Exploring the G-Quadruplex Formation of AS1411 Derivatives
Abstract
1. Introduction
2. Material and Methods
2.1. Oligonucleotides and Reagents
2.2. Förster Resonance Energy Transfer (FRET) Assays
2.2.1. FRET-Melting Competition (FRET-MC)
2.2.2. Isothermal-FRET (iso-FRET) Assay
2.3. Circular Dichroism (CD) Spectroscopy
2.4. UV Spectroscopy
2.4.1. Thermal Difference Spectra (TDS)
2.4.2. Isothermal Difference Spectra (IDS)
2.5. Non-Denaturing Polyacrylamide Gel Electrophoresis (PAGE) Analysis
2.6. Size Exclusion Chromatography (SEC)
2.7. Fluorescence Spectroscopy
2.8. Molecular Docking and Molecular Dynamics (MD) Simulations
2.9. Confocal Fluorescence Microscopy
2.10. Flow Cytometry
2.11. Statistical Analysis
3. Results and Discussion
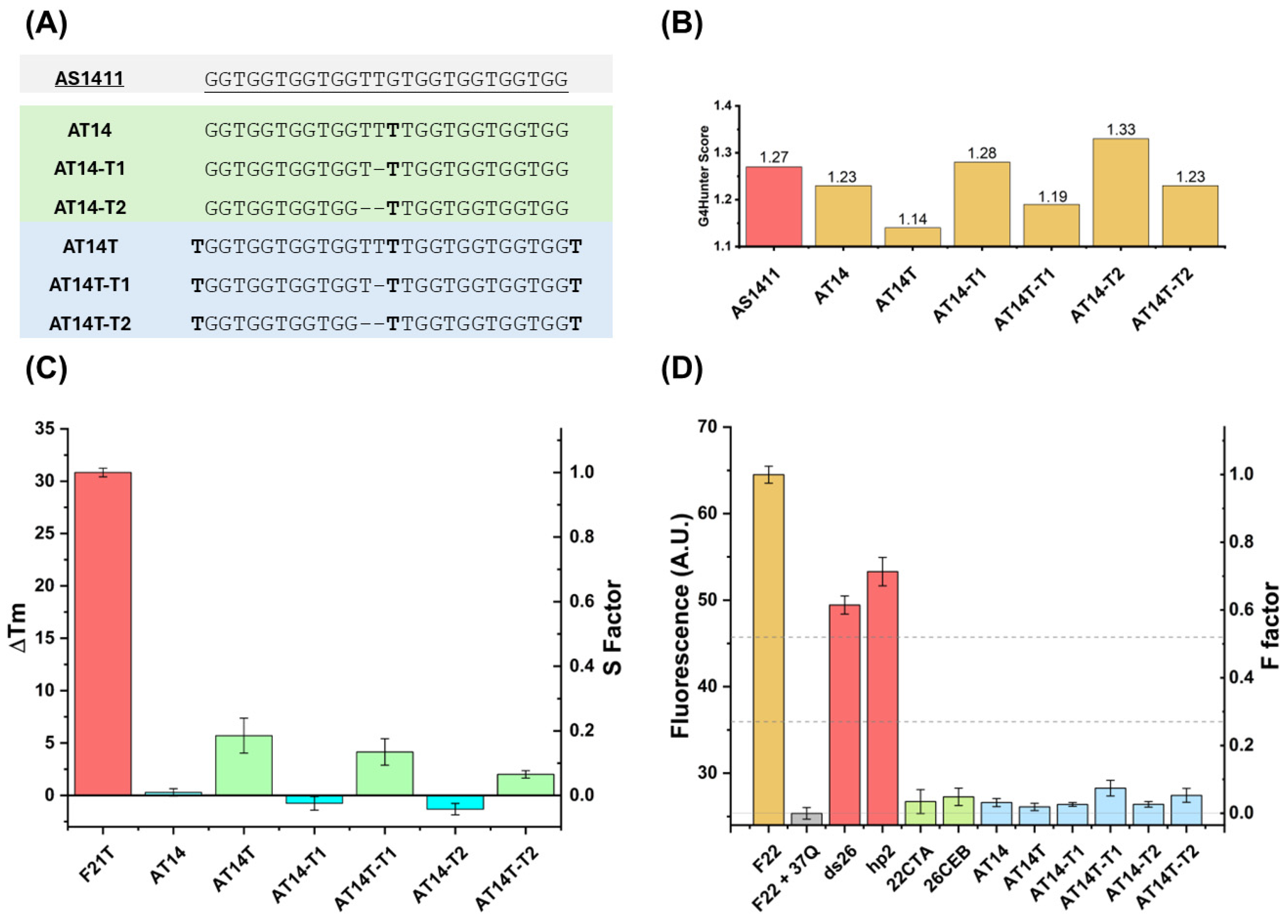
3.1. Design and G4 Formation Assessment by the Aptamer Derivatives
3.1.1. FRET-MC and iso-FRET
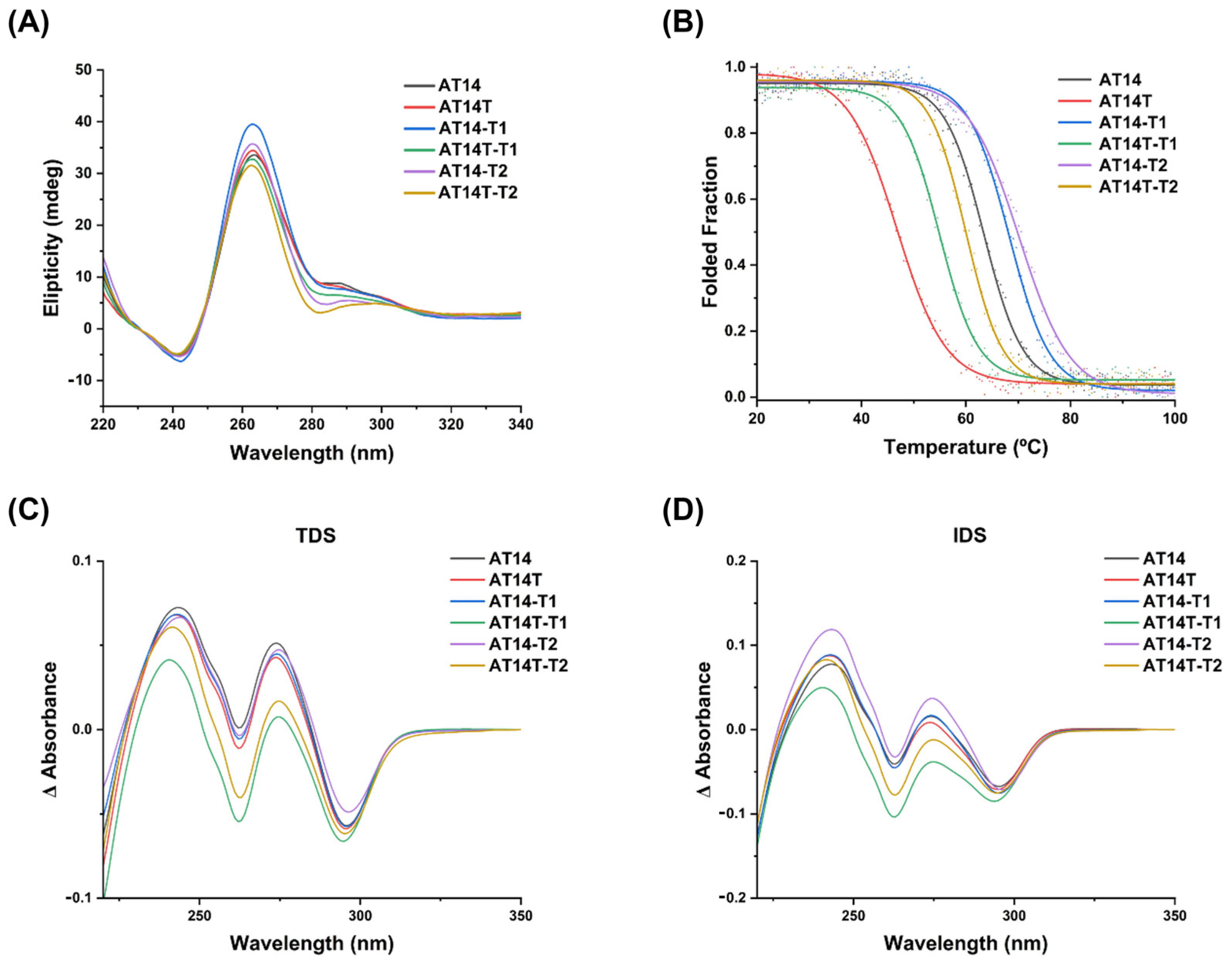
