Structure of a DNA Glycosylase Bound to a Nicked T:G Mismatch-Containing DNA
Abstract
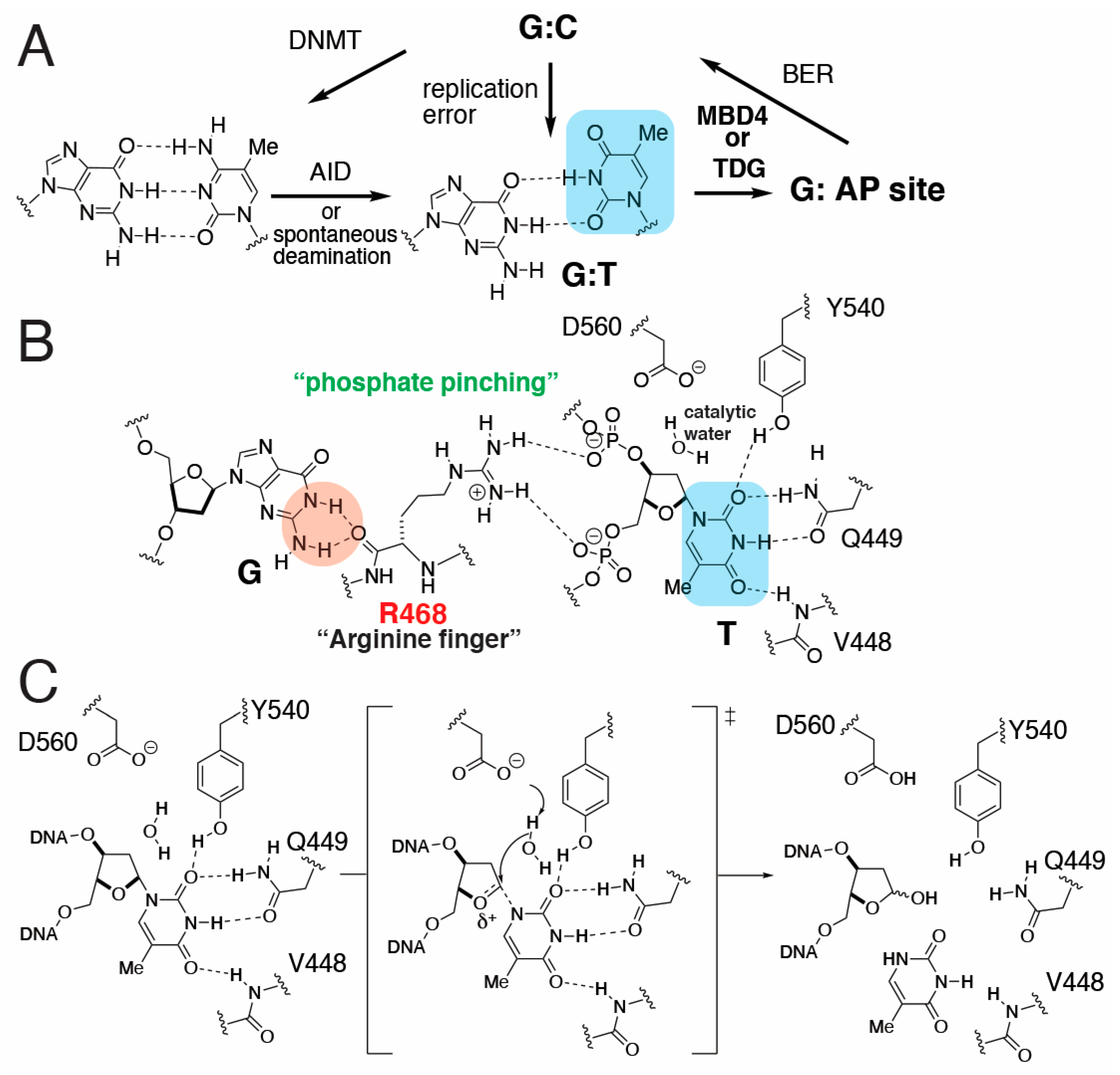
1. Introduction
2. Results and Discussion
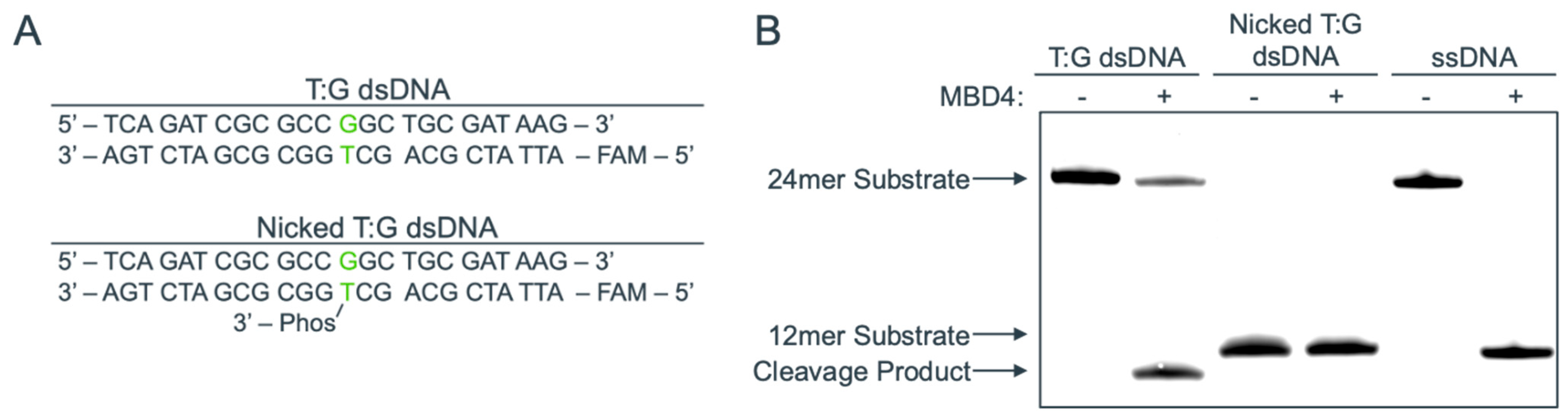
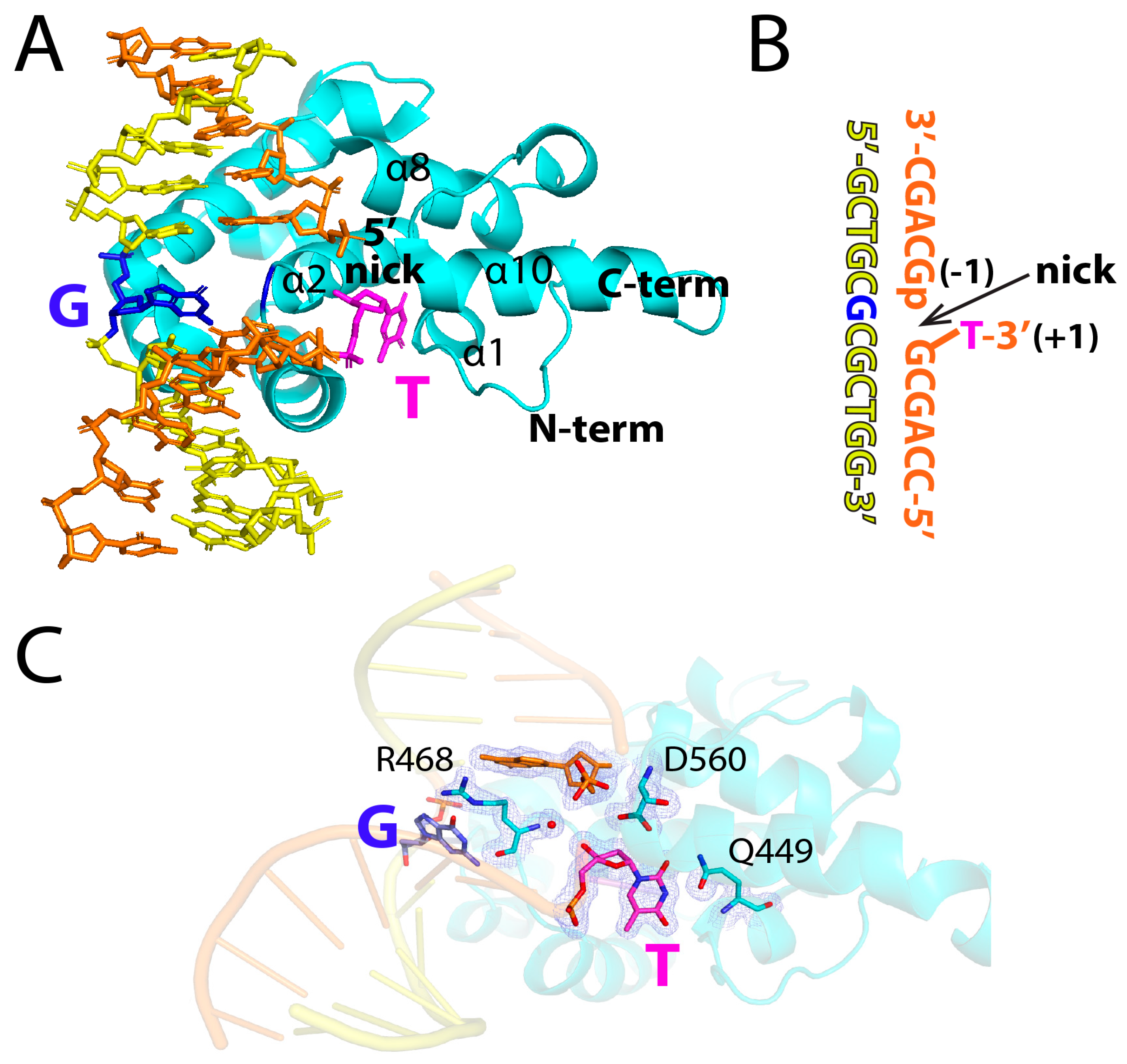
2.1. Structure of MBD4 in Complex with a Nicked T:G-Containing DNA
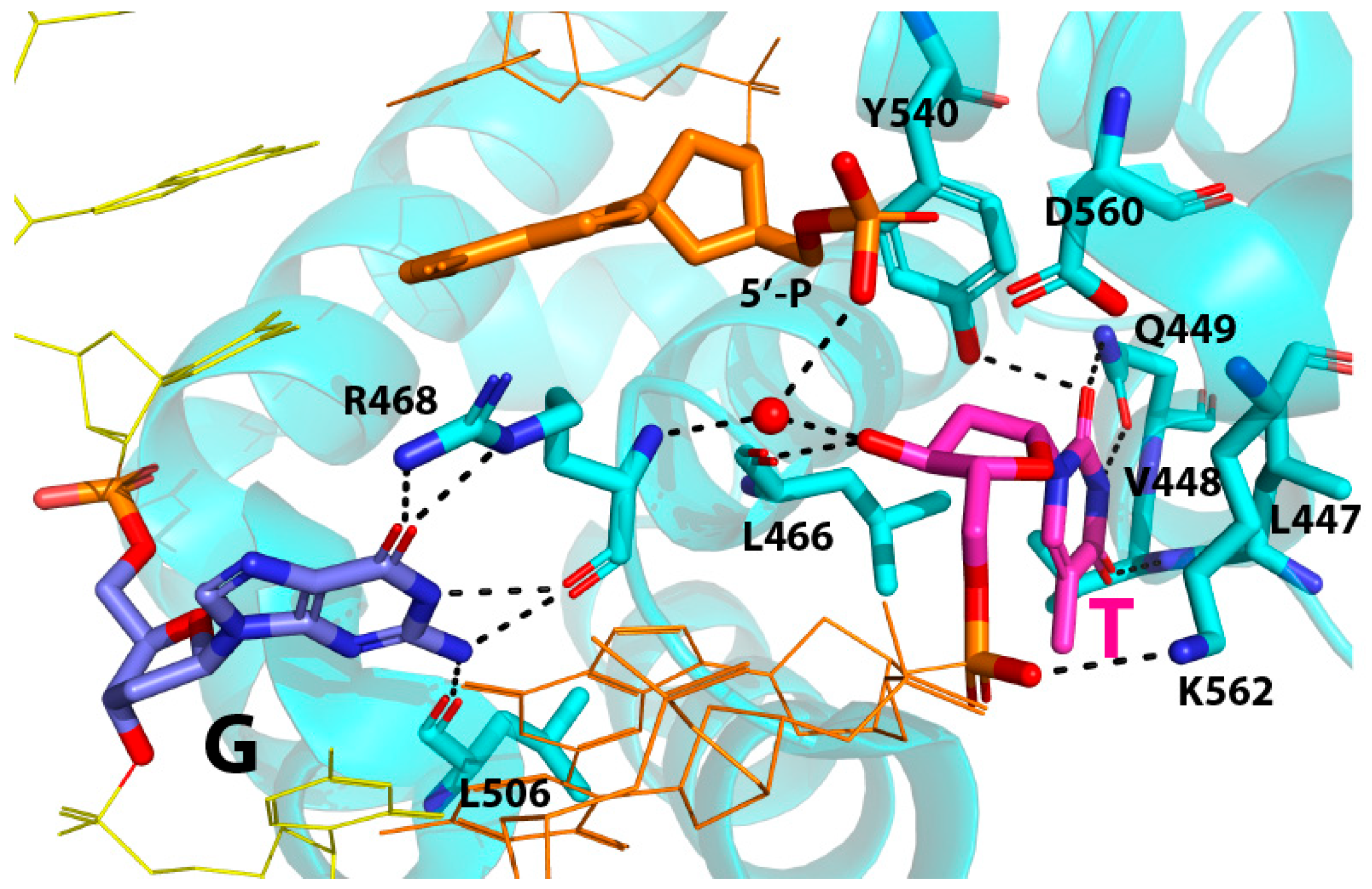
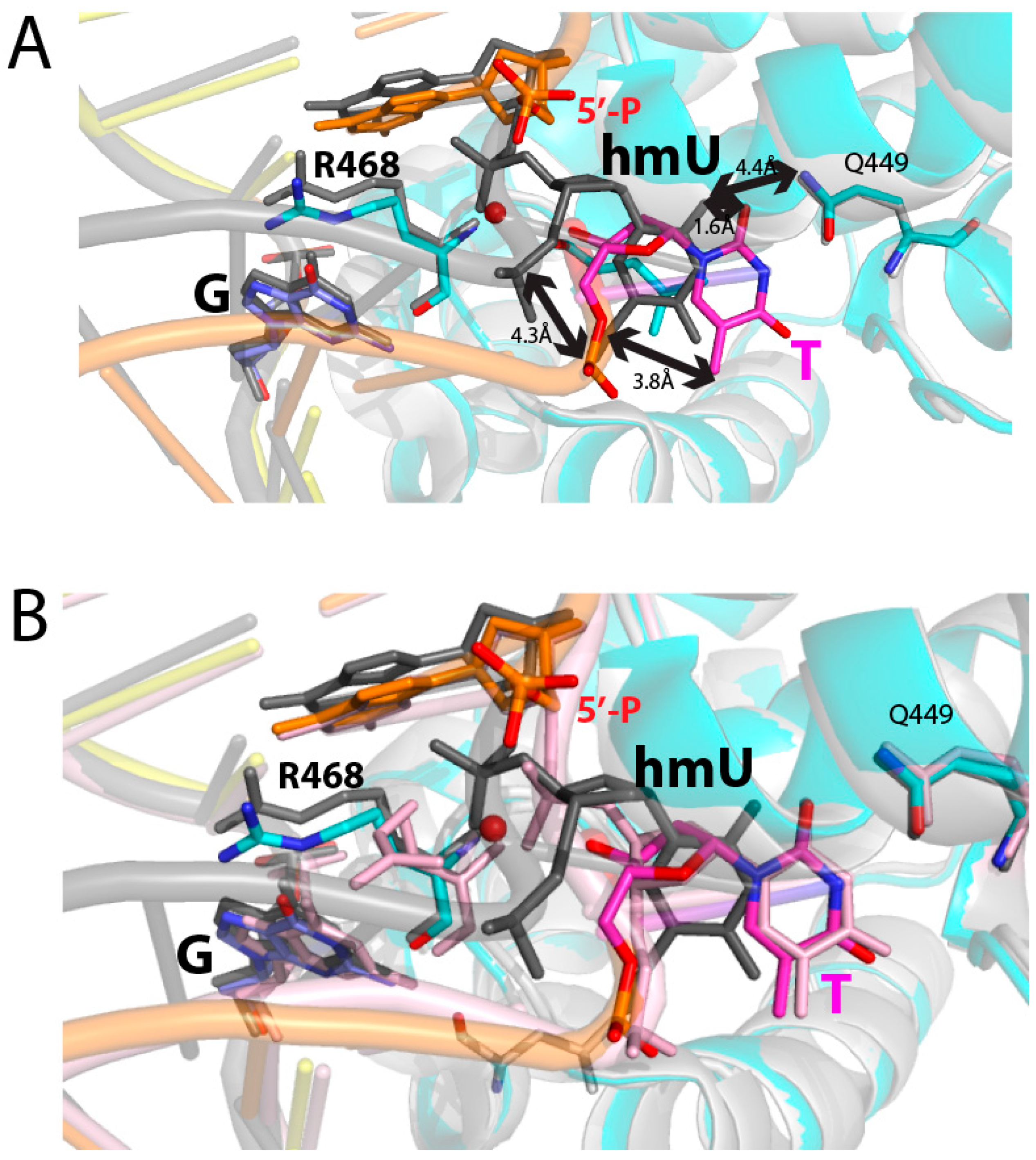
2.2. Binding Mode of a Nicked T:G-Containing DNA to MBD4 and the Origin of Its Inactivity
2.3. The Role of R468 in T:G Mismatch Recognition
2.4. The Role of R468 in the Base Extrusion Pathway
3. Materials and Methods
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arana, M.E.; Kunkel, T.A. Mutator phenotypes due to DNA replication infidelity. Semin. Cancer Biol. 2010, 20, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, T.A. The mutational specificity of DNA polymerase-beta during in vitro DNA synthesis. Production of frameshift, base substitution, and deletion mutations. J. Biol. Chem. 1985, 260, 5787–5796. [Google Scholar] [CrossRef] [PubMed]
- Bebenek, K.; Pedersen, L.C.; Kunkel, T.A. Replication infidelity via a mismatch with Watson-Crick geometry. Proc. Natl. Acad. Sci. USA 2011, 108, 1862–1867. [Google Scholar] [CrossRef]
- Wang, W.; Hellinga, H.W.; Beese, L.S. Structural evidence for the rare tautomer hypothesis of spontaneous mutagenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 17644–17648. [Google Scholar] [CrossRef] [PubMed]
