Implication of Potential Differential Roles of the Two Phosphoglucomutase Isoforms in the Protozoan Parasite Cryptosporidium parvum
Abstract
:1. Introduction
2. Results and Discussion
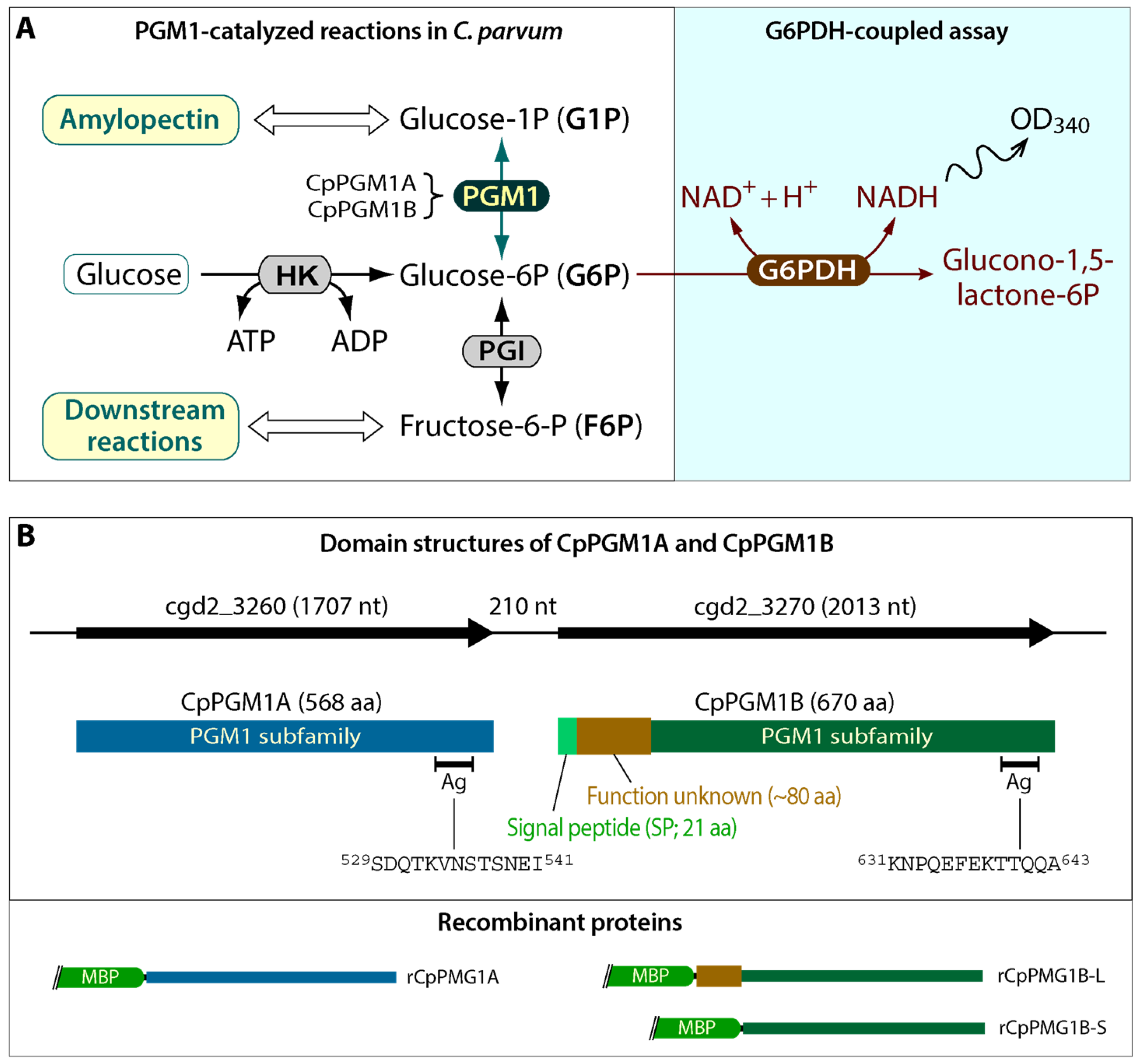
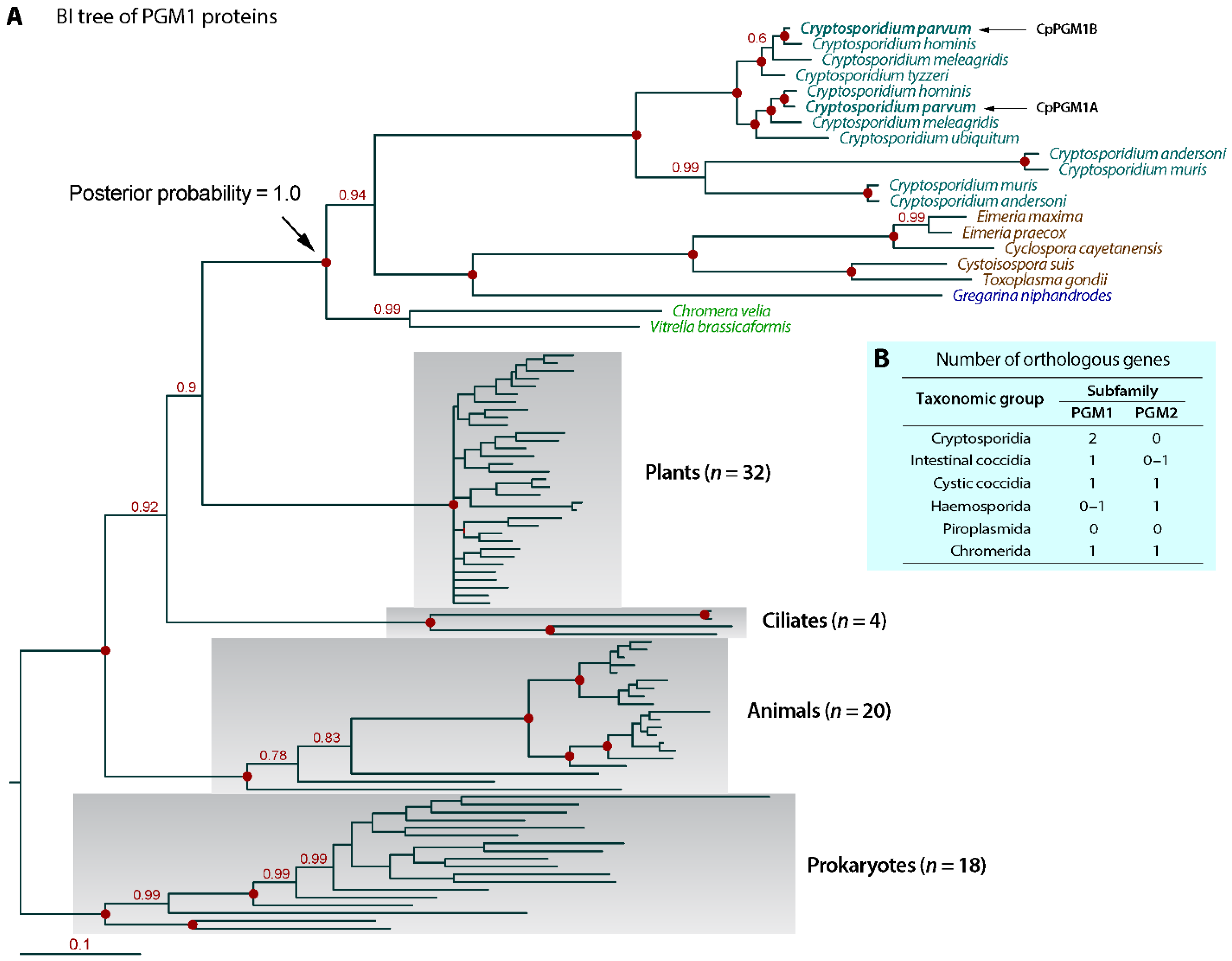
2.1. Cryptosporidium Possessed Two Tandemly Duplicated PGM1-Subfamily Genes Predicted to Encode a Cytosolic and a Non-Cytosolic Protein
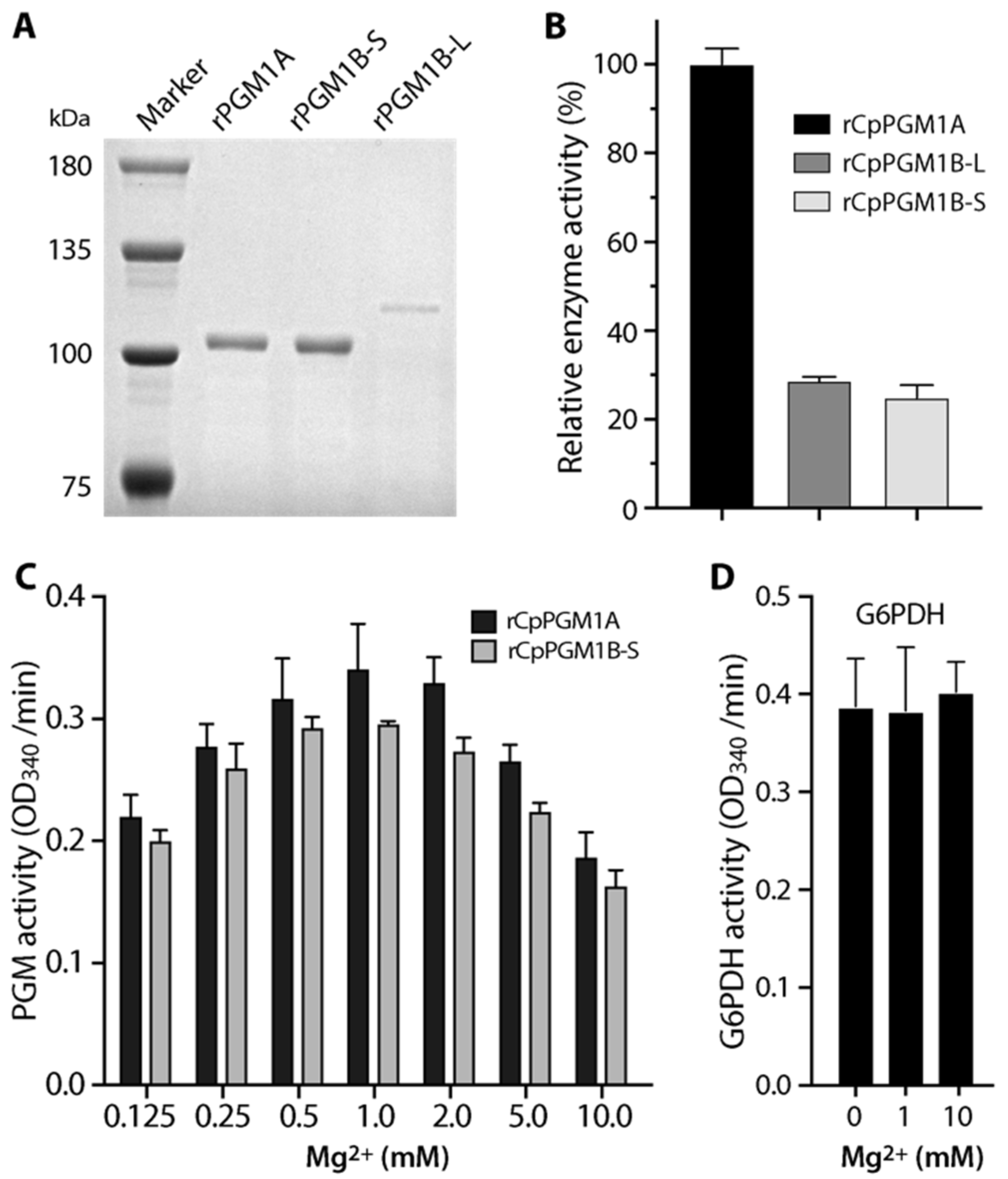
2.2. Both CpPGM1A and CpPGM1B Were Enzymatically Active
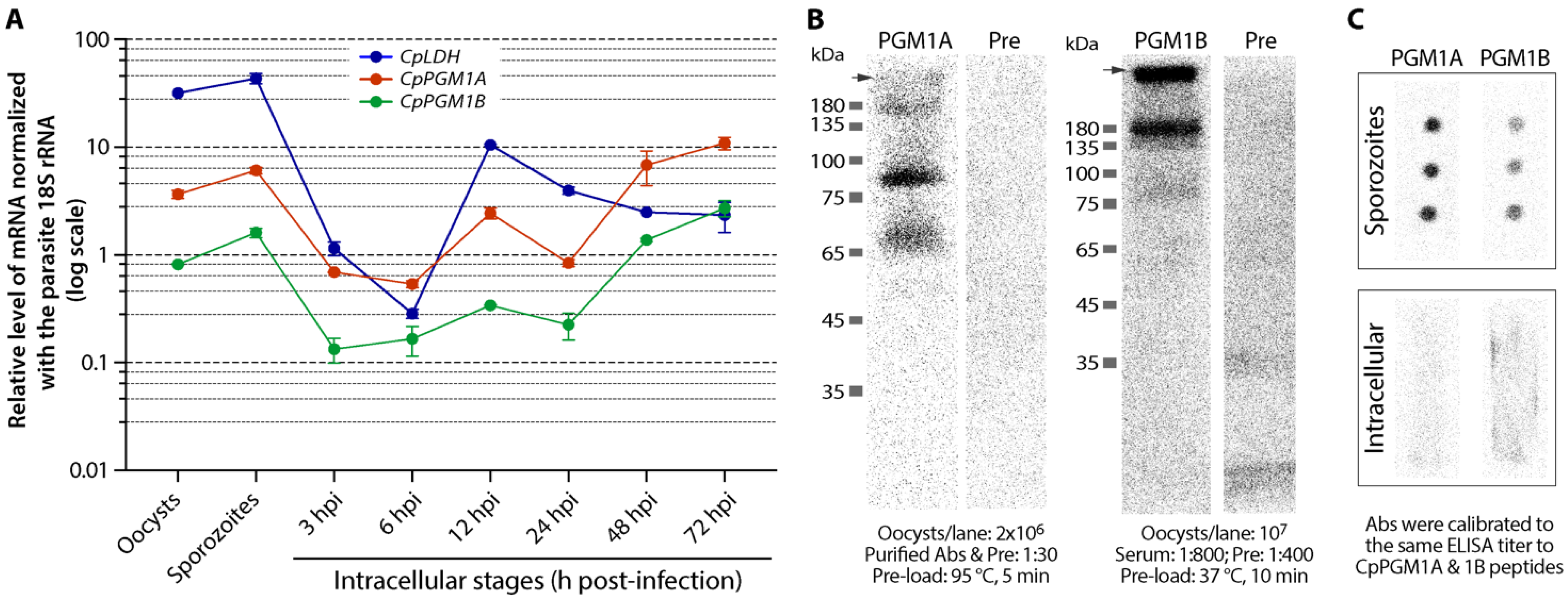
2.3. CpPGM1A and CpPGM1B Genes Were Differentially Expressed
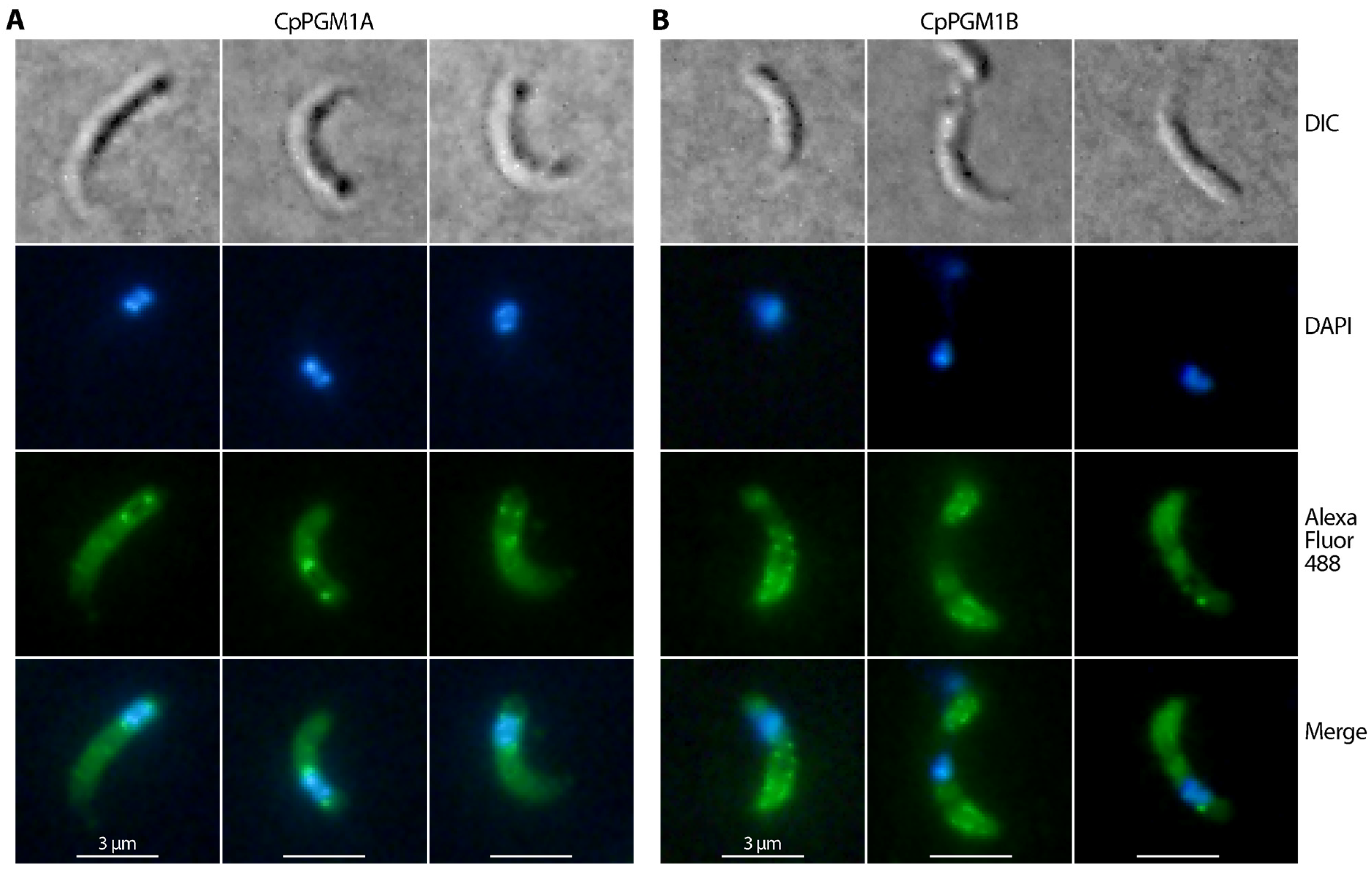
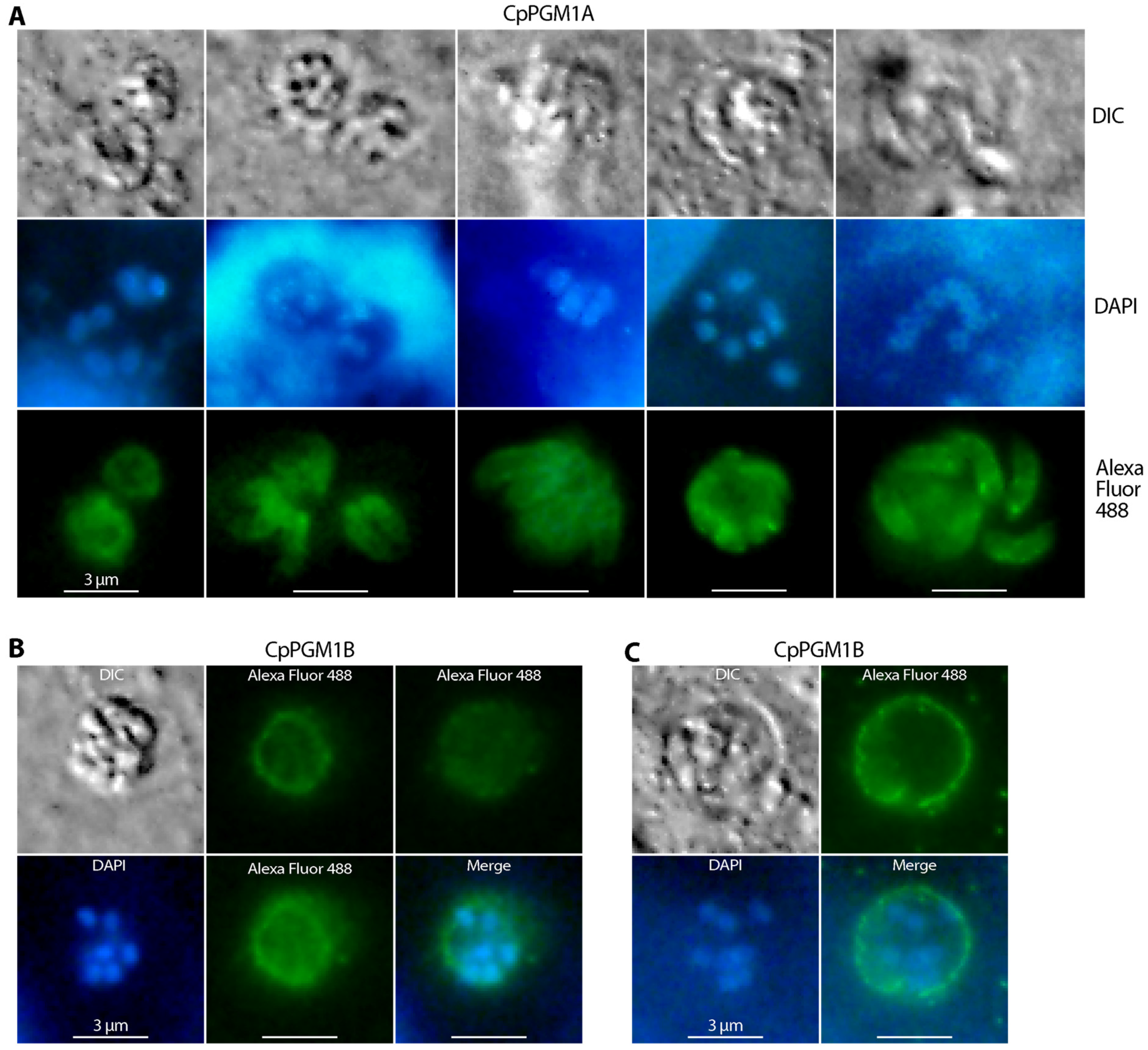
2.4. CpPGM1A and CpPGM1B Protein Were Differentially Distributed in the Sporozoites and Intracellular Parasites
3. Materials and Methods
3.1. The Parasite
3.2. Heterogenous Expression of Recombinant CpPGM1A and CpPGM1B Proteins
3.3. Determination of Enzyme Activity and Kinetics
3.4. Production of Polyclonal Antibodies against CpPGM1A and CpPGM1B
3.5. Western Blot and Dot Blot Analyses
3.6. Immunofluorescence Microscopic Assay (IFA)
3.7. Quantitative RT-PCR Detection of CpPGM1A and CpPGM1B Transcripts
3.8. Phylogenetic Reconstruction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Innes, E.A.; Chalmers, R.M.; Wells, B.; Pawlowic, M.C. A One Health Approach to Tackle Cryptosporidiosis. Trends Parasitol. 2020, 36, 290–303. [Google Scholar] [CrossRef] [Green Version]
