Conditional Control of CRISPR/Cas9 Function by Chemically Modified Oligonucleotides
Abstract
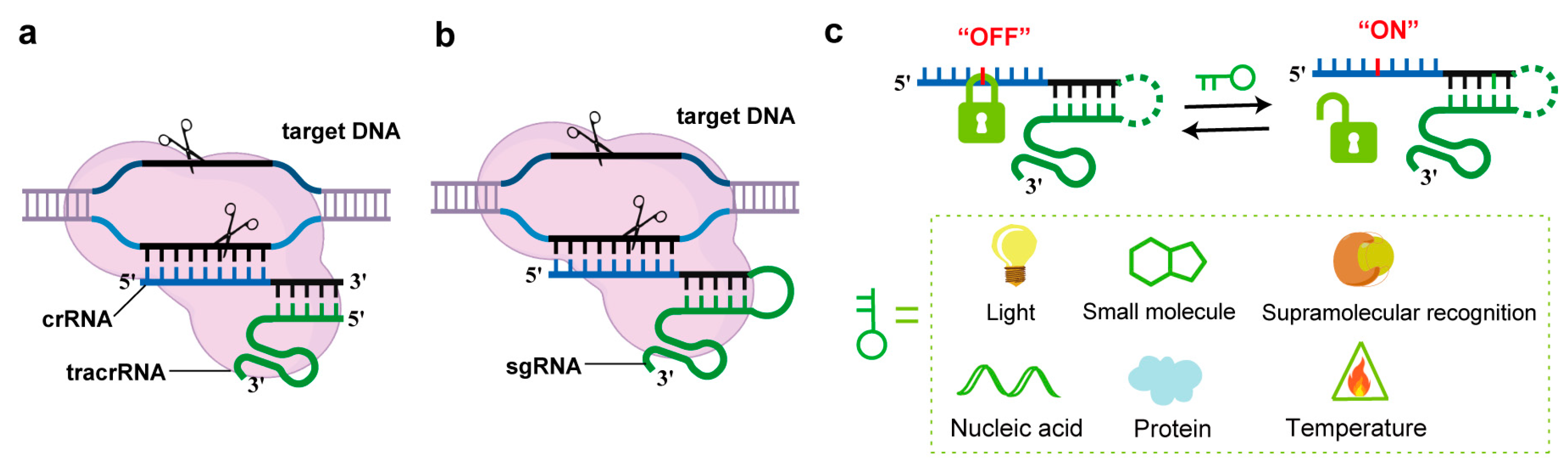
1. Introduction
2. Strategies for gRNA Regulation Based on Chemical Modifications of Oligonucleotides
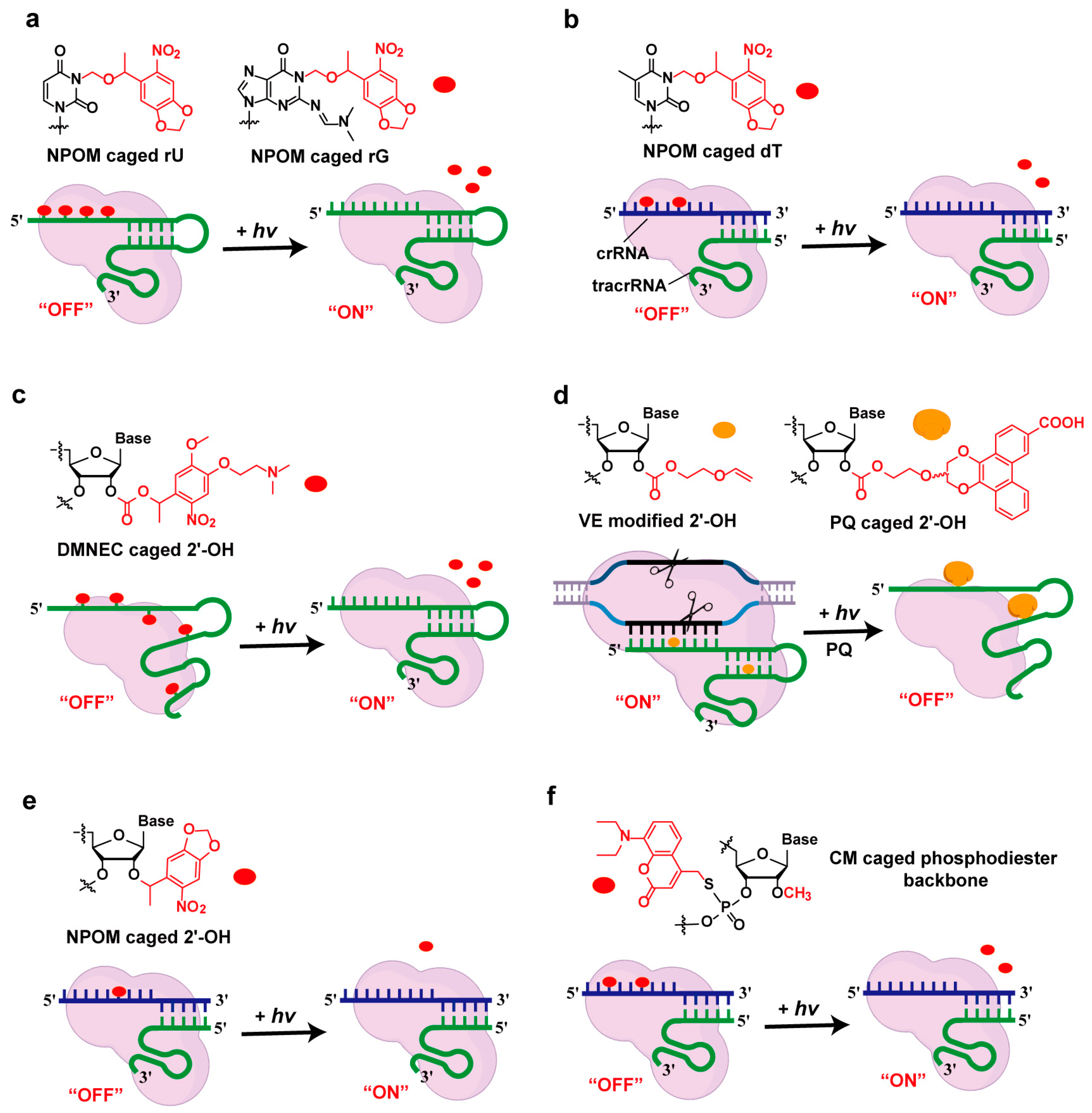
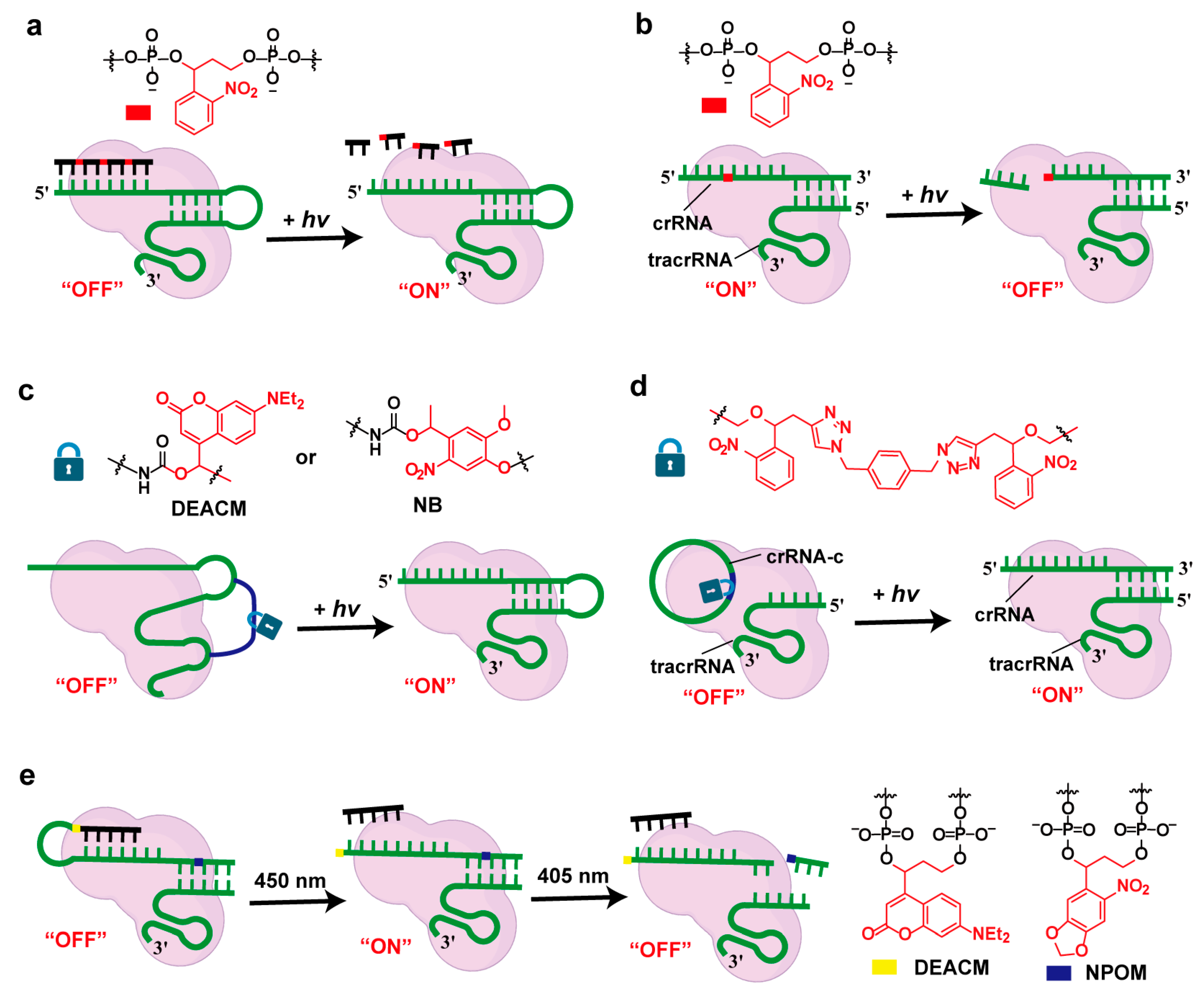
2.1. Light-Controlled Strategies
2.1.1. Photocaging Strategies
2.1.2. Photocleavage Strategies
2.2. Small-Molecule-Responsive Strategies
2.3. Supramolecular Recognition Strategies
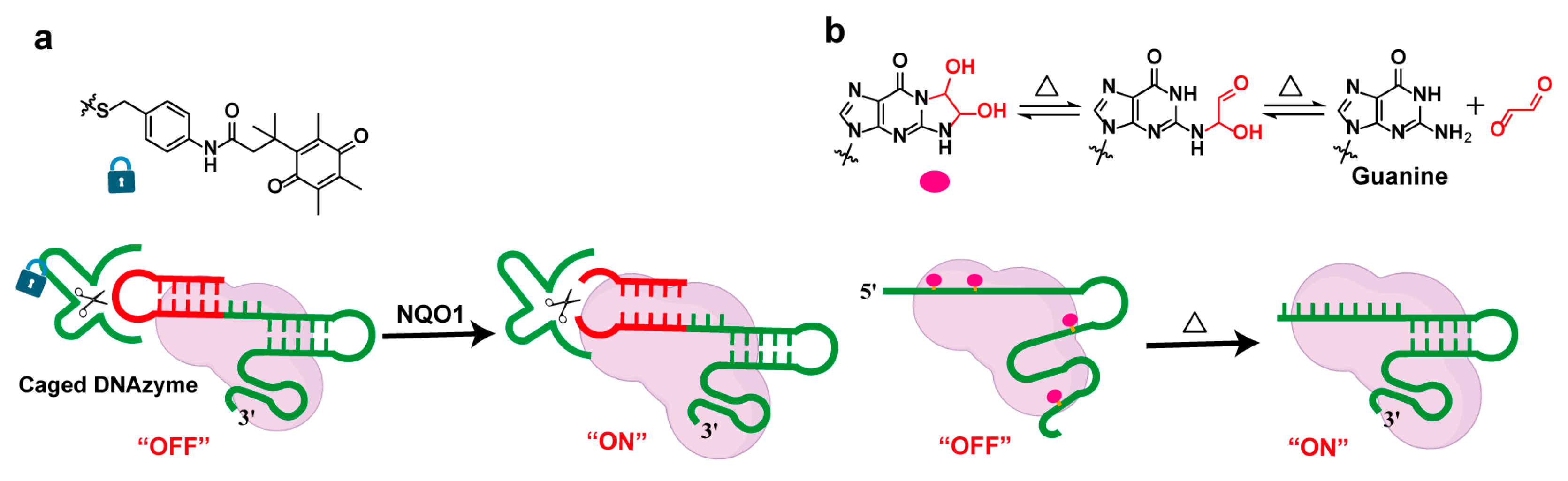
2.4. Other Condition-Responsive Strategies
2.4.1. Protein- or Oligonucleotide-Mediated Regulation Strategies
2.4.2. Temperature-Responsive Strategies
3. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Wiedenheft, B.; Sternberg, S.H.; Doudna, J.A. RNA-guided genetic silencing systems in bacteria and archaea. Nature 2012, 482, 331–338. [Google Scholar] [CrossRef]
- Mali, P.; Esvelt, K.M.; Church, G.M. Cas9 as a versatile tool for engineering biology. Nat. Methods 2013, 10, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Nishimasu, H.; Ran, F.A.; Hsu, P.D.; Konermann, S.; Shehata, S.I.; Dohmae, N.; Ishitani, R.; Zhang, F.; Nureki, O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 2014, 156, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef]
- Wang, F.; Wang, L.; Zou, X.; Duan, S.; Li, Z.; Deng, Z.; Luo, J.; Lee, S.Y.; Chen, S. Advances in CRISPR-Cas systems for RNA targeting, tracking and editing. Biotechnol. Adv. 2019, 37, 708–729. [Google Scholar] [CrossRef]
- Stellos, K.; Musunuru, K. Challenges and advances of CRISPR-Cas9 genome editing in therapeutics. Cardiovasc. Res. 2019, 115, e12–e14. [Google Scholar] [CrossRef]
- Barber, H.M.; Pater, A.A.; Gagnon, K.T.; Damha, M.J.; O’Reilly, D. Chemical engineering of CRISPR–Cas systems for therapeutic application. Nat. Rev. Drug Discov. 2024, 24, 209–230. [Google Scholar] [CrossRef]
- Yin, H.; Song, C.Q.; Suresh, S.; Wu, Q.; Walsh, S.; Rhym, L.H.; Mintzer, E.; Bolukbasi, M.F.; Zhu, L.J.; Kauffman, K.; et al. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol. 2017, 35, 1179–1187. [Google Scholar] [CrossRef]
- Hendel, A.; Bak, R.O.; Clark, J.T.; Kennedy, A.B.; Ryan, D.E.; Roy, S.; Steinfeld, I.; Lunstad, B.D.; Kaiser, R.J.; Wilkens, A.B.; et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015, 33, 985–989. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Zhu, J.; Su, X.; Lin, X.; Xu, L.; Xing, X. Employing pH-responsive RNA triplex to control CRISPR/Cas9-mediated gene manipulation in mammalian cells. Chin. Chem. Lett. 2024, 35, 109427. [Google Scholar] [CrossRef]