- Kimsey, I.J.; Szymanski, E.S.; Zahurancik, W.J.; Shakya, A.; Xue, Y.; Chu, C.C.; Sathyamoorthy, B.; Suo, Z.; Al-Hashimi, H.M. Dynamic basis for dG*dT misincorporation via tautomerization and ionization. Nature 2018, 554, 195–201. [Google Scholar] [CrossRef]
- Kimsey, I.J.; Petzold, K.; Sathyamoorthy, B.; Stein, Z.W.; Al-Hashimi, H.M. Visualizing transient Watson-Crick-like mispairs in DNA and RNA duplexes. Nature 2015, 519, 315–320. [Google Scholar] [CrossRef]
- Szymanski, E.S.; Kimsey, I.J.; Al-Hashimi, H.M. Direct NMR Evidence that Transient Tautomeric and Anionic States in dG.dT Form Watson-Crick-like Base Pairs. J. Am. Chem. Soc. 2017, 139, 4326–4329. [Google Scholar] [CrossRef]
- Osheroff, W.P.; Jung, H.K.; Beard, W.A.; Wilson, S.H.; Kunkel, T.A. The fidelity of DNA polymerase beta during distributive and processive DNA synthesis. J. Biol. Chem. 1999, 274, 3642–3650. [Google Scholar] [CrossRef]
- Iwanaga, A.; Ouchida, M.; Miyazaki, K.; Hori, K.; Mukai, T. Functional mutation of DNA polymerase beta found in human gastric cancer--inability of the base excision repair in vitro. Mutat. Res. 1999, 435, 121–128. [Google Scholar] [CrossRef]
- Roettger, M.P.; Bakhtina, M.; Tsai, M.D. Mismatched and matched dNTP incorporation by DNA polymerase beta proceed via analogous kinetic pathways. Biochemistry 2008, 47, 9718–9727. [Google Scholar] [CrossRef]