- Checkley, W.; White, A.C., Jr.; Jaganath, D.; Arrowood, M.J.; Chalmers, R.M.; Chen, X.M.; Fayer, R.; Griffiths, J.K.; Guerrant, R.L.; Hedstrom, L.; et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect. Dis. 2015, 15, 85–94. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Ryan, U.M.; Xiao, L. Genetic Diversity and Population Structure of Cryptosporidium. Trends Parasitol. 2018, 34, 997–1011. [Google Scholar] [CrossRef] [PubMed]
- Ryan, U.; Zahedi, A.; Paparini, A. Cryptosporidium in humans and animals—A one health approach to prophylaxis. Parasite Immunol. 2016, 38, 535–547. [Google Scholar] [CrossRef] [Green Version]
- Kotloff, K.L.; Nataro, J.P.; Blackwelder, W.C.; Nasrin, D.; Farag, T.H.; Panchalingam, S.; Wu, Y.; Sow, S.O.; Sur, D.; Breiman, R.F.; et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet 2013, 382, 209–222. [Google Scholar] [CrossRef]
- Schneider, A.; Wendt, S.; Lubbert, C.; Trawinski, H. Current pharmacotherapy of cryptosporidiosis: An update of the state-of-the-art. Expert Opin. Pharm. 2021, 22, 2337–2342. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Yin, J.; Cuny, G.D. Current status and challenges in drug discovery against the globally important zoonotic cryptosporidiosis. Anim. Dis. 2021, 1, 3. [Google Scholar] [CrossRef]
- Rider, S.D., Jr.; Zhu, G. Cryptosporidium: Genomic and biochemical features. Exp. Parasitol. 2010, 124, 2–9. [Google Scholar] [CrossRef] [Green Version]
- Zhu, G.; Keithly, J.S.; Philippe, H. What is the phylogenetic position of Cryptosporidium? Int. J. Syst. Evol. Microbiol. 2000, 50, 1673–1681. [Google Scholar] [CrossRef] [PubMed]