- Pelea, O.; Fulga, T.A.; Sauka-Spengler, T. RNA-responsive gRNAs for controlling CRISPR activity: Current advances, future directions, and potential applications. CRISPR J. 2022, 5, 642–659. [Google Scholar] [CrossRef] [PubMed]
- Allemailem, K.S.; Almatroudi, A.; Rahmani, A.H.; Alrumaihi, F.; Alradhi, A.E.; Alsubaiyel, A.M.; Algahtani, M.; Almousa, R.M.; Mahzari, A.; Sindi, A.A.A.; et al. Recent updates of the CRISPR/Cas9 genome editing system: Novel approaches to regulate its spatiotemporal control by genetic and physicochemical strategies. Int. J. Nanomed. 2024, 19, 5335–5363. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Y.; Han, L.; Che, Q.; Tan, J.; Zou, P.; Chen, Y. Photo-modulation of gene-editing enzymes CRISPR/Cas9 with bifunctional small-molecule ligands. Chin. J. Chem. 2023, 41, 3639–3644. [Google Scholar] [CrossRef]
- Prokhorova, D.V.; Kupryushkin, M.S.; Zhukov, S.A.; Zharkov, T.D.; Dovydenko, I.S.; Yakovleva, K.I.; Pereverzev, I.M.; Matveeva, A.M.; Pyshnyi, D.V.; Stepanov, G.A. Effect of the phosphoryl guanidine modification in chimeric DNA-RNA crRNAs on the activity of the CRISPR-Cas9 system in vitro. ACS Chem. Biol. 2024, 19, 1311–1319. [Google Scholar] [CrossRef]
- Hu, L.F.; Li, Y.X.; Wang, J.Z.; Zhao, Y.T.; Wang, Y. Controlling CRISPR-Cas9 by guide RNA engineering. Wiley Interdiscip. Rev. RNA 2023, 14, e1731. [Google Scholar] [CrossRef]
- Brown, W.; Zhou, W.; Deiters, A. Regulating CRISPR/Cas9 function through conditional guide RNA control. ChemBioChem 2021, 22, 63–72. [Google Scholar] [CrossRef]
- Cai, W.; Wang, M. Engineering nucleic acid chemistry for precise and controllable CRISPR/Cas9 genome editing. Sci. Bull. 2019, 64, 1841–1849. [Google Scholar] [CrossRef]
- Wang, X.; Wen, S.; Wu, Z.; Jiang, J.H. Orthogonal control of nucleic acid function via chemical caging-decaging strategies. ChemBioChem 2024, 25, e202400516. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Q.; Wang, J.; Tang, X. Chemical modification and transformation strategies of guide RNAs in CRISPR-Cas9 gene editing systems. ChemPlusChem 2021, 86, 587–600. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Deiters, A. Conditional control of CRISPR/Cas9 function. Angew. Chem. Int. Ed. 2016, 55, 5394–5399. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, C.; Zhang, J.; Lee, J.H.; Jiao, J.; Cheng, D.; Liu, L.; Kim, H.W.; Tao, Y.; Li, M. Spatiotemporal control of CRISPR/Cas9 gene editing. Signal Transduct. Target. Ther. 2021, 6, 238. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, Y.; Qiao, J.; Liu, Y. Recent advances in spatiotemporal control of the CRISPR/Cas9 system. Colloids Surf. B Biointerfaces 2025, 248, 114474. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Liu, L.; Jin, S.; Tota, E.; Li, Z.; Piao, X.; Zhang, X.; Fu, X.D.; Devaraj, N.K. Site-specific and enzymatic cross-linking of sgRNA enables wavelength-selectable photoactivated control of CRISPR gene editing. J. Am. Chem. Soc. 2022, 144, 4487–4495. [Google Scholar] [CrossRef]
- Sun, Y.J.; Chen, W.D.; Liu, J.; Li, J.J.; Zhang, Y.; Cai, W.Q.; Liu, L.; Tang, X.J.; Hou, J.; Wang, M.; et al. A conformational restriction strategy for the control of CRISPR/Cas gene editing with photoactivatable guide RNAs. Angew. Chem. Int. Ed. 2023, 62, e202212413. [Google Scholar] [CrossRef]
- Lei, H.; Zeng, T.; Ye, X.; Fan, R.; Xiong, W.; Tian, T.; Zhou, X. Chemical control of CRISPR gene editing via conditional diacylation crosslinking of guide RNAs. Adv. Sci. 2023, 10, e2206433. [Google Scholar] [CrossRef]
- Qi, Q.; Zhang, Y.; Xiong, W.; Liu, X.; Cui, S.; Ye, X.; Zhang, K.; Tian, T.; Xiang, Z. Norbornene-tetrazine ligation chemistry for controlling RNA-guided CRISPR systems. Chem. Sci. 2022, 13, 12577–12587. [Google Scholar] [CrossRef]
- Xiong, W.; Liu, X.; Qi, Q.; Ji, H.; Liu, F.; Zhong, C.; Liu, S.; Tian, T.; Zhou, X. Supramolecular CRISPR-OFF switches with host-guest chemistry. Nucleic Acids Res. 2022, 50, 1241–1255. [Google Scholar] [CrossRef]
- Shen, W.; Xiong, W.; Qi, Q.; Liu, X.; Xie, Z.; Zhang, Y.; Hou, J.; Tian, T.; Zhou, X. Regulating CRISPR/Cas9 using streptavidin-biotin interactions. Chin. J. Chem. 2024, 42, 1387–1393. [Google Scholar] [CrossRef]
- Jain, P.K.; Ramanan, V.; Schepers, A.G.; Dalvie, N.S.; Panda, A.; Fleming, H.E.; Bhatia, S.N. Development of light-activated CRISPR using guide RNAs with photocleavable protectors. Angew. Chem. Int. Ed. 2016, 55, 12440–12444. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wang, Z.; Jing, N.; Chen, W.; Tang, X. Photomodulation of caged RNA oligonucleotide functions in living systems. ChemPhotoChem 2020, 5, 12–21. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, C.-Q.; Zhang, Q.-L.; Shao, M.; Liu, Y.; Wang, L.-L.; Wang, Z.-Y.; Du, J.; Xu, L. Switching on/off of guide RNA by photoinduced strand displacement for functional control of CRISPR/Cas9. CCS Chem. 2024, 6, 1338–1351. [Google Scholar] [CrossRef]