- Lai, Y.; Jiang, Z.; Zhou, J.; Osemota, E.; Liu, Y. AP endonuclease 1 prevents the extension of a T/G mismatch by DNA polymerase beta to prevent mutations in CpGs during base excision repair. DNA Repair. 2016, 43, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Sjolund, A.B.; Senejani, A.G.; Sweasy, J.B. MBD4 and TDG: Multifaceted DNA glycosylases with ever expanding biological roles. Mutat. Res. 2013, 743–744, 12–25. [Google Scholar] [CrossRef]
- Jacobs, A.L.; Schar, P. DNA glycosylases: In DNA repair and beyond. Chromosoma 2012, 121, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Hendrich, B.; Hardeland, U.; Ng, H.H.; Jiricny, J.; Bird, A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature 1999, 401, 301–304. [Google Scholar] [CrossRef] [PubMed]
- Cortellino, S.; Turner, D.; Masciullo, V.; Schepis, F.; Albino, D.; Daniel, R.; Skalka, A.M.; Meropol, N.J.; Alberti, C.; Larue, L.; et al. The base excision repair enzyme MED1 mediates DNA damage response to antitumor drugs and is associated with mismatch repair system integrity. Proc. Natl. Acad. Sci. USA 2003, 100, 15071–15076. [Google Scholar] [CrossRef]
- Morera, S.; Grin, I.; Vigouroux, A.; Couve, S.; Henriot, V.; Saparbaev, M.; Ishchenko, A.A. Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA. Nucleic Acids Res. 2012, 40, 9917–9926. [Google Scholar] [CrossRef]
- Hashimoto, H.; Zhang, X.; Cheng, X. Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: Structural basis and implications for active DNA demethylation. Nucleic Acids Res. 2012, 40, 8276–8284. [Google Scholar] [CrossRef]
