Alpha and Omega Classification of β-Lactamase/Transpeptidase-like Superfamily Proteins Based on the Comparison of Their Structural Catalytic Cores
Abstract
1. Introduction
2. Results and Discussion
2.1. Creating a Dataset of the β-Lactamase/Transpeptidase-like Representative Structures
2.2. SCC Identification with the Example of β-Lactamase CTX-M-14
2.2.1. Conserved Local Motifs at the Basis of the SCC Identification
2.2.2. The “NucBase-Oxy” Zone
2.2.3. The NucBase-Omega Zone and Its Omega (Ω) Subzone
2.2.4. SCC as a Structural Association of the NucBase-Oxy and NucBase-Omega Zones and the A-tripeptide Link
| N | PDB ID | R (Å) | Protein | NucBase | Alpha | Omega | Oxy | Mediator | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Superfamily: β-lactamase/transpeptidase-like | |||||||||
| Family: β-lactamase/D-Ala carboxypeptidase | |||||||||
| N-like group (Class A) (79) | |||||||||
| SNN subgroup (67) | |||||||||
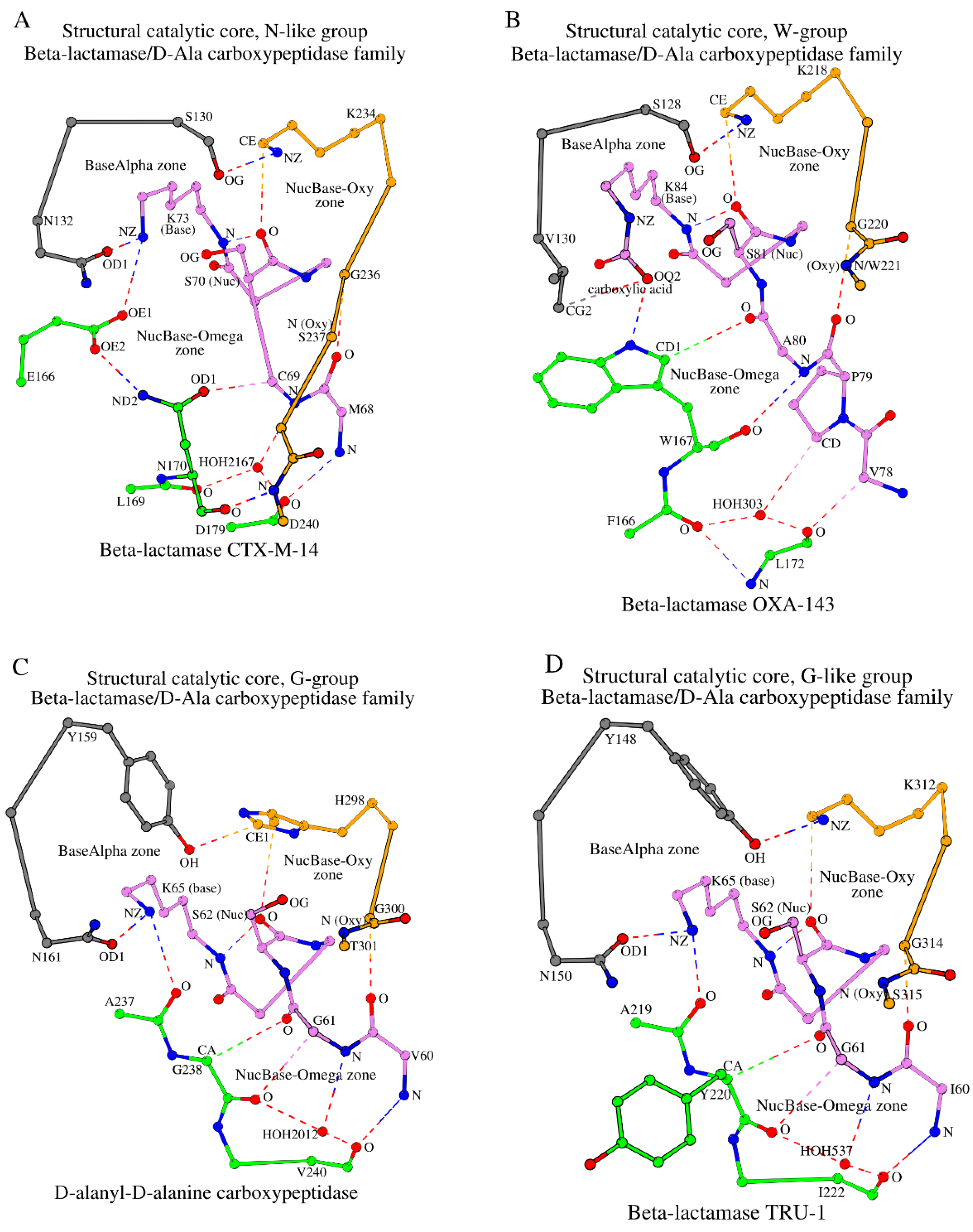
| 1 | 4UA6_A | 0.79 | β-lactamase CTX-M-14 | 68 MCSTSK 73 | 130 SDN 132 | E166, 169 LN••D 179 | 234 KTGSGD 240 | HOH2167 | [23] |
| SNS subgroup (5) | |||||||||
| 2 | 5F82_A | 0.96 | Carbapenemase GES-5 | 62 MGSTFK 67 | 125 SDN 127 | E161, 164 MS••D 174 | 229 KTGTCA 234 | HOH498 | [35] |
| SNG subgroup (2) | |||||||||
| 3 | 2QPN_A | 1.10 | Carbapenemase GES-1 | 62 MCSTFK 67 | 125 SDN 127 | E161, 164 MG••D 174 | 229 KTGTCA 234 | HOH338 | [36] |
| SSN subgroup (2) | |||||||||
| 4 | 7DDM_A | 1.20 | β-lactamase PenA39 | 68 FCSTFK 73 | 130 SDS 132 | E166, 169 LN••D 179 | 234 KTGTGD 240 | HOH476 | [37] |
| SGN subgroup (3) | |||||||||
| 5 | 5NJ2_A | 1.19 | β-lactamase BlaC | 68 FCSTFK 73 | 128 SDG 130 | E168, 171 LN••D 181 | 236 KTGTGD 242 | HOH547 | [38] |
| W-group (Class D) (45) | |||||||||
| SVW subgroup (36) | |||||||||
| 6 | 5IY2_B | 1.15 | β-lactamase OXA-143 | 78 VPASTFK 84 | 128 SAV 130 | 166 FW••L 172 | 218 KSGW 221 | HOH303 | [39] |
| SIW subgroup (5) | |||||||||
| 7 | 6W5E_A | 1.30 | β-lactamase BSU-2 | 98 TPQSTFK 104 | 149 SAI 151 | 187 FW••L 193 | 239 KTGT 242 | HOH450 | [40] |
