRPS system is more complex than previously thought. Tandem Te-domains were found in the C-terminus of OfaC, similar to several NRPSs from other Pseudomonas species.
2.2.6. Putisolvin Synthetase
Putisolvin is a LPBS synthesized by P. putida PL1445, which was isolated from soil heavily contaminated with polycyclic aromatic hydrocarbons. Putisolvin is a cyclic lipododecapeptide consisting of a 12-amino acid peptide linked to a hexanoic lipid by an ester linkage between the ninth serine residue and the C-terminal carboxyl group [58]. Putisolvin inhibits biofilm formation of other bacteria and exhibits zoosporicidal and antifungal activities [58,104]. Three genes (psoA, psoB, and psoC) were identified and shown to encode NRPS involved in putisolvin biosynthesis [105]. PsoA, PsoB, and PsoC contain two, seven, and three functional modules, respectively. The C-terminus in PsoC carries putative tandem Te-domains. Both domains harbor a highly conserved signature sequence (GXSXG) and the catalytic triad residues of Te-domains. Nine of the 12 amino acids are in the d-form, but no conventional E-domains were identified [105]. Analysis of specific sequence motifs in the T-domains suggested that the first nine T-domains in Pso synthetase are responsible for transferring d-amino acids [98]. Amino acid sequence analysis of the C-domains indicated that dual C/E domains are organized downstream of the first nine modules. Prediction of A-domain substrate specificity in the eleventh module indicates its preference for Val over Leu or Ile, which correlates well with the production ratios of putisolvin I and II [105].
2.2.7. Syringafactin Synthetase
Syringafactin is a novel linear lipooctapeptide produced by P. syringae pv. tomato DC3000. It contains an eight-amino acid linear peptide linked to a β-hydroxy fatty acid. The Val4 residue can be substituted with Leu or Ile. Syringafactin shows surfactant activity and is essential for the swarming motility of the producing strain, but its contribution to the pathogenicity has not been tested [68]. Syringafactin biosynthetic genes were identified from mining the P. syringae pv. tomato DC3000 genome. The gene clusters syfA and syfB encode three and five NRPS modules, respectively. The N-acyl domain present in the initiating module of SyfA indicated that syringafactin would contain an N-terminal fatty acid chain. SyfB contains tandem Te-domains at the C-terminus. The d/l-configuration of each residue has not been determined. However, based on the location of the dual C/E domains that are typically located downstream of d-amino acid–incorporating modules, the structure of syringafactin should be fatty acyl-d-Leu1-d-Leu2-d-Gln3-Leu4-d-Thr5-Val6-d-Leu7-Leu8, which differs from a previous report [68]. The N-terminal C-domain of SyfA shows the highest level of amino acid sequence similarity with the N-terminal C-domain of ArfA. It seems likely that protein domains corresponding to the first three modules of the arthrofactin NRPS are absent in syringafactin NRPS. This observation suggests that the syringafactin NRPS system in P. syringae pv. tomato DC3000 evolved from the arthrofactin system, after which three modules of the arthrofactin NRPS were deleted, resulting in the fusion of the N-terminus of ArfA with a portion of ArfB. Importantly, the deleted modules include the module that incorporates the threonyl residue that forms the ester linkage involved in cyclization of arthrofactin. Indeed, the structure of syringafactin is reported to be a linear form.
2.2.8. Entolysin Synthetase
Entolysin is a cyclic lipotetradecapeptide produced by an entomopathogenic bacterium Pseudomonas entomophila. This bacterium is able to infect and effectively kill various insects, and it is closely related to the saprophytic soil bacterium P. putida. Entolysin has a relatively small cyclic peptide moiety in which a lactone ring is formed between the tenth and the last amino acid. Entolysin is required for swarming motility and exhibits hemolytic and surfactant activity as described for other lipopeptides, but it does not participate in the virulence of the producing strain for killing Drosophila [65]. Three genes encoding entolysin synthetases were identified (etlA, etlB, and etlC). The deduced amino acid sequences are similar to other NRPSs and closely related to Pso synthetase in P. putida PCL1445. The etlA gene is not physically linked with etlB and etlC in the P. entomophila genome. This organization has also been reported for the viscosin and massetolide gene clusters. EtlA, EtlB, and EtlC comprise two, eight, and four functional modules of NRPS, all of which correspond to the number of amino acid residues in the product peptide. These modules are composed of typical domains. However, no cognate E-domains have been identified in EtlA/B/C. Amino acid sequence analysis indicated that all of the C-domains but C12 and C13 could function as dual C/E domains. In addition, the first C-domain of EtlA is similar to the N-acyl domain of other lipopeptides, and tandem Te-domains were identified in the C-terminus of EtlC [65].
3. Gene Regulation in Bacillus
Gene regulation of LPBSs produced by Bacillus spp. has been most intensively investigated within the surfactin biosynthesis system (Figure 3). Expression of the gene srfA is controlled by several peptide pheromones including ComX and Phr [106]. B. subtilis encodes eight Phr peptides (PhrA, PhrC [CSF], PhrE, PhrF, PhrG, PhrH, PhrI, and PhrK) and 11 aspartyl-phosphate phosphatase proteins (RapA to RapK). Each Phr peptide inhibits the activity of cotranscribed Rap protein. RapC, RapF, and RapK act as negative regulators of srfA [107]. ComX interacts with the membrane-bound histidine kinase ComP, which autophosphorylates upon stimulation and then transfers its phosphate to a serine residue in the response regulator ComA. The phosphorylated ComA binds to ComA boxes (T/GCGG-N4-CCGCA) upstream of the srfA promoter as a tetramer and initiates transcription of srfA [108,109]. Recently, it was found that ComA binds to a degenerate tripartite sequence consisting of three recognition elements (RE). RE1 and RE2 contain the inverted repeats previously characterized as part of the ComA-boxes. Meanwhile, RE3 is located downstream of RE1 and RE2 with a consensus sequence identical to that of RE1 [110]. In addition, mutation at three non-aspartate amino acids in the N-terminal portion of ComA decreases surfactin production [111]. These three amino acids may be involved in the phosphorylation mechanism. It was previously reported that glucose can stimulate the transcription of comA and consequently increases the expression of srfA [112,113]. Induction of srfA requires the oligopeptide permease Spo0K, which is involved in PhrC import [114]. On the other hand, expression of srfA is downregulated upon treatment with H2O2; this evidence led to identification of the H2O2 stress responsive regulator PerR. PerR positively regulates srfA expression by binding to PerR boxes located in the upstream region of ComA boxes in which H2O2 inhibits the DNA-binding activity of PerR [115]. Furthermore, the chaperon subunit ClpX and protease ClpP are required for the transcription of srfA at a step that follows ComP-dependent activation of ComA [116]. An additional transcription factor DegU also functions as a positive regulator for srfA transcription [117].

Figure 3.
The model of gene regulation involved in surfactin biosynthesis. Closed-head arrows indicate positive regulation whereas closed circles indicate negative regulation.
Overexpression of RapD, RapG, and RapH inhibits srfA transcription, and production of these Rap proteins is suppressed by RghR [118,119]. Mutation in sodA, which encodes superoxide dismutase, inhibits transcription of the comQXP quorum-sensing locus, thereby preventing srfA expression [120]. At high concentrations of amino acids such as Ile, Leu, and Val, CodY represses transcription of srfA by interacting specifically with the srfA promoter [121]. Like CodY, AbrB also negatively regulates srfA transcription. Expression of srfA is also repressed by the RNA polymerase-binding protein Spx [122]. On the other hand, the Spx-RNA polymerase interaction is required for positive transcriptional control of genes in response to thiol-oxidative stress [123]. Furthermore, 4′-phosphopantetheinyl transferase (Sfp/PPTase) is required for the activation of SrfA enzymes by converting the inactive apo-forms of the T-domains to the active holo-forms [124]. An acyltransferase SrfA-Te is also required in the initial step of transferring a hydroxyl fatty acid to the first amino acid in the peptide. The surfactin self-resistance protein, YerP, is required for surfactin exportation [125].
Regulation of the fengycin/plipastatin genes is positively controlled by DegQ, an enhancer of extracellular protease production [126]. degQ is a pleiotropic regulatory gene that controls the production of several hydrolytic enzymes [127]. Production of plipastatin is severely reduced in the degQ mutant, but no significant change is observed in surfactin production. A Sfp-like protein, Lpa-8, is also required for plipastatin production in B. subtilis YB8 [128]. Overexpression of degQ in B. subtilis 168 expressing lpa-8 yields a 10-fold increase in plipastatin production [126]. Recently, transcription analysis of fen in B. subtilis F29-3 demonstrated that RNA polymerase binds to the Aand T-rich sequences, called the UP element, which is located upstream of the fen promoter [129].