- Tzipori, S.; Widmer, G. The biology of Cryptosporidium. Contrib. Microbiol. 2000, 6, 1–32. [Google Scholar] [PubMed]
- Abrahamsen, M.S.; Templeton, T.J.; Enomoto, S.; Abrahante, J.E.; Zhu, G.; Lancto, C.A.; Deng, M.; Liu, C.; Widmer, G.; Tzipori, S.; et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 2004, 304, 441–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Zhang, H.; Guo, F.; Sun, M.; Zhu, G. A unique hexokinase in Cryptosporidium parvum, an apicomplexan pathogen lacking the Krebs cycle and oxidative phosphorylation. Protist 2014, 165, 701–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imada, M.; Kawashima, S.; Kanehisa, M.; Takeuchi, T.; Asai, T. Characterization of alpha-phosphoglucomutase isozymes from Toxoplasma gondii. Parasitol. Int. 2010, 59, 206–210. [Google Scholar] [CrossRef] [PubMed]
- Walther, T.; Baylac, A.; Alkim, C.; Vax, A.; Cordier, H.; Francois, J.M. The PGM3 gene encodes the major phosphoribomutase in the yeast Saccharomyces cerevisiae. FEBS Lett. 2012, 586, 4114–4118. [Google Scholar] [CrossRef]
- Tiwari, A.; Bhat, J.P. Molecular characterization reveals that YMR278w encoded protein is environmental stress response homologue of Saccharomyces cerevisiae PGM2. Biochem. Biochem. Biophys. Res. Commun. 2008, 366, 340–345. [Google Scholar] [CrossRef]
- Fazi, A.; Piacentini, M.P.; Piatti, E.; Accorsi, A. Purification and partial characterization of the phosphoglucomutase isozymes from human placenta. Prep. Biochem. 1990, 20, 219–240. [Google Scholar] [CrossRef] [PubMed]