- Trasvina-Arenas, C.H.; Demir, M.; Lin, W.J.; David, S.S. Structure, function and evolution of the Helix-hairpin-Helix DNA glycosylase superfamily: Piecing together the evolutionary puzzle of DNA base damage repair mechanisms. DNA Repair. 2021, 108, 103231. [Google Scholar] [CrossRef]
- Wong, E.; Yang, K.; Kuraguchi, M.; Werling, U.; Avdievich, E.; Fan, K.; Fazzari, M.; Jin, B.; Brown, A.M.; Lipkin, M.; et al. Mbd4 inactivation increases Cright-arrowT transition mutations and promotes gastrointestinal tumor formation. Proc. Natl. Acad. Sci. USA 2002, 99, 14937–14942. [Google Scholar] [CrossRef]
- Millar, C.B.; Guy, J.; Sansom, O.J.; Selfridge, J.; MacDougall, E.; Hendrich, B.; Keightley, P.D.; Bishop, S.M.; Clarke, A.R.; Bird, A. Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science 2002, 297, 403–405. [Google Scholar] [CrossRef]
- Wu, P.; Qiu, C.; Sohail, A.; Zhang, X.; Bhagwat, A.S.; Cheng, X. Mismatch repair in methylated DNA. Structure and activity of the mismatch-specific thymine glycosylase domain of methyl-CpG-binding protein MBD4. J. Biol. Chem. 2003, 278, 5285–5291. [Google Scholar] [CrossRef] [PubMed]
- Pidugu, L.S.; Bright, H.; Lin, W.J.; Majumdar, C.; Van Ostrand, R.P.; David, S.S.; Pozharski, E.; Drohat, A.C. Structural Insights into the Mechanism of Base Excision by MBD4. J. Mol. Biol. 2021, 433, 167097. [Google Scholar] [CrossRef]