- Hu, M.; Liu, R.; Qiu, Z.; Cao, F.; Tian, T.; Lu, Y.; Jiang, Y.; Zhou, X. Light-start CRISPR-Cas12a reaction with caged crRNA enables rapid and sensitive nucleic acid detection. Angew. Chem. Int. Ed. 2023, 62, e202300663. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Q.-L.; Liu, Y.; Wang, L.-L.; Wu, C.-Q.; Shao, M.; Xing, X.; Du, J.; Xu, L. Photo-controllable multiple and orthogonal regulation of gene expression by chemically modified oligonucleotides. Sci. China Chem. 2024, 68, 694–704. [Google Scholar] [CrossRef]
- Chen, W.D.; Liu, L.; Cheng, L. Functionally tunable star-shaped multivalent crRNAs for photocontrol CRISPR/Cas editing. Angew. Chem. Int. Ed. 2025, 14, e202506527. [Google Scholar]
- Deng, H.; Xu, H.; Wang, Y.; Jia, R.; Ma, X.; Feng, Y.; Chen, H. G-quadruplex-based CRISPR photoswitch for spatiotemporal control of genomic modulation. Nucleic Acids Res. 2023, 51, 4064–4077. [Google Scholar] [CrossRef]
- Yang, Z.; Guo, B.; Hu, C.; Tang, C.; Shen, Y.; Zhang, B.; Wang, F. Photoactivatable CRISPR/Cas9 lateral flow strip platform for one-pot rapid detection of squamous cell carcinoma antigen DNA in blood. Sens. Actuators B Chem. 2025, 434, 137607. [Google Scholar] [CrossRef]
- Taemaitree, L.; Brown, T. Shining light on CRISPR gene editing. ACS Cent. Sci. 2020, 6, 616–618. [Google Scholar] [CrossRef]
- Zhou, W.; Brown, W.; Bardhan, A.; Delaney, M.; Ilk, A.S.; Rauen, R.R.; Kahn, S.I.; Tsang, M.; Deiters, A. Spatiotemporal control of CRISPR/Cas9 function in cells and zebrafish using light-activated guide RNA. Angew. Chem. Int. Ed. 2020, 59, 8998–9003. [Google Scholar] [CrossRef]
- Moroz-Omori, E.V.; Satyapertiwi, D.; Ramel, M.C.; Hogset, H.; Sunyovszki, I.K.; Liu, Z.; Wojciechowski, J.P.; Zhang, Y.; Grigsby, C.L.; Brito, L.; et al. Photoswitchable gRNAs for spatiotemporally controlled CRISPR-Cas-based genomic regulation. ACS Cent. Sci. 2020, 6, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zou, R.S.; He, S.; Nihongaki, Y.; Li, X.; Razavi, S.; Wu, B.; Ha, T. Very fast CRISPR on demand. Science 2020, 368, 1265–1269. [Google Scholar] [CrossRef] [PubMed]
- Velema, W.A.; Kietrys, A.M.; Kool, E.T. RNA control by photoreversible acylation. J. Am. Chem. Soc. 2018, 140, 3491–3495. [Google Scholar] [CrossRef]