- Accorsi, A.; Piatti, E.; Piacentini, M.P.; Gini, S.; Fazi, A. Isoenzymes of phosphoglucomutase from human red blood cells: Isolation and kinetic properties. Prep. Biochem. 1989, 19, 251–271. [Google Scholar] [CrossRef]
- Csutora, P.; Karsai, A.; Nagy, T.; Vas, B.; Kovács, G.; Rideg, O.; Bogner, P.; Miseta, A. Lithium induces phosphoglucomutase activity in various tissues of rats and in bipolar patients. Int. J. Neuropsychopharmacol. 2006, 9, 613–619. [Google Scholar] [CrossRef] [Green Version]
- Csutora, P.; Strassz, A.; Boldizsar, F.; Nemeth, P.; Sipos, K.; Aiello, D.P.; Bedwell, D.M.; Miseta, A. Inhibition of phosphoglucomutase activity by lithium alters cellular calcium homeostasis and signaling in Saccharomyces cerevisiae. Am. J. Physiol. Cell Physiol. 2005, 289, C58–C67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Guo, F.; Zhu, G. Cryptosporidium Lactate Dehydrogenase Is Associated with the Parasitophorous Vacuole Membrane and Is a Potential Target for Developing Therapeutics. PLoS Pathog. 2015, 11, e1005250. [Google Scholar] [CrossRef]
- Zhang, H.; Guo, F.; Zhou, H.; Zhu, G. Transcriptome analysis reveals unique metabolic features in the Cryptosporidium parvum Oocysts associated with environmental survival and stresses. BMC Genom. 2012, 13, 647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X.; Yin, J.; Wang, D.; Li, X.; Zhang, Y.; Wang, C.; Zhang, Y.; Zhu, G. Discovery of New Microneme Proteins in Cryptosporidium parvum and Implication of the Roles of a Rhomboid Membrane Protein (CpROM1) in Host–Parasite Interaction. Front. Vet. Sci. 2021, 8, 778560. [Google Scholar] [CrossRef]