- Ouzon-Shubeita, H.; Jung, H.; Lee, M.H.; Koag, M.C.; Lee, S. Catalytic mechanism of the mismatch-specific DNA glycosylase methyl-CpG-binding domain 4. Biochem. J. 2020, 477, 1601–1612. [Google Scholar] [CrossRef] [PubMed]
- Ouzon-Shubeita, H.; Schmaltz, L.F.; Lee, S. Insights into the substrate discrimination mechanisms of methyl-CpG-binding domain 4. Biochem. J. 2021, 478, 1985–1997. [Google Scholar] [CrossRef]
- Barrett, T.E.; Savva, R.; Panayotou, G.; Barlow, T.; Brown, T.; Jiricny, J.; Pearl, L.H. Crystal structure of a G:T/U mismatch-specific DNA glycosylase: Mismatch recognition by complementary-strand interactions. Cell 1998, 92, 117–129. [Google Scholar] [CrossRef]
- Lau, A.Y.; Scharer, O.D.; Samson, L.; Verdine, G.L.; Ellenberger, T. Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: Mechanisms for nucleotide flipping and base excision. Cell 1998, 95, 249–258. [Google Scholar] [CrossRef]
- Hollis, T.; Ichikawa, Y.; Ellenberger, T. DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. EMBO J. 2000, 19, 758–766. [Google Scholar] [CrossRef]
- Hashimoto, H.; Hong, S.; Bhagwat, A.S.; Zhang, X.; Cheng, X. Excision of 5-hydroxymethyluracil and 5-carboxylcytosine by the thymine DNA glycosylase domain: Its structural basis and implications for active DNA demethylation. Nucleic Acids Res. 2012, 40, 10203–10214. [Google Scholar] [CrossRef] [PubMed]