| SLW subgroup (4) | |||||||||
| 8 | 6N1N_A | 1.60 | β-lactamase STD-1 | 62 LPASTFK 68 | 113 SAL 115 | 151 FW••L 157 | 203 KTGW 206 | HOH510 | [41] |
| W-group (5) | |||||||||
| SNW subgroup (4) | |||||||||
| 9 | 2IWB_A | 1.80 | Methicillin resistance mecR1 protein | 388 SPNSTYK 394 | 439 SVN 441 | 476 YW••L 482 | 528 KTGT 531 | HOH2115 | [42] |
| STW subgroup (1) | |||||||||
| 10 | 1NRF_A | 2.50 | Regulatory protein BlaR1 | 399 APASTYK 405 | 450 STT 452 | 487 YW••L 493 | 539 KTGT542 | HOH738 | [43] |
| G-group (23) | |||||||||
| YNG subgroup (4) | |||||||||
| 11 | 1YQS_A | 1.05 | D-alanyl-D-alanine carboxypeptidase | 60 VGSVTK 65 | 159 YSN 161 | 237 AG••V 240 | 298 HTGT 301 | HOH2012 | [44] |
| SNG subgroup (17) | |||||||||
| 12 | 5ZQA_A | 1.55 | Lmo2812 protein | 56 IASLSK 61 | 118 SAN 120 | 158 SG••A 167 | 222 KTGF 225 | HOH515 HOH418 | [45] |
| SCG subgroup (1) | |||||||||
| 13 | 1ES5_A | 1.40 | DD-transpeptidase | 33 TGSTTK 38 | 96 SGC 98 | 143 DG••N 150 | 213 KTGA 216 | HOH479 HOH347 | [46] |
| YSG subgroup (1) | |||||||||
| 14 | 1WYB_A | 1.80 | 6-aminohexanoate-dimer hydrolase | 110 LMSVSK 115 | 215 YCS 217 | 266 HG••V 269 | 342 GIGI 345 | CG/L109 CD1/L109 | [47] |
| G-like group (39) | |||||||||
| YNY subgroup (Class C) (33) | |||||||||
| 15 | 6FM6_A | 1.05 | β-lactamase TRU-1 | 60 IGSVSK 65 | 148 YSN 150 | 219 AY••I 222 | 312 KTGS 315 | HOH537 | [48] |
| YNA subgroup (1) | |||||||||
| 16 | 1EI5_A | 1.90 | D-aminopeptidase | 60 ICSVSK 65 | 153 YCN 153 | 225 DA••I 228 | 287 HGGA 290 | HOH531 | [49] |
| YLA subgroup (2) | |||||||||
| 17 | 1CI9_A | 1.80 | Esterase EstB | 73 LASVTK 78 | 181 YSL 183 | 274 GA••M 277 | 348 WGGV 351 | HOH1050 | [50] |
| YHQ subgroup (1) | |||||||||
| 18 | 4IVK_A | 1.80 | Carboxylesterase | 98 IYSMSK 103 | 218 YGH 220 | 295 GQ••M 298 | 381 WGGA 384 | HOH666 | [51] |
| YPH subgroup (1) | |||||||||
| 19 | 6KJC_A | 2.30 | Lovastatin esterase | 55 LASATK 60 | 170 YGP 172 | 252 GH••L 255 | 344 WGGG 347 | Y54 | [52] |