Gene regulation of the iturin family was first demonstrated within mycosubtilin-producing B. subtilis ATCC6633. Expression of the myc operon is independent of ComA, but still seems to be regulated via quorum sensing, as PhrC strongly stimulates expression. The sigma H factor, Spo0H, also influences expression of the myc operon, and addition of PhrC to the culture medium compensates for loss of Spo0H expression. Finally, the transition state regulator AbrB represses expression of myc, as deletion of abrB results in increased myc expression [130]. Further information regarding the gene regulation was obtained from the study of the bmy operon produced using the B. amyloliquefaciens FZB42. Expression of bmy is dependent on a single sigma A factor-dependent promoter and is positively controlled by the small regulatory protein DegQ, similar to fen. Similar to srfA, the global regulators DegU and ComA are required for the full transcriptional activation of bmy. DegU plays a key role because it binds directly to two sites located upstream of the bmy promoter. Moreover, post-transcriptional regulation of bacillomycin production is also suggested for both DegU and a transmembrane protein, YczE [131]. Like other lipopeptide synthetases in Bacillus, the Sfp-like protein, Lpa-14, is also required for iturin production [132]. Mutation of the Sfp-encoding gene simultaneously prevents B. amyloliquefaciens FZB42 from producing bacillomycin, fengycin, and surfactin [131].
4. Gene Regulation in Pseudomonas
Similar to ComP/ComA in Bacillus, a two-component system has been identified as the master transcriptional regulation system in Pseudomonads (Figure 4). Typically, this system consists of a sensor kinase GacS and response regulator GacA. GacS was first described in P. syringae pv. syringae B728a as an essential factor for lesion manifestation. Meanwhile, GacA was first identified as a global activator of antibiotic and cyanide production in P. fluorescens CHA0 [133]. It is proposed that upon interaction with the signal(s), GacS is activated by autophosphorylation and then GacA acts as a phosphoryl acceptor. After trans-phosphorylation, GacA activates transcription of the regulatory gene, which in turn controls the expression of target genes [133]. Based on bacterial two-hybrid analysis, the entire GacA molecule is necessary for GacA interaction with itself or GacS [134]. The GacS/GacA system positively controls the expression of genes required for the synthesis of lipopeptides (syringomycin, amphisin, putisolvin, and entolysin) because mutation in either gene impairs lipopeptide production [65,135–138]. A quorum-sensing system that triggers GacA/GacS phosphorylation during high cell density is essential for biosynthesis of the lipopeptide putisolvin [139]. In Gram-negative bacteria, the quorum-sensing system largely relies on the interaction of signaling molecule N-acyl homoserine lactones (AHLs) that are synthesized via LuxI protein with the transcriptional regulator LuxR. The quorum-sensing system in P. putida PCL1445 is composed of LuxI-homologous PpuI, LuxR-homologous PpuR, and RsaL. Expression of the genes ppuI and ppuR are required for the biosynthesis of AHLs, and mutation of these genes reduces putisolvin production. Meanwhile, overproduction of AHLs and putisolvin is observed in the rsaL mutant. This observation suggests that RsaL acts as a repressor of PpuI and PpuR. In contrast, biosynthesis of the lipopeptides massetolide, amphisin, and syringomycin is not regulated by AHL-based quorum sensing [48]. Downstream of the Gac system, the cognate transcriptional regulator LuxR regulates the production of several lipopeptides that bind to the operator of the NRPS genes. LuxR protein contains a DNA-binding helix-turn-helix motif in its C-terminal region. In Pseudomonas sp. MIS38, the LuxR-type transcription factor, ArfF, positively controls transcription of the gene arf [140]. LuxR-type protein is also implicated in the biosynthesis of entolysin, putisolvin, and syringafactin, and mutation of this gene results in the loss of lipopeptide production [65,68,105,141]. Two types of LuxR-type proteins are involved in biosynthesis of syringomycin, syringopeptin, and viscosin. SalA and SyrF are the LuxR-type proteins responsible for the production of two lipopeptides, syringomycin and syringopeptin, in P. syringae pv. syringae B301D. SalA is suggested to control transcription of SyrF, an apparent homolog of ArfF, and SyrF binds and trans-activates the target NRPS promoter [142]. Mutation of two LuxR-type proteins in P. fluorescens SBW25, ViscAR and ViscBCR, results in the reduction of NRPS gene transcription and loss of viscosin production [143]. Recently, heat shock proteins were shown to regulate biosynthesis of putisolvin and arthrofactin [140,141]. Mutation in the gene encoding DnaK, a HSP70 class heat shock protein, impairs putisolvin production in P. putida PCL1445. Together with DnaJ, DnaK regulates putisolvin synthesis at low temperature. Elimination of arthrofactin synthesis was identified following mutation of the gene encoding HtpG, a HSP90 class heat shock protein. However, normal expression of the arthrofactin synthetase genes is retained in the HtpG mutant. Thus, HtpG appears to be involved in the proper folding of positive transcription factors or in the assembly of the NRPS complex. The serine protease ClpP regulates massetolide biosynthesis in P. fluorescens SS101 via LuxR transcriptional regulators, and expression of ClpP is independent of regulation by the GacS/GacA system [144]. In contrast to surfactin synthesis, the chaperon subunit ClpX is not involved in the production of massetolide, and its gene is transcribed independently of clpP. Random mutagenesis in Pseudomonas MIS38 also led to identification of SpoT as a new regulator of arthrofactin synthesis [140]. Mutation in the SpoT-encoding gene prevents MIS38 from producing arthrofactin. SpoT is a (p)ppGpp synthetase/hydrolase responsible for cellular metabolism during nutritional starvation [145]. Epistasis analysis revealed that spoT positively regulates arthrofactin biosynthesis through arfF and arfB. Post-translational modification of NRPS in pseudomonads is also catalyzed by PPTase [146–148].

Figure 4.
The model of gene regulation involved in lipopeptide biosynthesis by Pseudomonas. Closed-head arrows indicate positive regulation whereas closed circles indicate negative regulation.
Exportation of lipopeptide in pseudomonads requires ATP-binding cassette (ABC) transporter systems, and the ABC transporter genes are then clustered together with a synthetase gene [48,65,68,98,105]. Exportation of syringomycin and syringopeptin requires two transporter systems known as SyrD and PseABC. SyrD is proposed to function as an ATP-driven efflux pump. Only trace quantities of both lipopeptides are produced by the syrD mutant, and the cells show significantly lower virulence [149, 150]. On the other hand, the tripartite ABC-type efflux transporter PseABC is homologous to the resistance-nodulation-cell division, RND, efflux system. Mutation in each of the genes pseABC results in a 40 to 60% decrease in both syringomycin and syringopeptin production [151]. The transporter system for arthrofactin was recently characterized in Pseudomonas MIS38 [152]. The genes encoding a putative periplasmic protein (ArfD) and a putative ABC transporter (ArfE) are located downstream of the arf gene cluster. Arthrofactin production is temporarily reduced in both mutants, but it eventually reached a similar level to that of MIS38 after 12 h cultivation. Furthermore, exportation of arthrofactin is almost completely blocked by ABC transporter inhibitors. This suggests that multiple ABC transporter systems can export arthrofactin and that accumulation of arthrofactin is toxic to the cells. Two genes that are predicted to encode homologs of the ABC transporters MacA and MacB in E. coli are located downstream of the putisolvin and entolysin synthetase genes [153]. Analysis of macA and macB mutations in P. putida PCL1445 shows a 70% decrease in putisolvin production [105]. Meanwhile, macA and macB mutations in P. entomophila result in almost a complete loss of entolysin production [65]. Therefore, we hypothesize that multiple ABC transporter systems generally play a role in transportation of LPBSs in pseudomonads.
5. Genetic Engineering of NRPS to Create Novel Products
A large number of LPBSs are synthesized by NRPSs that share similar modular architecture of their repetitive catalytic units and a similar assembly-line mechanism. However, recent genome analysis of the lipopeptide synthetases from both Bacillus and Pseudomonas strains revealed novel NRPS architecture that does not completely conform to the co-linearity rule, which encompasses multifunctional hybrid enzymes that can directly incorporate d-amino acid residues. In addition, most of the recently identified lipopeptide synthetases in Pseudomonas spp lack the cognate E-domains, but contain a dual C/E domain and also feature tandem Te-domains. These variations demonstrate natural versatility in evolving complex pathways for lipopeptide biosynthesis and may allow for artificial alteration of the protein template with the aim of reprogramming it to create novel compounds with improved properties.