- Maiti, A.; Morgan, M.T.; Pozharski, E.; Drohat, A.C. Crystal structure of human thymine DNA glycosylase bound to DNA elucidates sequence-specific mismatch recognition. Proc. Natl. Acad. Sci. USA 2008, 105, 8890–8895. [Google Scholar] [CrossRef]
- Horton, J.R.; Ratner, G.; Banavali, N.K.; Huang, N.; Choi, Y.; Maier, M.A.; Marquez, V.E.; MacKerell, A.D., Jr.; Cheng, X. Caught in the act: Visualization of an intermediate in the DNA base-flipping pathway induced by HhaI methyltransferase. Nucleic Acids Res. 2004, 32, 3877–3886. [Google Scholar] [CrossRef]
- Shigdel, U.K.; Ovchinnikov, V.; Lee, S.J.; Shih, J.A.; Karplus, M.; Nam, K.; Verdine, G.L. The trajectory of intrahelical lesion recognition and extrusion by the human 8-oxoguanine DNA glycosylase. Nat. Commun. 2020, 11, 4437. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Yang, W.; Karplus, M.; Verdine, G.L. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature 2005, 434, 612–618. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Radom, C.T.; Verdine, G.L. Trapping and structural elucidation of a very advanced intermediate in the lesion-extrusion pathway of hOGG1. J. Am. Chem. Soc. 2008, 130, 7784–7785. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Santos, W.L.; Verdine, G.L. Structure of a DNA glycosylase searching for lesions. Science 2006, 311, 1153–1157. [Google Scholar] [CrossRef]
- Qi, Y.; Spong, M.C.; Nam, K.; Banerjee, A.; Jiralerspong, S.; Karplus, M.; Verdine, G.L. Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme. Nature 2009, 462, 762–766. [Google Scholar] [CrossRef] [PubMed]