| SNM subgroup (1) | |||||||||
| 20 | 2BG1_A | 1.90 | Penicillin-binding protein 1b | 457 SPASTTK 463 | 516 SWN 518 | 555 PM••I 560 | 651 KTGT 654 | OG/S457 | [53] |
| Q-like group (Class A) (6) | |||||||||
| SNQ subgroup (5) | |||||||||
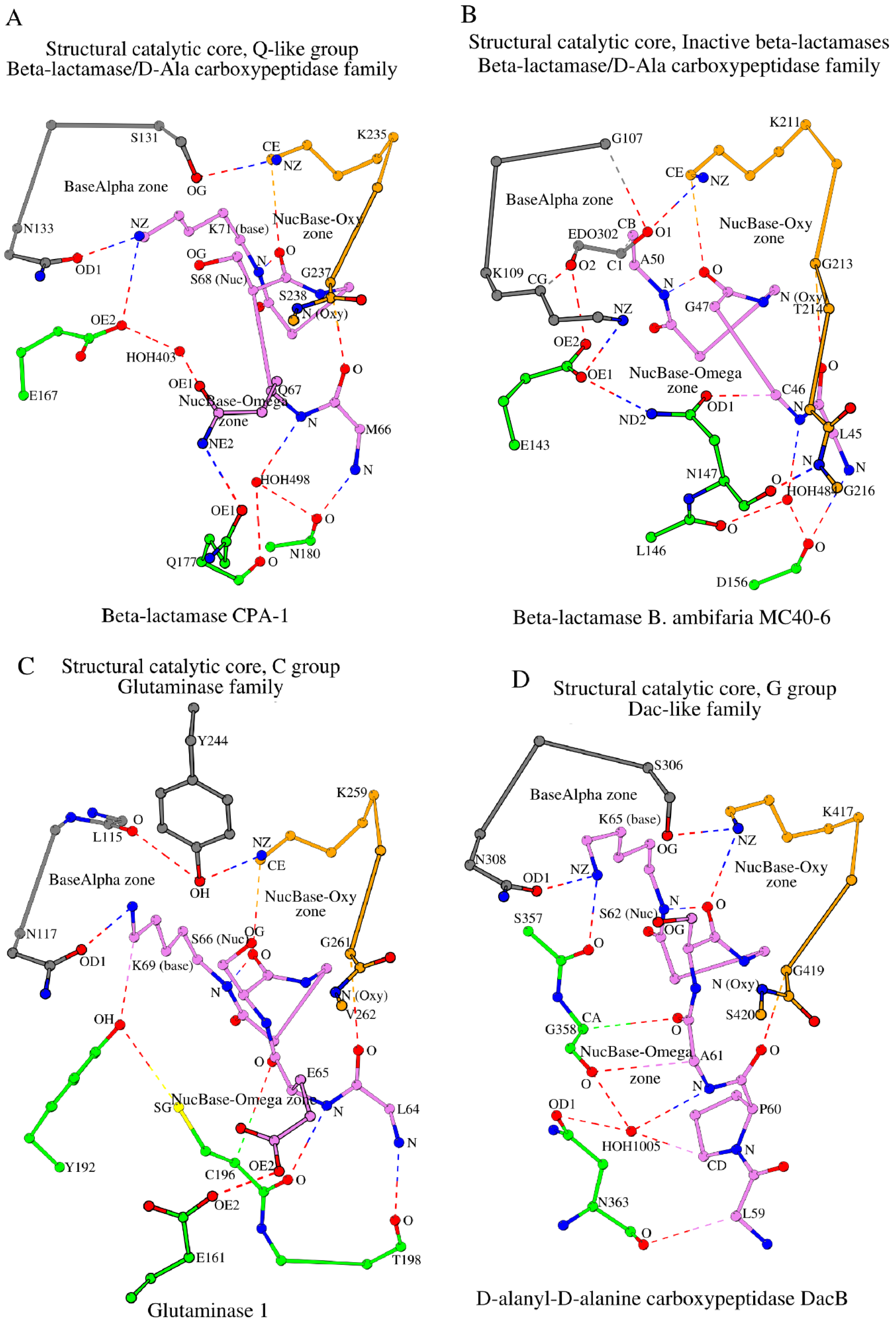
| 21 | 6V4W_A | 1.29 | β-lactamase CPA-1 | 66 MQSVFK 71 | 131 SDN 133 | E167, 177 –Q••N 180 | 235 KTGS 238 | HOH498 | [54] |
| SNT subgroup (1) | |||||||||
| 22 | 5TFQ_A | 1.07 | β-lactamase HGB-2 | 46 LLSVFK 51 | 112 SDN 114 | E148, 157 –T••N 160 | 215 KTGS 218 | HOH472 | [55] |
| Inactive β-lactamase group (2) | |||||||||
| GKN subgroup (2) | |||||||||
| 23 | 5IHV_A | 1.10 | β-lactamase B. ambifaria MC40-6 | 45 LCGTYA 50 | 107 GDK 109 | E143, 146 LN••D 156 | 211 KAGTGG 216 | HOH484 | [56] |
| Family: Glutaminase | |||||||||
| C-group (5) | |||||||||
| ONC subgroup (5) | |||||||||
| 24 | 1U60_A | 1.61 | Glutaminase 1 | 64 LESISK 69 | 115 LVN 117 | E161, Y192, 196 –C••T 198 | 259 KSGV 262 | N/A | [57] |
| Family: Dac-like | |||||||||
| G(Dac-like)-group (5) | |||||||||
| SNG subgroup (5) | |||||||||
| 25 | 2EX2_A | 1.55 | D-alanyl-D-alanine carboxypeptidase DacB | 59 LPASTQK 65 | 306 SDN 308 | 357 SG••N 363 | 417 KTGS 420 | HOH1005 | [58] |
2.3. The SCC in Proteins of the β-Lactamase/D-Ala Carboxypeptidase Family: Groups, Subgroups, and Classes
2.3.1. The N-like Group and Its Subgroups
2.3.2. The W-Group
2.3.3. The G-Group
2.3.4. The G-like Group
2.3.5. The Q-like Group
2.3.6. The Group of “Inactive” β-Lactamases, i.e., Those Unable to Perform Catalysis
2.4. Some Important Observations
- (1)
- Structural Catalytic Cores (SCCs) in N-like and W-groups have key differences. In the W-group, the appearance of key tryptophan (W) at position 170 instead of asparagine (N) at the equivalent position in the N-like group (see W167 in Figure 3B vs. N170 in Figure 3A), which is also reflected in the naming of the groups, leads to the two main structural changes in the protein SCC between the two groups. Due to the larger side-chain size and