- Rath, A.; Glibowicka, M.; Nadeau, V.G.; Chen, G.; Deber, C.M. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc. Natl. Acad. Sci. USA 2009, 106, 1760–1765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaur, J.; Bachhawat, A.K. A modified Western blot protocol for enhanced sensitivity in the detection of a membrane protein. Anal. Biochem. 2009, 384, 348–349. [Google Scholar] [CrossRef]
- Muller, S.; Diederichs, K.; Breed, J.; Kissmehl, R.; Hauser, K.; Plattner, H.; Welte, W. Crystal structure analysis of the exocytosis-sensitive phosphoprotein, pp63/parafusin (phosphoglucomutase), from Paramecium reveals significant conformational variability. J. Mol. Biol. 2002, 315, 141–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanderson, S.J.; Xia, D.; Prieto, H.; Yates, J.; Heiges, M.; Kissinger, J.C.; Bromley, E.; Lal, K.; Sinden, R.E.; Tomley, F.; et al. Determining the protein repertoire of Cryptosporidium parvum sporozoites. Proteomics 2008, 8, 1398–1414. [Google Scholar] [CrossRef] [Green Version]
- Snelling, W.J.; Lin, Q.; Moore, J.E.; Millar, B.C.; Tosini, F.; Pozio, E.; Dooley, J.S.; Lowery, C.J. Proteomics analysis and protein expression during sporozoite excystation of Cryptosporidium parvum (Coccidia, Apicomplexa). Mol. Cell Proteom. 2007, 6, 346–355. [Google Scholar] [CrossRef] [Green Version]
- Truong, Q.; Ferrari, B.C. Quantitative and qualitative comparisons of Cryptosporidium faecal purification procedures for the isolation of oocysts suitable for proteomic analysis. Int. J. Parasitol. 2006, 36, 811–819. [Google Scholar] [CrossRef]