Recently, new strategies for engineering NRPSs have been developed. The first successful report examined exchanging or replacing the minimal module (A- and T-domains) with the last Leu7 module within SrfA-C, a single-module with the simplest structure [14]. This module was deleted and replaced with several bacterial and fungal A/T-domains by homologous recombination. Construction of the gene encoding hybrid SrfA leads to the production of surfactin variants that retain their activity (Table 3). Using the same strategy, A/T-domain exchange was applied to multi-modular SrfA. Swapping of the Leu2 module results in an altered product whose peptide chain is shorter than that of wild-type [154]. However, this minimal module replacement resulted in a very low yield of variants. Therefore, Yakimov and coworkers (2000) developed a whole-module replacement within SrfA. A complete set of C/A/T-domains of the lichenysin A synthetase Gln1 module was introduced into the Glu1 module of SrfA. An altered product, surfactin [Gln1], was produced by the recombinant B. subtilis at a level similar to that of wild-type surfactin and it also exhibits stronger surface activity than surfactin [155]. The SrfA could also be modified by translocation of the C-terminal Te-domain to the end of the internal domains, thus resulting in new linear surfactin analogs [156]. Furthermore, the use of recombinant Te-domain to catalyze regiospecific cyclization of synthetic peptides or lipopeptides in vitro is a powerful approach for generating libraries of novel compounds with improved properties [157,158]. Another powerful approach for the genetic manipulation of NRPS templates is the directed mutation of the substrate specificity within the A-domain based on its specificity-conferring code [159]. Specificity-conferring codes of the Asp5 module within SrfA were adapted for the recognition of Asn. The engineered B. subtilis produces the new lipoheptapeptide, surfactin [Asn5] [13]. Along the same line, directed mutation of the Asp7 A-domain to Asn7 within CDA synthetase from Streptomyces coelicolar leads to production of the expected lipoundecapeptide containing Asn7 and an unexpected linear lipohexapeptide intermediate [160]. A reduction or increase in the number of peptide residues is an alternate approach to generate structural diversity of lipopeptides and glycopeptides. Deletion of an entire Leu2 module in SrfA causes secretion of the predicted surfactin variant with a smaller cyclic peptide. Furthermore, a novel lipohexapeptide is produced with a significantly high yield and enhanced antimicrobial activities [161, 162]. Amino acid insertion by module extension has been performed within the vancomycin-type glycopeptide antibiotic, balhimycin. Insertion of an entire d-hydroxyphenylglycine module into the balhimycin assembly line between modules 4 and 5 results in an elongated octapeptide product [163]. C-terminally truncated hexa- to di-peptide metabolites are also detected. An alternative approach was examined based on the function of communication-mediating (COM) domains that play an important role in the intermolecular communication within the NRPS system [164]. Swapping of COM domains have been exploited within SrfA and allow for biosynthesis of surfactin variants with different peptide chain lengths [165]. Using plasmid and transposon mutation of the srfA operon at specific and random positions, various intermediates from lipodipeptides to lipohexapeptides were identified by whole-cell MALDI-TOF MS analysis [166]. Although several NRPSs involved in lipopeptide production in pseudomonads have also been characterized, there is limited information available for rational modification of their NRPS modules in vivo. Utilizing a complementation method, Ackerley and Lamont (2004) examined the possibility for engineering pyoverdine synthetase (PvdD) in P. aeruginosa PAO1 to generate novel pyoverdine analogs. Introduction of the Thr-incorporating module from other species into the pvdD mutant restores pyoverdine production [167]. Recently, the researchers at Cubist Pharmaceuticals, Inc. have shown how these approaches can also be used for the production of lipopeptide variants by Streptomyces spp. with improved antibiotic activities [168,169].

Table 3.
Primary structure of engineered LPBSs produced by B. subtilis.
6. Conclusion
Gram-positive Bacillus and Gram-negative Pseudomonas strains produce a variety of lipopeptides with remarkable surface and biological activities. In contrast to the structural diversity of these lipopeptides, their biosynthetic mechanism is basically conserved. They are synthesized nonribosomally by a mega-peptide synthetase unit, NRPS, which is composed of several cooperating multifunctional modules, each capable of performing one cycle of peptide elongation. To become an active form, they are post-translationally modified by a PPTase and properly assembled by DnaJ/K and HtpG proteins. However, recent analysis of the lipopeptide synthetases suggests that there are several variants of NRPS architecture. Modification of NRPS by genetic engineering of the encoding genes is a promising method to produce useful variants. Accumulation of genetic information for lipopeptide synthetases should contribute to design biosurfactants with higher surface activity and/or novel features. Moreover, understanding of their biosynthetic pathways and genetic regulation mechanisms will facilitate not only uncovering the evolution of nonribosomal peptide synthesis mechanisms, but also the development of cost-effective methods for large-scale production of useful lipopeptides.
Acknowledgements
N.R. acknowledges BIOTEC, NSTDA for providing a General Research fund (BT-B-02-LM-BA- 5204). Experimental works on arthrofactin in M.M. laboratory was supported by The Takeda Science Foundation and KAKENHI (17510171).
References
- Georgiou, G; Lin, SC; Sharma, MM. Surface-active compounds from microorganisms. Biotechnology (NY) 1992, 10, 60–65. [Google Scholar]
- Desai, JD; Banat, IM. Microbial production of surfactants and their commercial potential. Microbiol. Mol. Biol. Rev 1997, 61, 47–64. [Google Scholar]
- Arima, K; Kakinuma, A; Tamura, G. Surfactin, a crystalline peptidelipid surfactant produced by Bacillus subtilis: isolation, characterization and its inhibition of fibrin clot formation. Biochem. Biophys. Res. Commun 1968, 31, 488–494. [Google Scholar]
- Nishikiori, T; Naganawa, H; Muraoka, Y; Aoyagi, T; Umezawa, H. Plipastatins: new inhibitors of phospholipase A2, produced by Bacillus cereus BMG302-fF67. III. Structural elucidation of plipastatins. J. Antibiot (Tokyo) 1986, 39, 755–761. [Google Scholar]
- Peypoux, F; Pommier, MT; Das, BC; Besson, F; Delcambe, L; Michel, G. Structures of bacillomycin D and bacillomycin L peptidolipid antibiotics from Bacillus subtilis. J. Antibiot (Tokyo) 1984, 37, 1600–1604. [Google Scholar]