additional interactions between tryptophan and the NucBase structural segment (Figure 3B vs. Figure 3A): (a) the intermediate role of E166 and the equivalent residues, which are invariant in the N-like group (Table 1), disappears; but instead (b) the oxyanion (Oxy) segment in the W-group is shorter as it lacks the two last residues and their respective interactions;
- (2)
- SCCs in G- and G-like groups are closer to the W-group rather than to the N-like group. In G- and G-like groups, there is no side chain in glycine at the key position 170 (G238 in Figure 3C; position numbering according to 4UA6). As a consequence, in these groups it is not the glycine at position 170 that participates in the NucBase-Omega interactions but the amino acid at position 169 (A237 in Figure 3C), which, similarly to as seen in the W-group, removes the necessity of E166 mediation (Figure 3A) and shortens the oxyanion (Oxy) segment. In this respect, the SCC of G- and G-like groups is structurally closer to the SCC of the W-group than to the N-like group;
- (3)
- SCCs in the Q-like group are closer to the N-like group rather than the W-group. In the Q-like group, there is glutamine at position 170 (Q178 in 6V4W; Table 1; position numbering according to 4UA6), which is not seen in the N-like group, but, similarly to the N-like group, this glutamine at position 170 interacts with the residue at the position preceding the catalytic nucleophile (for reference, see the interaction between N170 and C69 in the N-like group, Figure 3A). As a result, in the Q-like group, the intermediate glutamate appears (E167 in 6V4W; Table 1), which fulfills the role of E166 in N-like proteins (Figure 3A);
- (4)
- NucBase-Omega zones are different, NucBase-Oxy zones are similar. In β-lactamase/D-Ala carboxypeptidase family proteins, the main structural differences between SCCs in different groups are situated in their respective NucBase-Omega zones. At the same time, the NucBase-Oxy zones of the W-, G-, G-like, and Q-like groups are similar.