- Kadina, A.; Kietrys, A.M.; Kool, E.T. RNA cloaking by reversible acylation. Angew. Chem. Int. Ed. 2018, 57, 3059–3063. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wei, L.; Wang, J.Q.; Ji, H.; Xiong, W.; Liu, J.; Yin, P.; Tian, T.; Zhou, X. Light-driven activation of RNA-guided nucleic acid cleavage. ACS Chem. Biol. 2020, 15, 1455–1463. [Google Scholar] [CrossRef]
- Qi, Q.; Liu, X.; Xiong, W.; Zhang, K.; Shen, W.; Zhang, Y.; Xu, X.; Zhong, C.; Zhang, Y.; Tian, T.; et al. Reducing CRISPR-Cas9 off-target effects by optically controlled chemical modifications of guide RNA. Cell Chem. Biol. 2024, 31, 1839–1851. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Y.; Xie, F.; Lin, J.; Xu, L. Photocontrol of CRISPR/Cas9 function by site-specific chemical modification of guide RNA. Chem. Sci. 2020, 11, 11478–11484. [Google Scholar] [CrossRef]
- Gu, C.; Xiao, L.; Shang, J.; Xu, X.; He, L.; Xiang, Y. Chemical synthesis of stimuli-responsive guide RNA for conditional control of CRISPR-Cas9 gene editing. Chem. Sci. 2021, 12, 9934–9945. [Google Scholar] [CrossRef]
- Akhmetova, E.A.; Vokhtantsev, I.P.; Meschaninova, M.I.; Vorobyeva, M.A.; Zharkov, D.O.; Novopashina, D.S. Photocleavable guide RNA for photocontrolled CRISPR/Cas9 system. Russ. J. Bioorganic Chem. 2024, 50, 1314–1324. [Google Scholar] [CrossRef]
- Sakovina, L.; Vokhtantsev, I.; Akhmetova, E.; Vorobyeva, M.; Vorobjev, P.; Zharkov, D.O.; Novopashina, D. Photocleavable guide crRNAs for a light-controllable CRISPR/Cas9 system. Int. J. Mol. Sci. 2024, 25, 12392. [Google Scholar] [CrossRef]
- Liu, Q.; Deiters, A. Optochemical control of deoxyoligonucleotide function via a nucleobase-caging approach. Accounts Chem. Res. 2014, 47, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Zou, R.S.; Liu, Y.; Wu, B.; Ha, T. Cas9 deactivation with photocleavable guide RNAs. Mol. Cell 2021, 81, 1553–1565. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ling, X.; Su, X.; Zhang, S.; Wang, J.; Zhang, P.; Feng, W.; Zhu, Y.Y.; Liu, T.; Tang, X. Optical control of a CRISPR/Cas9 system for gene editing by using photolabile crRNA. Angew. Chem. Int. Ed. 2020, 59, 20895–20899. [Google Scholar] [CrossRef]
- Liu, X.; Xiong, W.; Qi, Q.; Zhang, Y.; Ji, H.; Cui, S.; An, J.; Sun, X.; Yin, H.; Tian, T.; et al. Rational guide RNA engineering for small-molecule control of CRISPR/Cas9 and gene editing. Nucleic Acids Res. 2022, 50, 4769–4783. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Lin, J.; Xu, L. Theophylline-induced synergic activation of guide RNA to control CRISPR/Cas9 function. Chem. Commun. 2021, 57, 5418–5421. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.; Xiong, W.; Li, M.; Qi, Q.; Liu, X.; Wang, S.; Tian, T.; Zhou, X. Enhanced control of RNA modification and CRISPR-Cas activity through redox-triggered disulfide cleavage. Bioorganic Med. Chem. 2024, 112, 117878. [Google Scholar] [CrossRef]