- Grangemard, I; Bonmatin, JM; Bernillon, J; Das, BC; Peypoux, F. Lichenysins G, a novel family of lipopeptide biosurfactants from Bacillus licheniformis IM 1307: production, isolation and structural evaluation by NMR and mass spectrometry. J. Antibiot (Tokyo) 1999, 52, 363–373. [Google Scholar]
- Yakimov, MM; Abraham, WR; Meyer, H; Laura, G; Golyshin, PN. Structural characterization of lichenysin A components by fast atom bombardment tandem mass spectrometry. Biochim. Biophys Acta 1999, 1438, 273–280. [Google Scholar]
- Morikawa, M; Hirata, Y; Imanaka, T. A study on the structure-function relationship of lipopeptide biosurfactants. Biochim. Biophys Acta 2000, 1488, 211–218. [Google Scholar]
- Nielsen, TH; Sorensen, D; Tobiasen, C; Andersen, JB; Christophersen, C; Givskov, M; Sorensen, J. Antibiotic and biosurfactant properties of cyclic lipopeptides produced by fluorescent Pseudomonas spp. from the sugar beet rhizosphere. Appl. Environ. Microbiol 2002, 68, 3416–3423. [Google Scholar]
- Sorensen, D; Nielsen, TH; Christophersen, C; Sorensen, J; Gajhede, M. Cyclic lipoundecapeptide amphisin from Pseudomonas sp. strain DSS73. Acta Crystallogr C 2001, 57, 1123–1124. [Google Scholar]
- Sorensen, D; Nielsen, TH; Sorensen, J; Christophersen, C. Cyclic lipoundecapeptide lokisin from Pseudomonas sp. strain DSS41. Tetrahedron Lett 2002, 43, 4421–4423. [Google Scholar]
- Ui, H; Miyake, T; Iinuma, H; Imoto, M; Naganawa, H; Hattori, S; Hamada, M; Takeuchi, T; Umezawa, S; Umezawa, K. Pholipeptin, a novel cyclic lipoundecapeptide from Pseudomonas fluorescens. J. Org. Chem 1997, 62, 103–108. [Google Scholar]
- Eppelmann, K; Stachelhaus, T; Marahiel, MA. Exploitation of the selectivity-conferring code of nonribosomal peptide synthetases for the rational design of novel peptide antibiotics. Biochemistry 2002, 41, 9718–9726. [Google Scholar]
- Stachelhaus, T; Schneider, A; Marahiel, MA. Rational design of peptide antibiotics by targeted replacement of bacterial and fungal domains. Science 1995, 269, 69–72. [Google Scholar]
- Koglin, A; Walsh, CT. Structural insights into nonribosomal peptide enzymatic assembly lines. Nat. Prod. Rep 2009, 26, 987–1000. [Google Scholar]
- Raaijmakers, JM; de Bruijn, I; Nybroe, O; Ongena, M. Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol Rev 2010. [Google Scholar] [CrossRef]
- Roongsawang, N; Thaniyavarn, J; Thaniyavarn, S; Kameyama, T; Haruki, M; Imanaka, T; Morikawa, M; Kanaya, S. Isolation and characterization of a halotolerant Bacillus subtilis BBK-1 which produces three kinds of lipopeptides: bacillomycin L, plipastatin, and surfactin. Extremophiles 2002, 6, 499–506. [Google Scholar]
- Koumoutsi, A; Chen, XH; Henne, A; Liesegang, H; Hitzeroth, G; Franke, P; Vater, J; Borriss, R. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J. Bacteriol 2004, 186, 1084–1096. [Google Scholar]
- Romero, D; de Vicente, A; Rakotoaly, RH; Dufour, SE; Veening, JW; Arrebola, E; Cazorla, FM; Kuipers, OP; Paquot, M; Perez-Garcia, A. The iturin and fengycin families of lipopeptides are key factors in antagonism of Bacillus subtilis toward Podosphaera fusca. Mol. Plant Microbe Interact 2007, 20, 430–440. [Google Scholar]
- Kim, PI; Ryu, J; Kim, YH; Chi, YT. Production of biosurfactant lipopeptides iturin A, fengycin and surfactin A from Bacillus subtilis CMB32 for control of Colletotrichum gloeosporioides. J. Microbiol. Biotechnol 2010, 20, 138–145. [Google Scholar]
- Yakimov, MM; Timmis, KN; Wray, V; Fredrickson, HL. Characterization of a new lipopeptide surfactant produced by thermotolerant and halotolerant subsurface Bacillus licheniformis BAS50. Appl. Environ. Microbiol 1995, 61, 1706–1713. [Google Scholar]
- Bonmatin, JM; Laprevote, O; Peypoux, F. Diversity among microbial cyclic lipopeptides: iturins and surfactins. Activity-structure relationships to design new bioactive agents. Comb. Chem High Throughput Screen 2003, 6, 541–556. [Google Scholar]
- Grangemard, I; Wallach, J; Maget-Dana, R; Peypoux, F. Lichenysin: a more efficient cation chelator than surfactin. Appl. Biochem. Biotechnol 2001, 90, 199–210. [Google Scholar]
- Rodrigues, L; Banat, IM; Teixeira, J; Oliveira, R. Biosurfactants: potential applications in medicine. J. Antimicrob. Chemother 2006, 57, 609–618. [Google Scholar] [Green Version]
- Bais, HP; Fall, R; Vivanco, JM. Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol 2004, 134, 307–319. [Google Scholar]
- Hofemeister, J; Conrad, B; Adler, B; Hofemeister, B; Feesche, J; Kucheryava, N; Steinborn, G; Franke, P; Grammel, N; Zwintscher, A; Leenders, F; Hitzeroth, G; Vater, J. Genetic analysis of the biosynthesis of nonribosomal peptide and polyketide-like antibiotics, iron uptake and biofilm formation by Bacillus subtilis A1/3. Mol. Genet Genomics 2004, 272, 363–378. [Google Scholar]
- Kearns, DB; Losick, R. Swarming motility in undomesticated Bacillus subtilis. Mol. Microbiol 2003, 49, 581–590. [Google Scholar]
- Julkowska, D; Obuchowski, M; Holland, IB; Seror, SJ. Comparative analysis of the development of swarming communities of Bacillus subtilis 168 and a natural wild type: critical effects of surfactin and the composition of the medium. J. Bacteriol 2005, 187, 65–76. [Google Scholar]
- Branda, SS; Gonzalez-Pastor, JE; Ben-Yehuda, S; Losick, R; Kolter, R. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci USA 2001, 98, 11621–11626. [Google Scholar]
- Mireles, JR; Toguchi, A; Harshey, RM. Salmonella enterica serovar typhimurium swarming mutants with altered biofilm-forming abilities: surfactin inhibits biofilm formation. J. Bacteriol 2001, 183, 5848–5854. [Google Scholar]
- Kakinuma, A; Ouchida, A; Shima, T; Sugino, H; Isono, M; Tamura, G; Arima, K. Conformation of the structure of surfactin by mass spectrometry. Agric. Biol. Chem 1969, 33, 1669–1671. [Google Scholar]
- Konz, D; Doekel, S; Marahiel, MA. Molecular and biochemical characterization of the protein template controlling biosynthesis of the lipopeptide lichenysin. J. Bacteriol 1999, 181, 133–140. [Google Scholar]
- Lin, SC; Minton, MA; Sharma, MM; Georgiou, G. Structural and immunological characterization of a biosurfactant produced by Bacillus licheniformis JF-2. Appl. Environ. Microbiol 1994, 60, 31–38. [Google Scholar]
- Jenny, K; Kappeli, O; Fiechter, A. Biosurfactants from Bacillus licheniformis: structural analysis and characterization. Appl. Microbiol. Biotechnol 1991, 36, 5–13. [Google Scholar]
- Horowitz, S; Gilbert, JN; Griffin, WM. Isolation and characterization of a surfactant produced by Bacillus licheniformis 86. J. Indus. Microbiol 1990, 6, 243–248. [Google Scholar]
- Naruse, N; Tenmyo, O; Kobaru, S; Kamei, H; Miyaki, T; Konishi, M; Oki, T. Pumilacidin, a complex of new antiviral antibiotics. Production, isolation, chemical properties, structure and biological activity. J Antibiot (Tokyo) 1990, 43, 267–280. [Google Scholar]
- Volpon, L; Besson, F; Lancelin, JM. NMR structure of antibiotics plipastatins A and B from Bacillus subtilis inhibitors of phospholipase A(2). FEBS Lett 2000, 485, 76–80. [Google Scholar]