2.5. SCC in Proteins of the Glutaminase Family (Example: Glutaminase 1)
2.6. SCC in Proteins of the Dac-like Family (Example: D-Alanyl-D-Alanine Carboxypeptidase DacB)
2.7. Overall Structural Comparison Between Groups of the β-Lactamase/Transpeptidase-like Superfamily Proteins and Molecular Function
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rawlings, N.D.; Barrett, A.J. MEROPS: The peptidase database. Nucleic Acids Res. 2000, 28, 323–325. [Google Scholar] [CrossRef] [PubMed]
- Chatonnet, A.; Perochon, M.; Velluet, E.; Marchot, P. The ESTHER database on alpha/beta hydrolase fold proteins—An overview of recent developments. Chem. Biol. Interact. 2023, 383, 110671. [Google Scholar] [CrossRef] [PubMed]
- Dodson, G.; Wlodawer, A. Catalytic triads and their relatives. Trends Biochem. Sci. 1998, 23, 347–352. [Google Scholar] [CrossRef]
- Polgár, L. The catalytic triad of serine peptidases. Cell. Mol. Life Sci. 2005, 62, 2161–2172. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Denesyuk, A.I.; Johnson, M.S.; Salo-Ahen, O.M.H.; Uversky, V.N.; Denessiouk, K. NBCZone: Universal three-dimensional construction of eleven amino acids near the catalytic nucleophile and base in the superfamily of (chymo)trypsin-like serine fold proteases. Int. J. Biol. Macromol. 2020, 153, 399–411. [Google Scholar] [CrossRef]
- Denesyuk, A.; Dimitriou, P.S.; Johnson, M.S.; Nakayama, T.; Denessiouk, K. The acid-base-nucleophile catalytic triad in ABH-fold enzymes is coordinated by a set of structural elements. PLoS ONE 2020, 15, e0229376. [Google Scholar] [CrossRef]
- Denessiouk, K.; Denesyuk, A.I.; Permyakov, S.E.; Permyakov, E.A.; Johnson, M.S.; Uversky, V.N. The active site of the SGNH hydrolase-like fold proteins: Nucleophile-oxyanion (Nuc-Oxy) and Acid-Base zones. Curr. Res. Struct. Biol. 2023, 7, 100123. [Google Scholar] [CrossRef]
- Denesyuk, A.I.; Denessiouk, K.; Johnson, M.S.; Uversky, V.N. Structural Catalytic Core in Subtilisin-like Proteins and Its Comparison to Trypsin-like Serine Proteases and Alpha/Beta-Hydrolases. Int. J. Mol. Sci. 2024, 25, 11858. [Google Scholar] [CrossRef]
- Kelly, J.A.; Kuzin, A.P. The refined crystallographic structure of a DD-peptidase penicillin-target enzyme at 1.6 Å resolution. J. Mol. Biol. 1995, 254, 223–236. [Google Scholar] [CrossRef]
- Silvaggi, N.R.; Anderson, J.W.; Brinsmade, S.R.; Pratt, R.F.; Kelly, J.A. The crystal structure of phosphonate-inhibited D-Ala-D-Ala peptidase reveals an analogue of a tetrahedral transition state. Biochemistry 2003, 42, 1199–1208. [Google Scholar] [CrossRef] [PubMed]
- Ekici, O.D.; Paetzel, M.; Dalbey, R.E. Unconventional serine proteases: Variations on the catalytic Ser/His/Asp triad configuration. Protein Sci. 2008, 17, 2023–2037. [Google Scholar] [CrossRef] [PubMed]
- Buller, A.R.; Townsend, C.A. Intrinsic evolutionary constraints on protease structure, enzyme acylation, and the identity of the catalytic triad. Proc. Natl. Acad. Sci. USA 2013, 110, E653–E661. [Google Scholar] [CrossRef] [PubMed]
- Andreeva, A.; Kulesha, E.; Gough, J.; Murzin, A.G. The SCOP database in 2020: Expanded classification of representative family and superfamily domains of known protein structures. Nucleic Acids Res. 2020, 48, D376–D382. [Google Scholar] [CrossRef]
- Leszczynski, J.F.; Rose, G.D. Loops in globular proteins: A novel category of secondary structure. Science 1986, 234, 849–855. [Google Scholar] [CrossRef]
- Goffin, C.; Ghuysen, J.M. Multimodular penicillin-binding proteins: An enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 1998, 62, 1079–1093. [Google Scholar] [CrossRef]
- Majiduddin, F.K.; Materon, I.C.; Palzkill, T.G. Molecular analysis of beta-lactamase structure and function. Int. J. Med. Microbiol. 2002, 292, 127–137. [Google Scholar] [CrossRef]
- Tooke, C.L.; Hinchliffe, P.; Bragginton, E.C.; Colenso, C.K.; Hirvonen, V.H.A.; Takebayashi, Y.; Spencer, J. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 2019, 431, 3472–3500. [Google Scholar] [CrossRef]
- Kaderabkova, N.; Bharathwaj, M.; Furniss, R.C.D.; Gonzalez, D.; Palmer, T.; Mavridou, D.A.I. The biogenesis of β-lactamase enzymes. Microbiology 2022, 168, 001217. [Google Scholar] [CrossRef]