- Ji, H.; Xiong, W.; Zhang, K.; Tian, T.; Zhou, X. Hydrogen peroxide-triggered chemical strategy for controlling CRISPR systems. Chem. Asian J. 2022, 17, e202200214. [Google Scholar] [CrossRef]
- Habibian, M.; McKinlay, C.; Blake, T.R.; Kietrys, A.M.; Waymouth, R.M.; Wender, P.A.; Kool, E.T. Reversible RNA acylation for control of CRISPR-Cas9 gene editing. Chem. Sci. 2019, 11, 1011–1016. [Google Scholar] [CrossRef]
- Wang, S.R.; Wu, L.Y.; Huang, H.Y.; Xiong, W.; Liu, J.; Wei, L.; Yin, P.; Tian, T.; Zhou, X. Conditional control of RNA-guided nucleic acid cleavage and gene editing. Nat. Commun. 2020, 11, 91. [Google Scholar] [CrossRef]
- Zeng, T.; Wu, Q.; Liu, Y.; Qi, Q.; Shen, W.; Gu, W.; Zhang, Y.; Xiong, W.; Xie, Z.; Qi, X.; et al. Unraveling the cleavage reaction of hydroxylamines with cyclopropenones considering biocompatibility. J. Am. Chem. Soc. 2024, 146, 35077–35089. [Google Scholar] [CrossRef]
- Velema, W.A.; Park, H.S.; Kadina, A.; Orbai, L.; Kool, E.T. Trapping transient RNA complexes by chemically reversible acylation. Angew. Chem. Int. Ed. 2020, 59, 22017–22022. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.R.; Huang, H.Y.; Liu, J.; Wei, L.; Wu, L.Y.; Xiong, W.; Yin, P.; Tian, T.; Zhou, X. The manipulation of RNA-guided nucleic acid cleavage with ninhydrin chemistry. Adv. Sci. 2020, 7, 1903770. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, B.L.; Guo, Z.; Bernardes, G.J.L. Inverse electron demand Diels-Alder reactions in chemical biology. Chem. Soc. Rev. 2017, 46, 4895–4950. [Google Scholar] [CrossRef] [PubMed]
- Hansell, C.F.; Espeel, P.; Stamenovic, M.M.; Barker, I.A.; Dove, A.P.; Du Prez, F.E.; O’Reilly, R.K. Additive-free clicking for polymer functionalization and coupling by tetrazine-norbornene chemistry. J. Am. Chem. Soc. 2011, 133, 13828–13831. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, Y.; Yu, L.; Wu, Z.; Jiang, J.H. Reversible acylation of RNA enables activatable biosensing. Anal. Chem. 2023, 95, 6490–6495. [Google Scholar] [CrossRef]
- Lei, H.; Fan, R.; Ye, X.; Xiong, W.; Cui, S.; Zeng, T.; Tian, T.; Zhou, X. A novel nucleobase modification strategy for controlling RNA-guided nucleic acid cleavage. CCS Chem. 2023, 5, 2933–2944. [Google Scholar] [CrossRef]