- Isogai, I; Takayama, S; Murakoshi, S; Suzuki, A. Structure of β-amino acids in antibiotics iturin A. Tetrahedron Lett 1982, 23, 3065–3068. [Google Scholar]
- Volpon, L; Tsan, P; Majer, Z; Vass, E; Hollosi, M; Noguera, V; Lancelin, JM; Besson, F. NMR structure determination of a synthetic analogue of bacillomycin Lc reveals the strategic role of L-Asn1 in the natural iturinic antibiotics. Spectrochim Acta Part A 2007, 67, 1374–1381. [Google Scholar]
- Duitman, EH; Hamoen, LW; Rembold, M; Venema, G; Seitz, H; Saenger, W; Bernhard, F; Reinhardt, R; Schmidt, M; Ullrich, C; Stein, T; Leenders, F; Vater, J. The mycosubtilin synthetase of Bacillus subtilis ATCC6633: a multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc. Natl. Acad. Sci USA 1999, 96, 13294–13299. [Google Scholar]
- Batrakov, SG; Rodionova, TA; Esipov, SE; Polyakov, NB; Sheichenko, VI; Shekhovtsova, NV; Lukin, SM; Panikov, NS; Nikolaev, YA. A novel lipopeptide, an inhibitor of bacterial adhesion, from the thermophilic and halotolerant subsurface Bacillus licheniformis strain 603. Biochim. Biophys Acta 2003, 1634, 107–115. [Google Scholar]
- Lee, SC; Kim, SH; Park, IH; Chung, SY; Choi, YL. Isolation and structural analysis of bamylocin A, novel lipopeptide from Bacillus amyloliquefaciens LP03 having antagonistic and crude oil-emulsifying activity. Arch. Microbiol 2007, 188, 307–312. [Google Scholar]
- He, H; Shen, B; Korshalla, J; Carter, GT. Circulocins, new antibacterial lipopeptides from Bacillus circulans, J2154. Tetrahedron 2001, 57, 1189–1195. [Google Scholar]
- Li, J; Jensen, SE. Nonribosomal biosynthesis of fusaricidins by Paenibacillus polymyxa PKB1 involves direct activation of a D-amino acid. Chem. Biol 2008, 15, 118–127. [Google Scholar]
- Hathout, Y; Ho, YP; Ryzhov, V; Demirev, P; Fenselau, C. Kurstakins: a new class of lipopeptides isolated from Bacillus thuringiensis. J. Nat. Prod 2000, 63, 1492–1496. [Google Scholar]
- de Bruijn, I; de Kock, MJ; Yang, M; de Waard, P; van Beek, TA; Raaijmakers, JM. Genome-based discovery, structure prediction and functional analysis of cyclic lipopeptide antibiotics in Pseudomonas species. Mol. Microbiol 2007, 63, 417–428. [Google Scholar]
- Nielsen, TH; Christophersen, C; Anthoni, U; Sorensen, J. Viscosinamide, a new cyclic depsipeptide with surfactant and antifungal properties produced by Pseudomonas fluorescens DR54. J. Appl. Microbiol 1999, 87, 80–90. [Google Scholar]
- de Bruijn, I; de Kock, MJ; de Waard, P; van Beek, TA; Raaijmakers, JM. Massetolide A biosynthesis in Pseudomonas fluorescens. J. Bacteriol 2008, 190, 2777–2789. [Google Scholar]
- Quail, JW; Ismail, N; Pedras, MS; Boyetchko, SM. Pseudophomins A and B, a class of cyclic lipodepsipeptides isolated from a Pseudomonas species. Acta Crystallogr C 2002, 58, 268–271. [Google Scholar]
- Sinnaeve, D; Hendrickx, PM; Van Hemel, J; Peys, E; Kieffer, B; Martins, JC. The solution structure and self-association properties of the cyclic lipodepsipeptide pseudodesmin A support its pore-forming potential. Chemistry 2009, 15, 12653–12662. [Google Scholar]
- Guenzi, E; Galli, G; Grgurina, I; Gross, DC; Grandi, G. Characterization of the syringomycin synthetase gene cluster. A link between prokaryotic and eukaryotic peptide synthetases. J. Biol. Chem 1998, 273, 32857–32863. [Google Scholar]
- Sorensen, KN; Kim, KH; Takemoto, JY. In vitro antifungal and fungicidal activities and erythrocyte toxicities of cyclic lipodepsinonapeptides produced by Pseudomonas syringae pv. syringae. Antimicrob. Agents Chemother 1996, 40, 2710–2713. [Google Scholar]
- Ballio, A; Bossa, F; Collina, A; Gallo, M; Iacobellis, NS; Paci, M; Pucci, P; Scaloni, A; Segre, A; Simmaco, M. Structure of syringotoxin, a bioactive metabolite of Pseudomonas syringae pv. syringae. FEBS Lett 1990, 269, 377–380. [Google Scholar]
- Harrison, L; Teplow, DB; Rinaldi, M; Strobel, G. Pseudomycins, a family of novel peptides from Pseudomonas syringae possessing broad-spectrum antifungal activity. J. Gen. Microbiol 1991, 137, 2857–2865. [Google Scholar]
- Scaloni, A; Dalla Serra, M; Amodeo, P; Mannina, L; Vitale, RM; Segre, AL; Cruciani, O; Lodovichetti, F; Greco, ML; Fiore, A; Gallo, M; D’Ambrosio, C; Coraiola, M; Menestrina, G; Graniti, A; Fogliano, V. Structure, conformation and biological activity of a novel lipodepsipeptide from Pseudomonas corrugata: cormycin A. Biochem. J 2004, 384, 25–36. [Google Scholar]
- Morikawa, M; Daido, H; Takao, T; Murata, S; Shimonishi, Y; Imanaka, T. A new lipopeptide biosurfactant produced by Arthrobacter sp. strain MIS38. J. Bacteriol 1993, 175, 6459–6466. [Google Scholar]
- Henriksen, A; Anthoni, U; Nielsen, TH; Sorensen, J; Christophersen, C; Gajhede, M. Cyclic lipoundecapeptide tensin from Pseudomonas fluorescens strain 96.578. Acta Crystallogr C 2000, 56 Pt 1, 113–115. [Google Scholar]
- Kuiper, I; Lagendijk, EL; Pickford, R; Derrick, JP; Lamers, GE; Thomas-Oates, JE; Lugtenberg, BJ; Bloemberg, GV. Characterization of two Pseudomonas putida lipopeptide biosurfactants, putisolvin I and II, which inhibit biofilm formation and break down existing biofilms. Mol. Microbiol 2004, 51, 97–113. [Google Scholar]
- Bassarello, C; Lazzaroni, S; Bifulco, G; Lo Cantore, P; Iacobellis, NS; Riccio, R; Gomez- Paloma, L; Evidente, A. Tolaasins A-E, five new lipodepsipeptides produced by Pseudomonas tolaasii. J. Nat. Prod 2004, 67, 811–816. [Google Scholar]
- Ballio, A; Bossa, F; Camoni, L; Di Giorgio, D; Flamand, MC; Maraite, H; Nitti, G; Pucci, P; Scaloni, A. Structure of fuscopeptins, phytotoxic metabolites of Pseudomonas fuscovaginae. FEBS Lett 1996, 381, 213–216. [Google Scholar]
- Emanuele, MC; Scaloni, A; Lavermicocca, P; Jacobellis, NS; Camoni, L; Di Giorgio, D; Pucci, P; Paci, M; Segre, A; Ballio, A. Corpeptins, new bioactive lipodepsipeptides from cultures of Pseudomonas corrugata. FEBS Lett 1998, 433, 317–320. [Google Scholar]
- Ballio, A; Barra, D; Bossa, F; Collina, A; Grgurina, I; Marino, G; Moneti, G; Paci, M; Pucci, P; Segre, A; et al. Syringopeptins, new phytotoxic lipodepsipeptides of Pseudomonas syringae pv. syringae. FEBS Lett 1991, 291, 109–112. [Google Scholar]
- Ballio, A; Bossa, F; Di Giorgio, D; Di Nola, A; Manetti, C; Paci, M; Scaloni, A; Segre, AL. Solution conformation of the Pseudomonas syringae pv. syringae phytotoxic lipodepsipeptide syringopeptin 25-A. Two-dimensional NMR, distance geometry and molecular dynamics. Eur. J. Biochem 1995, 234, 747–758. [Google Scholar]
- Scaloni, A; Camoni, L; di Giorgio, D; Scortichini, M; Cozzolino, R; Ballio, A. A new syringopeptin produced by a Pseudomonas syringae pv syringae strain isolated from diseased twigs of laurel. Physiol. Mol. Plant Pathol 1997, 51, 259–264. [Google Scholar]
- Vallet-Gely, I; Novikov, A; Augusto, L; Liehl, P; Bolbach, G; Pechy-Tarr, M; Cosson, P; Keel, C; Caroff, M; Lemaitre, B. Association of hemolytic activity of Pseudomonas entomophila, a versatile soil bacterium, with cyclic lipopeptide production. Appl. Environ. Microbiol 2010, 76, 910–921. [Google Scholar]