- Banerjee, S.; Pieper, U.; Kapadia, G.; Pannell, L.K.; Herzberg, O. Role of the omega-loop in the activity, substrate specificity, and structure of class A beta-lactamase. Biochemistry 1998, 37, 3286–3296. [Google Scholar] [CrossRef]
- Szarecka, A.; Lesnock, K.R.; Ramirez-Mondragon, C.A.; Nicholas, H.B., Jr.; Wymore, T. The Class D beta-lactamase family: Residues governing the maintenance and diversity of function. Protein Eng. Des. Sel. 2011, 24, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Egorov, A.; Rubtsova, M.; Grigorenko, V.; Uporov, I.; Veselovsky, A. The Role of the Ω-Loop in Regulation of the Catalytic Activity of TEM-Type β-Lactamases. Biomolecules 2019, 9, 854. [Google Scholar] [CrossRef] [PubMed]
- Nichols, D.A.; Hargis, J.C.; Sanishvili, R.; Jaishankar, P.; Defrees, K.; Smith, E.W.; Wang, K.K.; Prati, F.; Renslo, A.R.; Woodcock, H.L.; et al. Ligand-Induced Proton Transfer and Low-Barrier HB Revealed by X-ray Crystallography. J. Am. Chem. Soc. 2015, 137, 8086–8095. [Google Scholar] [CrossRef]
- Ambler, R.P. The structure of beta-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1980, 289, 321–331. [Google Scholar] [CrossRef]
- Hall, B.G.; Barlow, M. Structure-based phylogenies of the serine beta-lactamases. J. Mol. Evol. 2003, 57, 255–260. [Google Scholar] [CrossRef]
- Naas, T.; Oueslati, S.; Bonnin, R.A.; Dabos, M.L.; Zavala, A.; Dortet, L.; Retailleau, P.; Iorga, B.I. Beta-lactamase database (BLDB)—Structure and function. J. Enzyme Inhib. Med. Chem. 2017, 32, 917–919. [Google Scholar] [CrossRef]
- Agarwal, V.; Yadav, T.C.; Tiwari, A.; Varadwaj, P. Detailed investigation of catalytically important residues of class A β-lactamase. J. Biomol. Struct. Dyn. 2023, 41, 2046–2073. [Google Scholar] [CrossRef]
- Sun, T.; Bethel, C.R.; Bonomo, R.A.; Knox, J.R. Inhibitor-resistant class A beta-lactamases: Consequences of the Ser130-to-Gly mutation seen in Apo and tazobactam structures of the SHV-1 variant. Biochemistry 2004, 43, 14111–14117. [Google Scholar] [CrossRef]
- Jacob, F.; Joris, B.; Lepage, S.; Dusart, J.; Frère, J.M. Role of the conserved amino acids of the ‘SDN’ loop (Ser130, Asp131 and Asn132) in a class A beta-lactamase studied by site-directed mutagenesis. Biochem. J. 1990, 271, 399–406. [Google Scholar] [CrossRef]
- Herzberg, O. Refined crystal structure of beta-lactamase from Staphylococcus aureus PC1 at 2.0 Å resolution. J. Mol. Biol. 1991, 217, 701–719. [Google Scholar] [CrossRef]
- Derewenda, Z.S. C-H Groups as Donors in Hydrogen Bonds: A Historical Overview and Occurrence in Proteins and Nucleic Acids. Int. J. Mol. Sci. 2023, 24, 13165. [Google Scholar] [CrossRef]
- Derewenda, Z.S.; Derewenda, U.; Kobos, P.M. (His)C epsilon-H…O=C < hydrogen bond in the active sites of serine hydrolases. J. Mol. Biol. 1994, 241, 83–93. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Hutchinson, E.G.; Michie, A.D.; Wallace, A.C.; Jones, M.L.; Thornton, J.M. PDBsum: A Web-based database of summaries and analyses of all PDB structures. Trends Biochem. Sci. 1997, 22, 488–490. [Google Scholar] [CrossRef]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995, 8, 127–134. [Google Scholar] [CrossRef]
- Smith, C.A.; Nossoni, Z.; Toth, M.; Stewart, N.K.; Frase, H.; Vakulenko, S.B. Role of the Conserved Disulfide Bridge in Class A Carbapenemases. J. Biol. Chem. 2016, 291, 22196–22206. [Google Scholar] [CrossRef]
- Smith, C.A.; Caccamo, M.; Kantardjieff, K.A.; Vakulenko, S. Structure of GES-1 at atomic resolution: Insights into the evolution of carbapenamase activity in the class A extended-spectrum beta-lactamases. Acta Crystallogr. D Biol. Crystallogr. 2007, 63 Pt 9, 982–992. [Google Scholar] [CrossRef]
- Nukaga, M.; Becka, S.A.; Zeiser, E.T.; Hoshino, Y.; LiPuma, J.J.; Papp-Wallace, K.M. Frameshift Mutations in Genes Encoding PBP3 and PBP4 Trigger an Unusual, Extreme Beta-Lactam Resistance Phenotype in Burkholderia multivorans. 2021. Available online: https://www.wwpdb.org/pdb?id=pdb_00007ddm (accessed on 10 October 2024).