- Qi, Q.; Liu, X.; Fu, F.; Shen, W.; Cui, S.; Yan, S.; Zhang, Y.; Du, Y.; Tian, T.; Zhou, X. Utilizing epigenetic modification as a reactive handle to regulate RNA function and CRISPR-based gene regulation. J. Am. Chem. Soc. 2023, 145, 11678–11689. [Google Scholar] [CrossRef]
- Zhang, K.; Shen, W.; Zhao, Y.; Xu, X.; Liu, X.; Qi, Q.; Huang, S.; Tian, T.; Zhou, X. Strategic base modifications refine RNA function and reduce CRISPR–Cas9 off-targets. Nucleic Acids Res. 2025, 53, gkaf082. [Google Scholar] [CrossRef]
- Xiao, L.; Wang, L.L.; Wu, C.Q.; Li, H.; Zhang, Q.L.; Wang, Y.; Xu, L. Controllable DNA hybridization by host-guest complexation-mediated ligand invasion. Nat. Commun. 2022, 13, 5936. [Google Scholar] [CrossRef]
- Kankanamalage, D.; Tran, J.H.T.; Beltrami, N.; Meng, K.; Zhou, X.; Pathak, P.; Isaacs, L.; Burin, A.L.; Ali, M.F.; Jayawickramarajah, J. DNA strand displacement driven by host-guest interactions. J. Am. Chem. Soc. 2022, 144, 16502–16511. [Google Scholar] [CrossRef]
- Lin, J.; Wang, W.J.; Wang, Y.; Liu, Y.; Xu, L. Building endogenous gene connections through RNA self-assembly controlled CRISPR/Cas9 function. J. Am. Chem. Soc. 2021, 143, 19834–19843. [Google Scholar] [CrossRef]
- Wang, W.J.; Lin, J.; Wu, C.Q.; Luo, A.L.; Xing, X.; Xu, L. Establishing artificial gene connections through RNA displacement-assembly-controlled CRISPR/Cas9 function. Nucleic Acids Res. 2023, 51, 7691–7703. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Liu, Y.; Lai, P.; Ye, H.; Xu, L. Conditional guide RNA through two intermediate hairpins for programmable CRISPR/Cas9 function: Building regulatory connections between endogenous RNA expressions. Nucleic Acids Res. 2020, 48, 11773–11784. [Google Scholar] [CrossRef]
- Li, Y.; Teng, X.; Zhang, K.; Deng, R.; Li, J. RNA strand displacement responsive CRISPR/Cas9 system for mRNA sensing. Anal. Chem. 2019, 91, 3989–3996. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Tan, K.; Wang, H.; Shang, J.; Wan, Y.; Liu, X.; Weng, X.; Wang, F. Orthogonal demethylase-activated deoxyribozyme for intracellular imaging and gene regulation. J. Am. Chem. Soc. 2021, 143, 6895–6904. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Liu, J.; Chen, X.; Mao, L.; Wang, M. Orthogonal chemical activation of enzyme-inducible CRISPR/Cas9 for cell-selective genome editing. J. Am. Chem. Soc. 2022, 144, 22272–22280. [Google Scholar] [CrossRef]