- Slapeta, J.; Keithly, J.S. Cryptosporidium parvum mitochondrial-type HSP70 targets homologous and heterologous mitochondria. Eukaryot. Cell 2004, 3, 483–494. [Google Scholar] [CrossRef] [Green Version]
- Keithly, J.S.; Langreth, S.G.; Buttle, K.F.; Mannella, C.A. Electron tomographic and ultrastructural analysis of the Cryptosporidium parvum relict mitochondrion, its associated membranes, and organelles. J. Eukaryot. Microbiol. 2005, 52, 132–140. [Google Scholar] [CrossRef]
- Tetley, L.; Brown, S.M.A.; McDonald, V.; Coombs, G.H. Ultrastructural analysis of the sporozoite of Cryptosporidium parvum. Microbiology 1998, 144, 3249–3255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsubayashi, M.; Ando, H.; Kimata, I.; Nakagawa, H.; Furuya, M.; Tani, H.; Sasai, K. Morphological changes and viability of Cryptosporidium parvum sporozoites after excystation in cell-free culture media. Parasitology 2010, 137, 1861–1866. [Google Scholar] [CrossRef] [PubMed]
- Aldeyarbi, H.M.; Karanis, P. The fine structure of sexual stage development and sporogony of Cryptosporidium parvum in cell-free culture. Parasitology 2016, 143, 749–761. [Google Scholar] [CrossRef]
- Kono, M.; Prusty, D.; Parkinson, J.; Gilberger, T.W. The apicomplexan inner membrane complex. Front. Biosci. 2013, 18, 982–992. [Google Scholar] [CrossRef]