- Elings, W.; Tassoni, R.; van der Schoot, S.A.; Luu, W.; Kynast, J.P.; Dai, L.; Blok, A.J.; Timmer, M.; Florea, B.I.; Pannu, N.S.; et al. Phosphate Promotes the Recovery of Mycobacterium tuberculosis β-Lactamase from Clavulanic Acid Inhibition. Biochemistry 2017, 56, 6257–6267. [Google Scholar] [CrossRef]
- Toth, M.; Smith, C.A.; Antunes, N.T.; Stewart, N.K.; Maltz, L.; Vakulenko, S.B. The role of conserved surface hydrophobic residues in the carbapenemase activity of the class D β-lactamases. Acta Crystallogr. D Struct. Biol. 2017, 73 Pt 8, 692–701. [Google Scholar] [CrossRef]
- Stewart, N.K.; Bhattacharya, M.; Toth, M.; Smith, C.A.; Vakulenko, S.B. A surface loop modulates activity of the Bacillus class D β-lactamases. J. Struct. Biol. 2020, 211, 107544. [Google Scholar] [CrossRef]
- Michalska, K.; Tesar, C.; Endres, M.; Joachimiak, A.; Satchell, K.J. Crystal Structure of Class D Beta-Lactamase from Sebaldella termitidis ATCC 33386. 2018. Available online: https://www.wwpdb.org/pdb?id=pdb_00006n1n (accessed on 10 October 2024).
- Marrero, A.; Mallorquí-Fernández, G.; Guevara, T.; García-Castellanos, R.; Gomis-Rüth, F.X. Unbound and acylated structures of the MecR1 extracellular antibiotic-sensor domain provide insights into the signal-transduction system that triggers methicillin resistance. J. Mol. Biol. 2006, 361, 506–521. [Google Scholar] [CrossRef]
- Kerff, F.; Charlier, P.; Colombo, M.L.; Sauvage, E.; Brans, A.; Frère, J.M.; Joris, B.; Fonzé, E. Crystal structure of the sensor domain of the BlaR penicillin receptor from Bacillus licheniformis. Biochemistry 2003, 42, 12835–12843. [Google Scholar] [CrossRef] [PubMed]
- Llinás, A.; Ahmed, N.; Cordaro, M.; Laws, A.P.; Frère, J.M.; Delmarcelle, M.; Silvaggi, N.R.; Kelly, J.A.; Page, M.I. Inactivation of bacterial DD-peptidase by beta-sultams. Biochemistry 2005, 44, 7738–7746. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.H.; Cha, H.J.; Kim, Y.G. Crystal Structures of Penicillin-Binding Protein D2 from Listeria monocytogenes and Structural Basis for Antibiotic Specificity. Antimicrob. Agents Chemother. 2018, 62, e00796-18. [Google Scholar] [CrossRef]
- Fonze, E.; Charlier, P. S216A Mutant of Streptomyces K15 DD-Transpeptidase. 2000. Available online: https://www.wwpdb.org/pdb?id=pdb_00001es5 (accessed on 10 October 2024).
- Negoro, S.; Ohki, T.; Shibata, N.; Mizuno, N.; Wakitani, Y.; Tsurukame, J.; Matsumoto, K.; Kawamoto, I.; Takeo, M.; Higuchi, Y. Structure of 6-Aminohexanoate-Dimer Hydrolase. 2005. Available online: https://www.wwpdb.org/pdb?id=pdb_00001wyb (accessed on 10 October 2024).