- Gross, H; Stockwell, VO; Henkels, MD; Nowak-Thompson, B; Loper, JE; Gerwick, WH. The genomisotopic approach: a systematic method to isolate products of orphan biosynthetic gene clusters. Chem. Biol 2007, 14, 53–63. [Google Scholar]
- Janek, T; Lukaszewicz, M; Rezanka, T; Krasowska, A. Isolation and characterization of two new lipopeptide biosurfactants produced by Pseudomonas fluorescens BD5 isolated from water from the Arctic Archipelago of Svalbard. Bioresour. Technol 2010, 101, 6118–6123. [Google Scholar]
- Berti, AD; Greve, NJ; Christensen, QH; Thomas, MG. Identification of a biosynthetic gene cluster and the six associated lipopeptides involved in swarming motility of Pseudomonas syringae pv. tomato DC3000. J. Bacteriol 2007, 189, 6312–6323. [Google Scholar]
- Cosmina, P; Rodriguez, F; de Ferra, F; Grandi, G; Perego, M; Venema, G; van Sinderen, D. Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol. Microbiol 1993, 8, 821–831. [Google Scholar]
- Roongsawang, N; Lim, SP; Washio, K; Takano, K; Kanaya, S; Morikawa, M. Phylogenetic analysis of condensation domains in the nonribosomal peptide synthetases. FEMS Microbiol. Lett 2005, 252, 143–151. [Google Scholar]
- Kraas, FI; Helmetag, V; Wittmann, M; Strieker, M; Marahiel, MA. Functional dissection of surfactin synthetase initiation module reveals insights into the mechanism of lipoinitiation. Chem. Biol 2010, 17, 872–880. [Google Scholar]
- Schneider, A; Marahiel, MA. Genetic evidence for a role of thioesterase domains, integrated in or associated with peptide synthetases, in non-ribosomal peptide biosynthesis in Bacillus subtilis. Arch. Microbiol 1998, 169, 404–410. [Google Scholar]
- Schwarzer, D; Mootz, HD; Linne, U; Marahiel, MA. Regeneration of misprimed nonribosomal peptide synthetases by type II thioesterases. Proc. Natl. Acad. Sci USA 2002, 99, 14083–14088. [Google Scholar]
- Yeh, E; Kohli, RM; Bruner, SD; Walsh, CT. Type II thioesterase restores activity of a NRPS module stalled with an aminoacyl-S-enzyme that cannot be elongated. ChemBioChem 2004, 5, 1290–1293. [Google Scholar]
- Menkhaus, M; Ullrich, C; Kluge, B; Vater, J; Vollenbroich, D; Kamp, RM. Structural and functional organization of the surfactin synthetase multienzyme system. J. Biol. Chem 1993, 268, 7678–7684. [Google Scholar]
- Steller, S; Sokoll, A; Wilde, C; Bernhard, F; Franke, P; Vater, J. Initiation of surfactin biosynthesis and the role of the SrfD-thioesterase protein. Biochemistry 2004, 43, 11331–11343. [Google Scholar]
- Vanittanakom, N; Loeffler, W; Koch, U; Jung, G. Fengycin-a novel antifungal lipopeptide antibiotic produced by Bacillus subtilis F-29-3. J. Antibiot (Tokyo) 1986, 39, 888–901. [Google Scholar]
- Rivardo, F; Turner, RJ; Allegrone, G; Ceri, H; Martinotti, MG. Anti-adhesion activity of two biosurfactants produced by Bacillus spp. prevents biofilm formation of human bacterial pathogens. Appl. Microbiol. Biotechnol 2009, 83, 541–553. [Google Scholar]
- Thaniyavarn, J; Roongsawang, N; Kameyama, T; Haruki, M; Imanaka, T; Morikawa, M; Kanaya, S. Production and characterization of biosurfactants from Bacillus licheniformis F2.2. Biosci. Biotechnol. Biochem 2003, 67, 1239–1244. [Google Scholar]
- Wu, CY; Chen, CL; Lee, YH; Cheng, YC; Wu, YC; Shu, HY; Gotz, F; Liu, ST. Nonribosomal synthesis of fengycin on an enzyme complex formed by fengycin synthetases. J. Biol. Chem 2007, 282, 5608–5616. [Google Scholar]
- Leclere, V; Marti, R; Bechet, M; Fickers, P; Jacques, P. The lipopeptides mycosubtilin and surfactin enhance spreading of Bacillus subtilis strains by their surface-active properties. Arch. Microbiol 2006, 186, 475–483. [Google Scholar]
- Tsuge, K; Akiyama, T; Shoda, M. Cloning, sequencing, and characterization of the iturin A operon. J. Bacteriol 2001, 183, 6265–6273. [Google Scholar]
- Moyne, AL; Cleveland, TE; Tuzun, S. Molecular characterization and analysis of the operon encoding the antifungal lipopeptide bacillomycin D. FEMS Microbiol. Lett 2004, 234, 43–49. [Google Scholar]
- Hansen, DB; Bumpus, SB; Aron, ZD; Kelleher, NL; Walsh, CT. The loading module of mycosubtilin: an adenylation domain with fatty acid selectivity. J. Am. Chem. Soc 2007, 129, 6366–6367. [Google Scholar]
- Aron, ZD; Fortin, PD; Calderone, CT; Walsh, CT. FenF: servicing the mycosubtilin synthetase assembly line in trans. ChemBioChem 2007, 8, 613–616. [Google Scholar]
- Aron, ZD; Dorrestein, PC; Blackhall, JR; Kelleher, NL; Walsh, CT. Characterization of a new tailoring domain in polyketide biogenesis: the amine transferase domain of MycA in the mycosubtilin gene cluster. J. Am. Chem. Soc 2005, 127, 14986–14987. [Google Scholar]
- Dittmann, J; Wenger, RM; Kleinkauf, H; Lawen, A. Mechanism of cyclosporin A biosynthesis. Evidence for synthesis via a single linear undecapeptide precursor. J. Biol. Chem 1994, 269, 2841–2846. [Google Scholar]
- Walton, JD. Two enzymes involved in biosynthesis of the host-selective phytotoxin HC-toxin. Proc. Natl. Acad. Sci USA 1987, 84, 8444–8447. [Google Scholar]
- Balibar, CJ; Vaillancourt, FH; Walsh, CT. Generation of D amino acid residues in assembly of arthrofactin by dual condensation/epimerization domains. Chem. Biol 2005, 12, 1189–1200. [Google Scholar]
- Gross, H; Loper, JE. Genomics of secondary metabolite production by Pseudomonas spp. Nat. Prod. Rep 2009, 26, 1408–1446. [Google Scholar]
- Hutchison, ML; Gross, DC. Lipopeptide phytotoxins produced by Pseudomonas syringae pv. syringae: comparison of the biosurfactant and ion channel-forming activities of syringopeptin and syringomycin. Mol. Plant Microbe Interact 1997, 10, 347–354. [Google Scholar]
- Guenzi, E; Galli, G; Grgurina, I; Pace, E; Ferranti, P; Grandi, G. Coordinate transcription and physical linkage of domains in surfactin synthetase are not essential for proper assembly and activity of the multienzyme complex. J. Biol. Chem 1998, 273, 14403–14410. [Google Scholar]
- Vaillancourt, FH; Yin, J; Walsh, CT. SyrB2 in syringomycin E biosynthesis is a nonheme FeII alpha-ketoglutarate- and O2-dependent halogenase. Proc. Natl. Acad. Sci USA 2005, 102, 10111–10116. [Google Scholar]
- Singh, GM; Vaillancourt, FH; Yin, J; Walsh, CT. Characterization of SyrC, an aminoacyltransferase shuttling threonyl and chlorothreonyl residues in the syringomycin biosynthetic assembly line. Chem. Biol 2007, 14, 31–40. [Google Scholar]
- Singh, GM; Fortin, PD; Koglin, A; Walsh, CT. beta-Hydroxylation of the aspartyl residue in the phytotoxin syringomycin E: characterization of two candidate hydroxylases AspH and SyrP in Pseudomonas syringae. Biochemistry 2008, 47, 11310–11320. [Google Scholar]
- Scholz-Schroeder, BK; Soule, JD; Gross, DC. The sypA, sypB, and sypC synthetase genes encode twenty-two modules involved in the nonribosomal peptide synthesis of syringopeptin by Pseudomonas syringae pv. syringae B301D. Mol. Plant Microbe Interact 2003, 16, 271–280. [Google Scholar]
- Ikegami, T. Personal communication, Osaka University: Osaka, Japan, 2004.