- Dunn, A.R.; Kad, N.M.; Nelson, S.R.; Warshaw, D.M.; Wallace, S.S. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Res. 2011, 39, 7487–7498. [Google Scholar] [CrossRef]
- Blainey, P.C.; van Oijen, A.M.; Banerjee, A.; Verdine, G.L.; Xie, X.S. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl. Acad. Sci. USA 2006, 103, 5752–5757. [Google Scholar] [CrossRef]
- O’Brien, P.J.; Ellenberger, T. The Escherichia coli 3-methyladenine DNA glycosylase AlkA has a remarkably versatile active site. J. Biol. Chem. 2004, 279, 26876–26884. [Google Scholar] [CrossRef]
- Bruner, S.D.; Norman, D.P.; Verdine, G.L. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature 2000, 403, 859–866. [Google Scholar] [CrossRef] [PubMed]
- Fromme, J.C.; Banerjee, A.; Huang, S.J.; Verdine, G.L. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature 2004, 427, 652–656. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Collaborative Computational Project, number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994, 50, 760–763. [Google Scholar] [CrossRef] [PubMed]
- Winn, M.D.; Murshudov, G.N.; Papiz, M.Z. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003, 374, 300–321. [Google Scholar] [CrossRef] [PubMed]

| PDB Code | wt MBD4-Nicked DNA (5CHZ) |
|---|---|
| Data Collection | |
| Space Group | P212121 |
| Cell constants | |
| a (Å) | 41.886 |