3.1.2. CD Studies
3.1.3. UV Spectroscopy Analysis
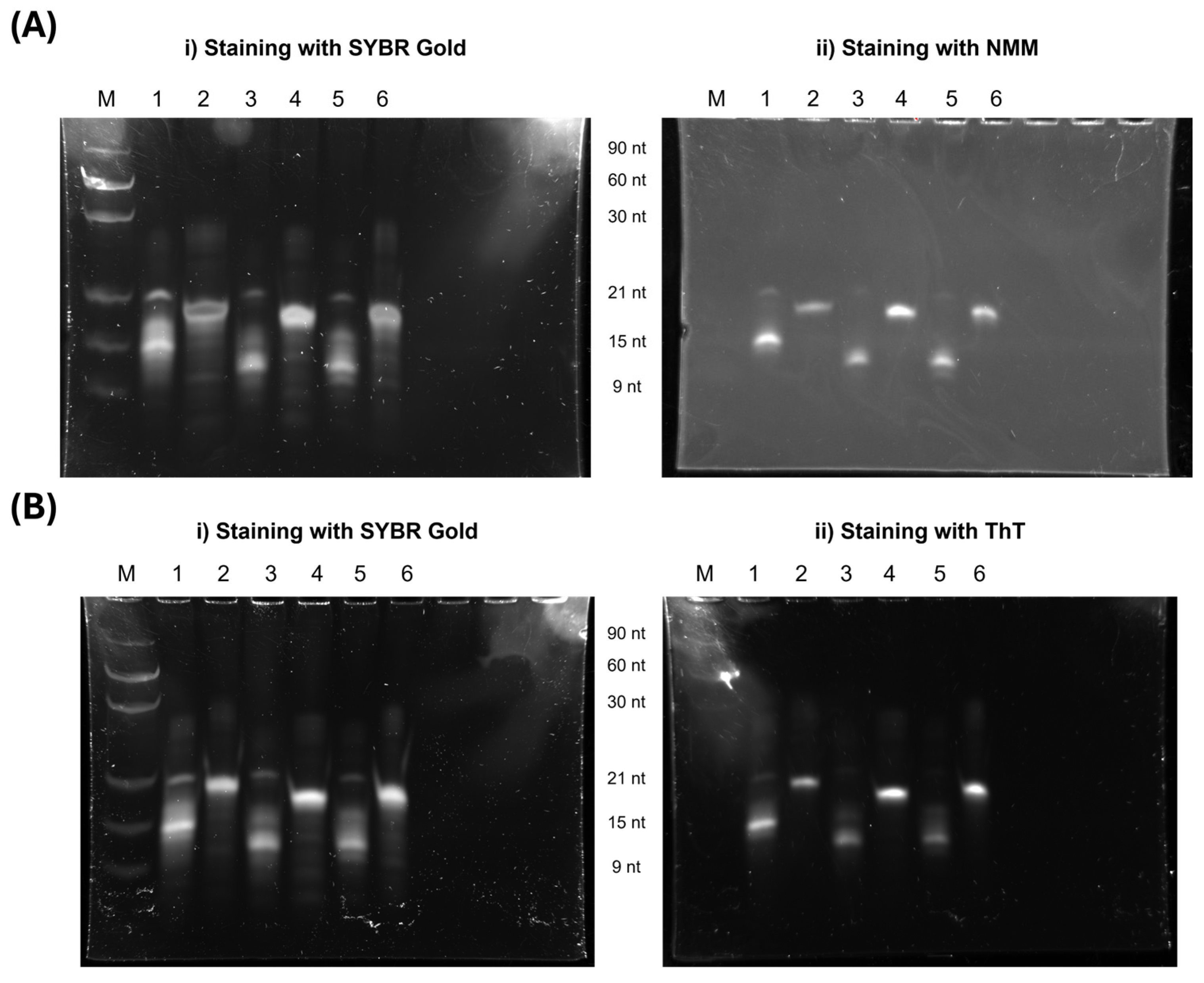
3.1.4. PAGE and SEC Analysis
3.2. Interaction Studies Between Aptamer Derivatives and NCL
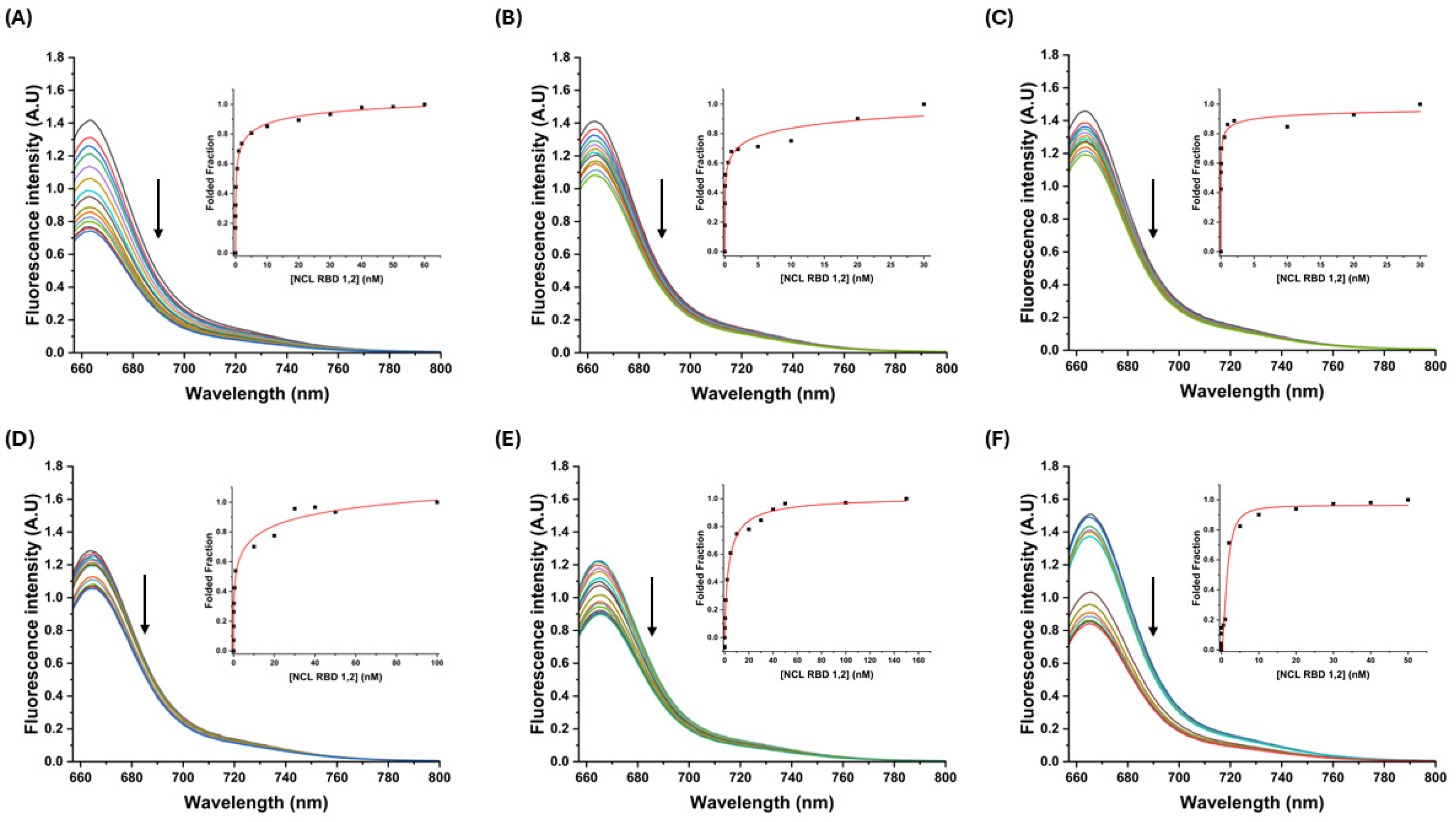
3.2.1. Fluorescence Titrations
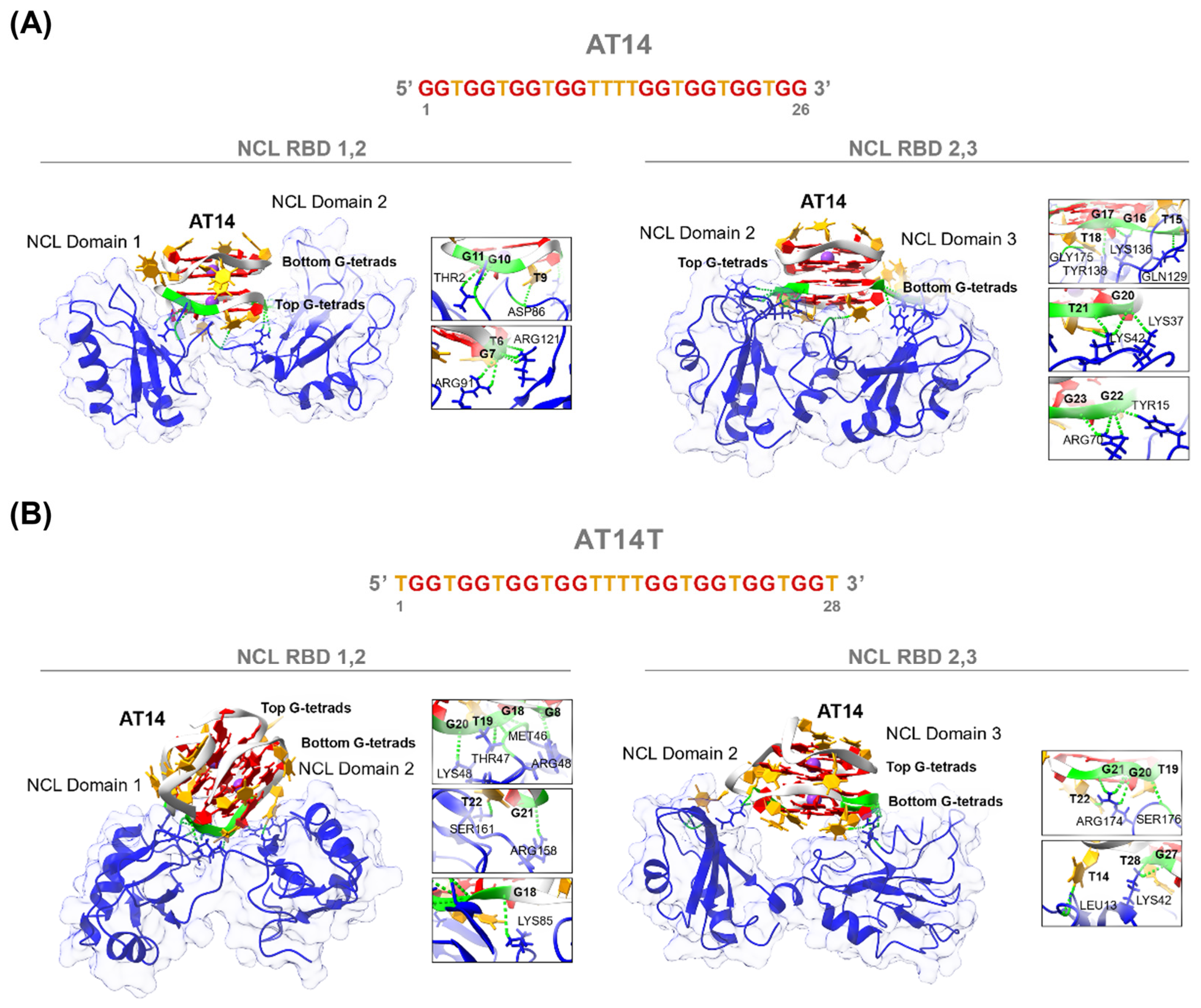