- Roongsawang, N; Hase, K; Haruki, M; Imanaka, T; Morikawa, M; Kanaya, S. Cloning and characterization of the gene cluster encoding arthrofactin synthetase from Pseudomonas sp. MIS38. Chem. Biol 2003, 10, 869–880. [Google Scholar]
- Andersen, JB; Koch, B; Nielsen, TH; Sorensen, D; Hansen, M; Nybroe, O; Christophersen, C; Sorensen, J; Molin, S; Givskov, M. Surface motility in Pseudomonas sp. DSS73 is required for efficient biological containment of the root-pathogenic microfungi Rhizoctonia solani and Pythium ultimum. Microbiology 2003, 149, 37–46. [Google Scholar]
- Roongsawang, N; Washio, K; Morikawa, M. Hokkaido University: Hokkaido, Japan; Unpublished work; 2007. [Google Scholar]
- Roongsawang, N; Washio, K; Morikawa, M. In vivo characterization of tandem C-terminal thioesterase domains in arthrofactin synthetase. ChemBioChem 2007, 8, 501–512. [Google Scholar]
- Lim, SP; Roongsawang, N; Washio, K; Morikawa, M. Functional analysis of a pyoverdine synthetase from Pseudomonas sp. MIS38. Biosci. Biotechnol. Biochem 2007, 71, 2002–2009. [Google Scholar]
- Saini, HS; Barragan-Huerta, BE; Lebron-Paler, A; Pemberton, JE; Vazquez, RR; Burns, AM; Marron, MT; Seliga, CJ; Gunatilaka, AA; Maier, RM. Efficient purification of the biosurfactant viscosin from Pseudomonas libanensis strain M9-3 and its physicochemical and biological properties. J. Nat. Prod 2008, 71, 1011–1015. [Google Scholar]
- Kruijt, M; Tran, H; Raaijmakers, JM. Functional, genetic and chemical characterization of biosurfactants produced by plant growth-promoting Pseudomonas putida 267. J. Appl. Microbiol 2009, 107, 546–556. [Google Scholar]
- Dubern, JF; Coppoolse, ER; Stiekema, WJ; Bloemberg, GV. Genetic and functional characterization of the gene cluster directing the biosynthesis of putisolvin I and II in Pseudomonas putida strain PCL1445. Microbiology 2008, 154, 2070–2083. [Google Scholar]
- Magnuson, R; Solomon, J; Grossman, AD. Biochemical and genetic characterization of a competence pheromone from Bacillus subtilis. Cell 1994, 77, 207–216. [Google Scholar]
- Auchtung, JM; Lee, CA; Grossman, AD. Modulation of the ComA-dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides. J. Bacteriol 2006, 188, 5273–5285. [Google Scholar]
- Roggiani, M; Dubnau, D. ComA, a phosphorylated response regulator protein of Bacillus subtilis, binds to the promoter region of srfA. J. Bacteriol 1993, 175, 3182–3187. [Google Scholar]
- Yakimov, MM; Golyshin, PN. ComA-dependent transcriptional activation of lichenysin A synthetase promoter in Bacillus subtilis cells. Biotechnol. Prog 1997, 13, 757–761. [Google Scholar]
- Griffith, KL; Grossman, AD. A degenerate tripartite DNA-binding site required for activation of ComA-dependent quorum response gene expression in Bacillus subtilis. J. Mol. Biol 2008, 381, 261–275. [Google Scholar]
- Wang, X; Luo, C; Liu, Y; Nie, Y; Zhang, R; Chen, Z. Three non-aspartate amino acid mutations in the ComA Response regulator receiver motif severely decrease surfactin production, competence development and spore formation in Bacillus subtilis. J. Microbiol. Biotechnol 2010, 20, 301–310. [Google Scholar]
- Nakano, MM; Zuber, P. Cloning and characterization of srfB, a regulatory gene involved in surfactin production and competence in Bacillus subtilis. J. Bacteriol 1989, 171, 5347–5353. [Google Scholar]
- Kim, SB; Shin, BS; Choi, SK; Kim, CK; Park, SH. Involvement of acetyl phosphate in the in vivo activation of the response regulator ComA in Bacillus subtilis. FEMS Microbiol. Lett 2001, 195, 179–183. [Google Scholar]
- Cosby, WM; Vollenbroich, D; Lee, OH; Zuber, P. Altered srf expression in Bacillus subtilis resulting from changes in culture pH is dependent on the Spo0K oligopeptide permease and the ComQX system of extracellular control. J. Bacteriol 1998, 180, 1438–1445. [Google Scholar]
- Hayashi, K; Ohsawa, T; Kobayashi, K; Ogasawara, N; Ogura, M. The H2O2 stress-responsive regulator PerR positively regulates srfA expression in Bacillus subtilis. J. Bacteriol 2005, 187, 6659–6667. [Google Scholar]
- Nakano, MM; Zhu, Y; Liu, J; Reyes, DY; Yoshikawa, H; Zuber, P. Mutations conferring amino acid residue substitutions in the carboxy-terminal domain of RNA polymerase alpha can suppress clpX and clpP with respect to developmentally regulated transcription in Bacillus subtilis. Mol. Microbiol 2000, 37, 869–884. [Google Scholar]
- Mader, U; Antelmann, H; Buder, T; Dahl, MK; Hecker, M; Homuth, G. Bacillus subtilis functional genomics: genome-wide analysis of the DegS-DegU regulon by transcriptomics and proteomics. Mol. Genet Genomics 2002, 268, 455–467. [Google Scholar]
- Hayashi, K; Kensuke, T; Kobayashi, K; Ogasawara, N; Ogura, M. Bacillus subtilis RghR (YvaN) represses rapG and rapH, which encode inhibitors of expression of the srfA operon. Mol. Microbiol 2006, 59, 1714–1729. [Google Scholar]
- Ogura, M; Fujita, Y. Bacillus subtilis rapD, a direct target of transcription repression by RghR, negatively regulates srfA expression. FEMS Microbiol. Lett 2007, 268, 73–80. [Google Scholar]
- Ohsawa, T; Tsukahara, K; Sato, T; Ogura, M. Superoxide stress decreases expression of srfA through inhibition of transcription of the comQXP quorum-sensing locus in Bacillus subtilis. J. Biochem 2006, 139, 203–211. [Google Scholar]
- Serror, P; Sonenshein, AL. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J. Bacteriol 1996, 178, 5910–5915. [Google Scholar]
- Nakano, S; Nakano, MM; Zhang, Y; Leelakriangsak, M; Zuber, P. A regulatory protein that interferes with activator-stimulated transcription in bacteria. Proc. Natl. Acad. Sci USA 2003, 100, 4233–4238. [Google Scholar]
- Reyes, DY; Zuber, P. Activation of transcription initiation by Spx: formation of transcription complex and identification of a cis-acting element required for transcriptional activation. Mol. Microbiol 2008, 69, 765–779. [Google Scholar]
- Quadri, LE; Weinreb, PH; Lei, M; Nakano, MM; Zuber, P; Walsh, CT. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry 1998, 37, 1585–1595. [Google Scholar]
- Tsuge, K; Ohata, Y; Shoda, M. Gene yerP, involved in surfactin self-resistance in Bacillus subtilis. Antimicrob. Agents Chemother 2001, 45, 3566–3573. [Google Scholar]
- Tsuge, K; Ano, T; Hirai, M; Nakamura, Y; Shoda, M. The genes degQ, pps, and lpa-8 (sfp) are responsible for conversion of Bacillus subtilis 168 to plipastatin production. Antimicrob. Agents Chemother 1999, 43, 2183–2192. [Google Scholar]
- Msadek, T; Kunst, F; Klier, A; Rapoport, G. DegS-DegU and ComP-ComA modulator-effector pairs control expression of the Bacillus subtilis pleiotropic regulatory gene degQ. J. Bacteriol 1991, 173, 2366–2377. [Google Scholar]
- Tsuge, K; Ano, T; Shoda, M. Isolation of a gene essential for biosynthesis of the lipopeptide antibiotics plipastatin B1 and surfactin in Bacillus subtilis YB8. Arch. Microbiol 1996, 165, 243–251. [Google Scholar]
- Ke, WJ; Chang, BY; Lin, TP; Liu, ST. Activation of the promoter of the fengycin synthetase operon by the UP element. J. Bacteriol 2009, 191, 4615–4623. [Google Scholar]
- Duitman, EH; Wyczawski, D; Boven, LG; Venema, G; Kuipers, OP; Hamoen, LW. Novel methods for genetic transformation of natural Bacillus subtilis isolates used to study the regulation of the mycosubtilin and surfactin synthetases. Appl. Environ. Microbiol 2007, 73, 3490–3496. [Google Scholar]
- Koumoutsi, A; Chen, XH; Vater, J; Borriss, R. DegU and YczE positively regulate the synthesis of bacillomycin D by Bacillus amyloliquefaciens strain FZB42. Appl. Environ. Microbiol 2007, 73, 6953–6964. [Google Scholar]
- Huang, CC; Ano, T; Shoda, M. Nucleotide sequence and characteristics of the gene, lpa-14, responsible for biosynthesis of the lipopeptide antibiotics iturinA and surfactin from Bacillus subtilis RB14. J. Ferment. Bioeng 1993, 76, 445–450. [Google Scholar]
- Heeb, S; Haas, D. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant Microbe Interact 2001, 14, 1351–1363. [Google Scholar]
- Workentine, ML; Chang, L; Ceri, H; Turner, RJ. The GacS-GacA two-component regulatory system of Pseudomonas fluorescens: a bacterial two-hybrid analysis. FEMS Microbiol. Lett 2009, 292, 50–56. [Google Scholar]