| b | 54.875 |
| c | 104.522 |
| Resolution (Å) a | 39–1.83 (1.89–1.83) |
| Rmerge b | 0.071 (0.544) |
| <I/σ> | 30.7 (3.15) |
| Completeness (%) | 97.7 (96.8) |
| Redundancy | 6.9 (6.8) |
| Refinement | |
| Rworkc/Rfreed (%) | 18.6/21.8 |
| Unique reflections | 20,686 |
| Mean B factor (Å2) | |
| Protein | 26.65 |
| Ligand | 17.97 |
| Solvent | 36.02 |
| Ramachandran plot | |
| Most favored (%) | 99.3 |
| Additional allowed (%) | 0.7 |
| RMSD | |
| Bond lengths (Å) | 0.018 |
| Bond angles (°) | 1.81 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ouzon-Shubeita, H.; Barnes, R.; Schmaltz, L.F.; Lee, S. Structure of a DNA Glycosylase Bound to a Nicked T:G Mismatch-Containing DNA. Molecules 2025, 30, 2083. https://doi.org/10.3390/molecules30092083
Ouzon-Shubeita H, Barnes R, Schmaltz LF, Lee S. Structure of a DNA Glycosylase Bound to a Nicked T:G Mismatch-Containing DNA. Molecules. 2025; 30(9):2083. https://doi.org/10.3390/molecules30092083
Chicago/Turabian StyleOuzon-Shubeita, Hala, Rebecca Barnes, Lillian F. Schmaltz, and Seongmin Lee. 2025. "Structure of a DNA Glycosylase Bound to a Nicked T:G Mismatch-Containing DNA" Molecules 30, no. 9: 2083. https://doi.org/10.3390/molecules30092083
APA StyleOuzon-Shubeita, H., Barnes, R., Schmaltz, L. F., & Lee, S. (2025). Structure of a DNA Glycosylase Bound to a Nicked T:G Mismatch-Containing DNA. Molecules, 30(9), 2083. https://doi.org/10.3390/molecules30092083