- Pomel, S.; Luk, F.C.; Beckers, C.J. Host cell egress and invasion induce marked relocations of glycolytic enzymes in Toxoplasma gondii tachyzoites. PLoS Pathog. 2008, 4, e1000188. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Wyroba, E.; Satir, B.H. RNAi knockdown of parafusin inhibits the secretory pathway. Eur. J. Cell Biol. 2011, 90, 844–853. [Google Scholar] [CrossRef]
- Zhao, H.; Satir, B.H. Parafusin is a membrane and vesicle associated protein that cycles at exocytosis. Eur. J. Cell Biol. 1998, 75, 46–53. [Google Scholar] [CrossRef]
- Satir, B.H.; Wyroba, E.; Liu, L.; Lethan, M.; Satir, P.; Christensen, S.T. Evolutionary implications of localization of the signaling scaffold protein parafusin to both cilia and the nucleus. Cell Biol. Int. 2015, 39, 136–145. [Google Scholar] [CrossRef]
- Matthiesen, S.H.; Shenoy, S.M.; Kim, K.; Singer, R.H.; Satir, B.H. Role of the parafusin orthologue, PRP1, in microneme exocytosis and cell invasion in Toxoplasma gondii. Cell Microbiol. 2003, 5, 613–624. [Google Scholar] [CrossRef] [Green Version]
- Saha, S.; Coleman, B.I.; Dubey, R.; Blader, I.J.; Gubbels, M.J. Two Phosphoglucomutase Paralogs Facilitate Ionophore-Triggered Secretion of the Toxoplasma Micronemes. Msphere 2017, 2, e00521-17. [Google Scholar] [CrossRef] [Green Version]
- Arrowood, M.J. Cryptosporidium Oocyst Purification Using Discontinuous Gradient Centrifugation. Methods Mol. Biol. 2020, 2052, 43–59. [Google Scholar] [CrossRef]
- Lateef, S.S.; Gupta, S.; Jayathilaka, L.P.; Krishnanchettiar, S.; Huang, J.S.; Lee, B.S. An improved protocol for coupling synthetic peptides to carrier proteins for antibody production using DMF to solubilize peptides. J. Biomol. Tech. 2007, 18, 173–176. [Google Scholar] [PubMed]