- Willis, DK; Holmstadt, JJ; Kinscherf, TG. Genetic evidence that loss of virulence associated with gacS or gacA mutations in Pseudomonas syringae B728a does not result from effects on alginate production. Appl. Environ. Microbiol 2001, 67, 1400–1403. [Google Scholar]
- Koch, B; Nielsen, TH; Sorensen, D; Andersen, JB; Christophersen, C; Molin, S; Givskov, M; Sorensen, J; Nybroe, O. Lipopeptide production in Pseudomonas sp. strain DSS73 is regulated by components of sugar beet seed exudate via the Gac two-component regulatory system. Appl. Environ. Microbiol 2002, 68, 4509–4516. [Google Scholar]
- Nielsen, TH; Nybroe, O; Koch, B; Hansen, M; Sorensen, J. Genes involved in cyclic lipopeptide production are important for seed and straw colonization by Pseudomonas sp. strain DSS73. Appl. Environ. Microbiol 2005, 71, 4112–4116. [Google Scholar]
- Dubern, JF; Bloemberg, GV. Influence of environmental conditions on putisolvins I and II production in Pseudomonas putida strain PCL1445. FEMS Microbiol. Lett 2006, 263, 169–175. [Google Scholar]
- Dubern, JF; Lugtenberg, BJ; Bloemberg, GV. The ppuI-rsaL-ppuR quorum-sensing system regulates biofilm formation of Pseudomonas putida PCL1445 by controlling biosynthesis of the cyclic lipopeptides putisolvins I and II. J. Bacteriol 2006, 188, 2898–2906. [Google Scholar]
- Washio, K; Lim, SP; Roongsawang, N; Morikawa, M. Identification and characterization of the genes responsible for the production of the cyclic lipopeptide arthrofactin by Pseudomonas sp. MIS38. Biosci. Biotechnol. Biochem 2010, 74, 992–999. [Google Scholar]
- Dubern, JF; Lagendijk, EL; Lugtenberg, BJ; Bloemberg, GV. The heat shock genes dnaK, dnaJ, and grpE are involved in regulation of putisolvin biosynthesis in Pseudomonas putida PCL1445. J. Bacteriol 2005, 187, 5967–5976. [Google Scholar]
- Wang, N; Lu, SE; Records, AR; Gross, DC. Characterization of the transcriptional activators SalA and SyrF, Which are required for syringomycin and syringopeptin production by Pseudomonas syringae pv. syringae. J. Bacteriol 2006, 188, 3290–3298. [Google Scholar]
- de Bruijn, I; Raaijmakers, JM. Diversity and functional analysis of LuxR-type transcriptional regulators of cyclic lipopeptide biosynthesis in Pseudomonas fluorescens. Appl. Environ. Microbiol 2009, 75, 4753–4761. [Google Scholar]
- de Bruijn, I; Raaijmakers, JM. Regulation of cyclic lipopeptide biosynthesis in Pseudomonas fluorescens by the ClpP protease. J. Bacteriol 2009, 191, 1910–1923. [Google Scholar]
- Srivatsan, A; Wang, JD. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr. Opin. Microbiol 2008, 11, 100–105. [Google Scholar]
- Finking, R; Solsbacher, J; Konz, D; Schobert, M; Schafer, A; Jahn, D; Marahiel, MA. Characterization of a new type of phosphopantetheinyl transferase for fatty acid and siderophore synthesis in Pseudomonas aeruginosa. J. Biol. Chem 2002, 277, 50293–50302. [Google Scholar]
- Barekzi, N; Joshi, S; Irwin, S; Ontl, T; Schweizer, HP. Genetic characterization of pcpS, encoding the multifunctional phosphopantetheinyl transferase of Pseudomonas aeruginosa. Microbiology 2004, 150, 795–803. [Google Scholar]
- Seidle, HF; Couch, RD; Parry, RJ. Characterization of a nonspecific phosphopantetheinyl transferase from Pseudomonas syringae pv. syringae FF5. Arch. Biochem. Biophys 2006, 446, 167–174. [Google Scholar]
- Quigley, NB; Mo, YY; Gross, DC. SyrD is required for syringomycin production by Pseudomonas syringae pv. syringae and is related to a family of ATP-binding secretion proteins. Mol. Microbiol 1993, 9, 787–801. [Google Scholar]
- Grgurina, I; Gross, DC; Iacobellis, NS; Lavermicocca, P; Takemoto, JY; Benincasa, M. Phytotoxin production by Pseudomonas syringae pv. syringae: Syringopeptin production by syr mutants defective in biosynthesis or secretion of syringomycin. FEMS Microbiol. Lett 1996, 138, 35–39. [Google Scholar]
- Kang, H; Gross, DC. Characterization of a resistance-nodulation-cell division transporter system associated with the syr-syp genomic island of Pseudomonas syringae pv. syringae. Appl. Environ. Microbiol 2005, 71, 5056–5065. [Google Scholar]
- Lim, SP; Roongsawang, N; Washio, K; Morikawa, M. Flexible exportation mechanisms of arthrofactin in Pseudomonas sp. MIS38. J. Appl. Microbiol 2009, 107, 157–166. [Google Scholar]
- Kobayashi, N; Nishino, K; Yamaguchi, A. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J. Bacteriol 2001, 183, 5639–5644. [Google Scholar]
- Schneider, A; Stachelhaus, T; Marahiel, MA. Targeted alteration of the substrate specificity of peptide synthetases by rational module swapping. Mol. Gen. Genet 1998, 257, 308–318. [Google Scholar]
- Yakimov, MM; Giuliano, L; Timmis, KN; Golyshin, PN. Recombinant acylheptapeptide lichenysin: high level of production by Bacillus subtilis cells. J. Mol. Microbiol. Biotechnol 2000, 2, 217–224. [Google Scholar]
- de Ferra, F; Rodriguez, F; Tortora, O; Tosi, C; Grandi, G. Engineering of peptide synthetases. Key role of the thioesterase-like domain for efficient production of recombinant peptides. J. Biol. Chem 1997, 272, 25304–25309. [Google Scholar]
- Trauger, JW; Kohli, RM; Mootz, HD; Marahiel, MA; Walsh, CT. Peptide cyclization catalysed by the thioesterase domain of tyrocidine synthetase. Nature 2000, 407, 215–218. [Google Scholar]
- Kohli, RM; Trauger, JW; Schwarzer, D; Marahiel, MA; Walsh, CT. Generality of peptide cyclization catalyzed by isolated thioesterase domains of nonribosomal peptide synthetases. Biochemistry 2001, 40, 7099–7108. [Google Scholar]
- Stachelhaus, T; Mootz, HD; Marahiel, MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol 1999, 6, 493–505. [Google Scholar]
- Uguru, GC; Milne, C; Borg, M; Flett, F; Smith, CP; Micklefield, J. Active-site modifications of adenylation domains lead to hydrolysis of upstream nonribosomal peptidyl thioester intermediates. J. Am. Chem. Soc 2004, 126, 5032–5033. [Google Scholar]
- Mootz, HD; Kessler, N; Linne, U; Eppelmann, K; Schwarzer, D; Marahiel, MA. Decreasing the ring size of a cyclic nonribosomal peptide antibiotic by in-frame module deletion in the biosynthetic genes. J. Am. Chem. Soc 2002, 124, 10980–10981. [Google Scholar]
- Symmank, H; Franke, P; Saenger, W; Bernhard, F. Modification of biologically active peptides: production of a novel lipohexapeptide after engineering of Bacillus subtilis surfactin synthetase. Protein Eng 2002, 15, 913–921. [Google Scholar]
- Butz, D; Schmiederer, T; Hadatsch, B; Wohlleben, W; Weber, T; Sussmuth, RD. Module extension of a non-ribosomal peptide synthetase of the glycopeptide antibiotic balhimycin produced by Amycolatopsis balhimycina. ChemBioChem 2008, 9, 1195–1200. [Google Scholar]
- Hahn, M; Stachelhaus, T. Harnessing the potential of communication-mediating domains for the biocombinatorial synthesis of nonribosomal peptides. Proc. Natl. Acad. Sci USA 2006, 103, 275–280. [Google Scholar]
- Chiocchini, C; Linne, U; Stachelhaus, T. In vivo biocombinatorial synthesis of lipopeptides by COM domain-mediated reprogramming of the surfactin biosynthetic complex. Chem. Biol 2006, 13, 899–908. [Google Scholar]
- Vater, J; Wilde, C; Kell, H. In situ detection of the intermediates in the biosynthesis of surfactin, a lipoheptapeptide from Bacillus subtilis OKB 105, by whole-cell cell matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in combination with mutant analysis. Rapid. Commun. Mass Spectrom 2009, 23, 1493–1498. [Google Scholar]
- Ackerley, DF; Lamont, IL. Characterization and genetic manipulation of peptide synthetases in Pseudomonas aeruginosa PAO1 in order to generate novel pyoverdines. Chem. Biol 2004, 11, 971–980. [Google Scholar]
- Baltz, RH. Biosynthesis and genetic engineering of lipopeptides in Streptomyces roseosporus. Methods Enzymol 2009, 458, 511–531. [Google Scholar]
- Nguyen, KT; He, X; Alexander, DC; Li, C; Gu, JQ; Mascio, C; Van Praagh, A; Mortin, L; Chu, M; Silverman, JA; Brian, P; Baltz, RH. Genetically engineered lipopeptide antibiotics related to A54145 and daptomycin with improved properties. Antimicrob. Agents Chemother 2010, 54, 1404–1413. [Google Scholar]
© 2011 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).