- Pozzi, C.; Di Pisa, F.; De Luca, F.; Benvenuti, M.; Docquier, J.D.; Mangani, S. Atomic-Resolution Structure of a Class C β-Lactamase and Its Complex with Avibactam. ChemMedChem 2018, 13, 1437–1446. [Google Scholar] [CrossRef]
- Bompard-Gilles, C.; Remaut, H.; Villeret, V.; Prangé, T.; Fanuel, L.; Delmarcelle, M.; Joris, B.; Frère, J.; Van Beeumen, J. Crystal structure of a D-aminopeptidase from Ochrobactrum anthropi, a new member of the ‘penicillin-recognizing enzyme’ family. Structure 2000, 8, 971–980. [Google Scholar] [CrossRef]
- Wagner, U.G.; Petersen, E.I.; Schwab, H.; Kratky, C. EstB from Burkholderia gladioli: A novel esterase with a beta-lactamase fold reveals steric factors to discriminate between esterolytic and beta-lactam cleaving activity. Protein Sci. 2002, 11, 467–478. [Google Scholar] [CrossRef]
- Cha, S.S.; An, Y.J.; Jeong, C.S.; Kim, M.K.; Jeon, J.H.; Lee, C.M.; Lee, H.S.; Kang, S.G.; Lee, J.H. Structural basis for the β-lactamase activity of EstU1, a family VIII carboxylesterase. Proteins 2013, 81, 2045–2051. [Google Scholar] [CrossRef]
- Liang, Y.; Lu, X. Structural insights into the catalytic mechanism of lovastatin hydrolase. J. Biol. Chem. 2020, 295, 1047–1055. [Google Scholar] [CrossRef]
- Macheboeuf, P.; Di Guilmi, A.M.; Job, V.; Vernet, T.; Dideberg, O.; Dessen, A. Active site restructuring regulates ligand recognition in class A penicillin-binding proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 577–582. [Google Scholar] [CrossRef]
- Tan, K.; Welk, L.; Endres, M.; Joachimiak, A. The Crystal Structure of a Beta-Lactamase from Chitinophaga pinensis DSM 2588. 2019. Available online: https://www.wwpdb.org/pdb?id=pdb_00006v4w (accessed on 10 October 2024).
- Nocek, B.; Hatzos-Skintges, C.; Babnigg, G.; Joachimiak, A. Crystal Structure of a Representative of Class A Beta-Lactamase from Bacteroides cellulosilyticus DSM 14838. 2016. Available online: https://www.wwpdb.org/pdb?id=pdb_00005tfq (accessed on 10 October 2024).
- Mayclin, S.J.; Abendroth, J.; Lorimer, D.D.; Edwards, T.E. Crystal Structure of a Beta-Lactamase from Burkholderia ambifaria. 2016. Available online: https://www.wwpdb.org/pdb?id=pdb_00005ihv (accessed on 10 October 2024).
- Brown, G.; Singer, A.; Proudfoot, M.; Skarina, T.; Kim, Y.; Chang, C.; Dementieva, I.; Kuznetsova, E.; Gonzalez, C.F.; Joachimiak, A.; et al. Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis. Biochemistry 2008, 47, 5724–5735. [Google Scholar] [CrossRef] [PubMed]
- Kishida, H.; Unzai, S.; Roper, D.I.; Lloyd, A.; Park, S.Y.; Tame, J.R. Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics. Biochemistry 2006, 45, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Clementel, D.; Del Conte, A.; Monzon, A.M.; Camagni, G.F.; Minervini, G.; Piovesan, D.; Tosatto, S.C.E. RING 3.0: Fast generation of probabilistic residue interaction networks from structural ensembles. Nucleic Acids Res. 2022, 50, W651–W656. [Google Scholar] [CrossRef] [PubMed]
- Kraulis, P.J. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 1991, 24, 946–950. [Google Scholar] [CrossRef]
- Holm, L.; Laiho, A.; Törönen, P.; Salgado, M. DALI shines a light on remote homologs: One hundred discoveries. Protein Sci. 2023, 32, e4519. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2025. Nucleic Acids Res. 2025, 53, D609–D617. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Denesyuk, A.I.; Denessiouk, K.; Johnson, M.S.; Uversky, V.N. Alpha and Omega Classification of β-Lactamase/Transpeptidase-like Superfamily Proteins Based on the Comparison of Their Structural Catalytic Cores. Molecules 2025, 30, 2019. https://doi.org/10.3390/molecules30092019
Denesyuk AI, Denessiouk K, Johnson MS, Uversky VN. Alpha and Omega Classification of β-Lactamase/Transpeptidase-like Superfamily Proteins Based on the Comparison of Their Structural Catalytic Cores. Molecules. 2025; 30(9):2019. https://doi.org/10.3390/molecules30092019
Chicago/Turabian StyleDenesyuk, Alexander I., Konstantin Denessiouk, Mark S. Johnson, and Vladimir N. Uversky. 2025. "Alpha and Omega Classification of β-Lactamase/Transpeptidase-like Superfamily Proteins Based on the Comparison of Their Structural Catalytic Cores" Molecules 30, no. 9: 2019. https://doi.org/10.3390/molecules30092019
APA StyleDenesyuk, A. I., Denessiouk, K., Johnson, M. S., & Uversky, V. N. (2025). Alpha and Omega Classification of β-Lactamase/Transpeptidase-like Superfamily Proteins Based on the Comparison of Their Structural Catalytic Cores. Molecules, 30(9), 2019. https://doi.org/10.3390/molecules30092019