- Zhang, H.; Zhu, G. Quantitative RT-PCR assay for high-throughput screening (HTS) of drugs against the growth of Cryptosporidium parvum in vitro. Front. Microbiol. 2015, 6, 991. [Google Scholar] [CrossRef] [Green Version]
- Mauzy, M.J.; Enomoto, S.; Lancto, C.A.; Abrahamsen, M.S.; Rutherford, M.S. The Cryptosporidium parvum transcriptome during in vitro development. PLoS ONE 2012, 7, e31715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, F.; Zhang, H.; Payne, H.R.; Zhu, G. Differential Gene Expression and Protein Localization of Cryptosporidium parvum Fatty Acyl-CoA Synthetase Isoforms. J. Eukaryot. Microbiol. 2016, 63, 233–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nie, J.; Yin, J.; Wang, D.; Wang, C.; Zhu, G. Implication of Potential Differential Roles of the Two Phosphoglucomutase Isoforms in the Protozoan Parasite Cryptosporidium parvum. Pathogens 2022, 11, 21. https://doi.org/10.3390/pathogens11010021
Nie J, Yin J, Wang D, Wang C, Zhu G. Implication of Potential Differential Roles of the Two Phosphoglucomutase Isoforms in the Protozoan Parasite Cryptosporidium parvum. Pathogens. 2022; 11(1):21. https://doi.org/10.3390/pathogens11010021
Chicago/Turabian StyleNie, Jiawen, Jigang Yin, Dongqiang Wang, Chenchen Wang, and Guan Zhu. 2022. "Implication of Potential Differential Roles of the Two Phosphoglucomutase Isoforms in the Protozoan Parasite Cryptosporidium parvum" Pathogens 11, no. 1: 21. https://doi.org/10.3390/pathogens11010021