- Burnett, W.V. Northern blotting of RNA denatured in glyoxal without buffer recirculation. Biotechniques 1997, 22, 668–671. [Google Scholar] [CrossRef]
- Knutson, S.D.; Sanford, A.A.; Swenson, C.S.; Korn, M.M.; Manuel, B.A.; Heemstra, J.M. Thermoreversible control of nucleic acid structure and function with glyoxal caging. J. Am. Chem. Soc. 2020, 142, 17766–17781. [Google Scholar] [CrossRef]
- Clydesdale, G.J.; Dandie, G.W.; Muller, H.K. Ultraviolet light induced injury: Immunological and inflammatory effects. Immunol. Cell Biol. 2001, 79, 547–568. [Google Scholar] [CrossRef]
- Zhou, Y.; Kong, D.; Wang, X.; Yu, G.; Wu, X.; Guan, N.; Weber, W.; Ye, H. A small and highly sensitive red/far-red optogenetic switch for applications in mammals. Nat. Biotechnol. 2022, 40, 262–272. [Google Scholar] [CrossRef]
- Tan, P.; He, L.; Huang, Y.; Zhou, Y. Optophysiology: Illuminating cell physiology with optogenetics. Physiol. Rev. 2022, 102, 1263–1325. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, S.; Guo, Y.; Hu, P.; Shi, J. Magnetothermal-activated gene editing strategy for enhanced tumor cell apoptosis. J. Nanobiotechnology 2024, 22, 450. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Huang, Z.; Liu, Y.; He, P.; Wang, Y.; Yan, L.; Wang, X.; Gao, S.; Zhou, X.; Yoon, C.W.; et al. Ultrasound control of genomic regulatory toolboxes for cancer immunotherapy. Nat. Commun. 2024, 15, 10444. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Liu, L.; Cheng, L. Conditionally activated cross-linked crRNAs for CRISPR/Cas12a based nucleic acid detection. ACS Synth. Biol. 2025, 14, 94–100. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Liu, Y.; Song, H.; Zhang, X.; Wang, Y. Conditional Control of CRISPR/Cas9 Function by Chemically Modified Oligonucleotides. Molecules 2025, 30, 1956. https://doi.org/10.3390/molecules30091956
Wang L, Liu Y, Song H, Zhang X, Wang Y. Conditional Control of CRISPR/Cas9 Function by Chemically Modified Oligonucleotides. Molecules. 2025; 30(9):1956. https://doi.org/10.3390/molecules30091956
Chicago/Turabian StyleWang, Liangliang, Yan Liu, Hongjun Song, Xue Zhang, and Yang Wang. 2025. "Conditional Control of CRISPR/Cas9 Function by Chemically Modified Oligonucleotides" Molecules 30, no. 9: 1956. https://doi.org/10.3390/molecules30091956
APA StyleWang, L., Liu, Y., Song, H., Zhang, X., & Wang, Y. (2025). Conditional Control of CRISPR/Cas9 Function by Chemically Modified Oligonucleotides. Molecules, 30(9), 1956. https://doi.org/10.3390/molecules30091956