3.2.2. Computational Studies
3.3. In Cell Studies
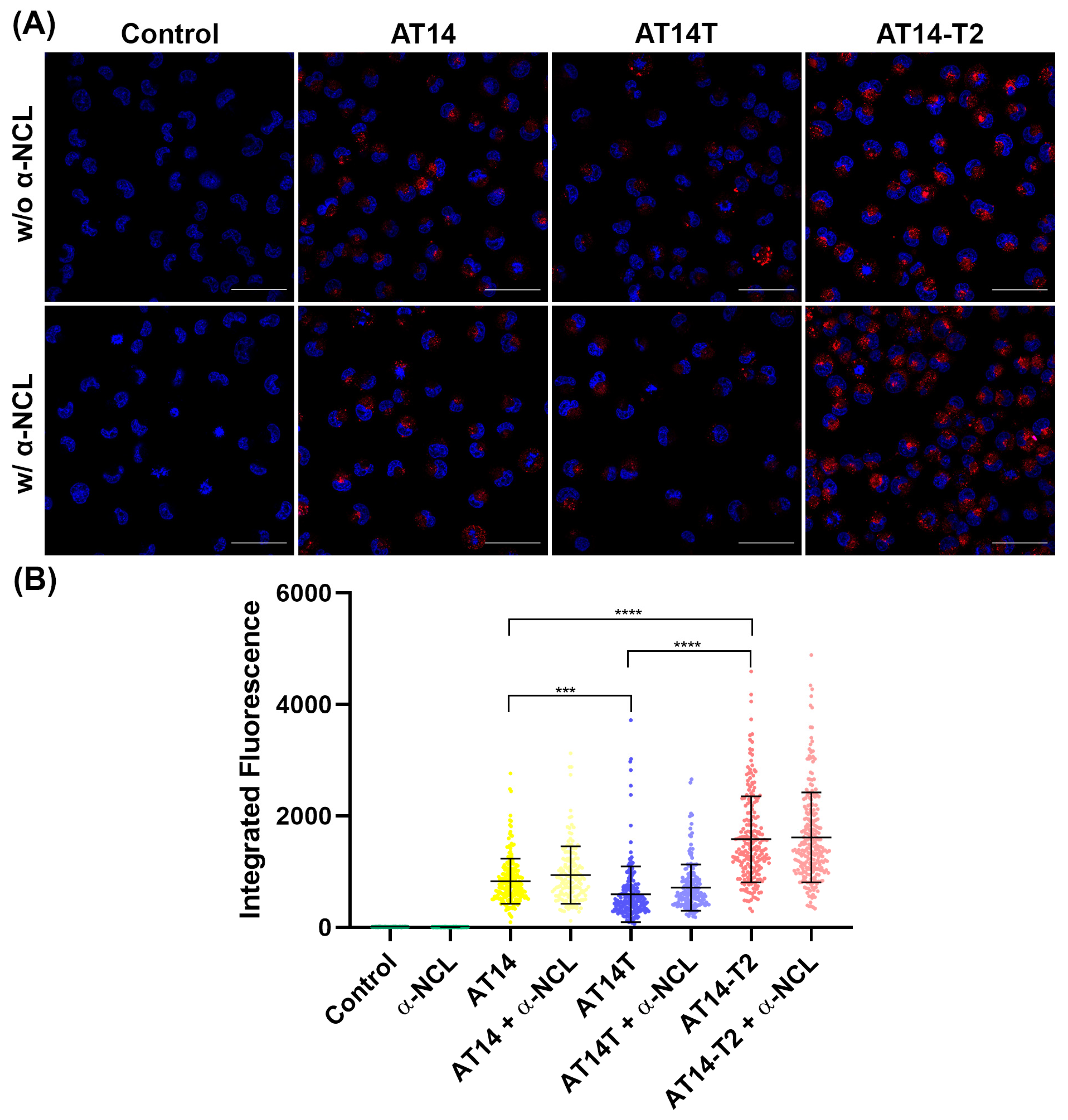
3.3.1. Internalization by Confocal Microscopy
3.3.2. Uptake by Flow Cytometry
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gamboa, J.; Lourenço, P.; Cruz, C.; Gallardo, E. Aptamers for the Delivery of Plant-Based Compounds: A Review. Pharmaceutics 2024, 16, 541. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.F.; Ling, M.; Kacherovsky, N.; Pun, S.H. Aptamers 101, aptamer discovery and in vitro applications in biosensors and separations. Chem. Sci. 2023, 14, 4961–4978. [Google Scholar] [CrossRef] [PubMed]
- Di Mauro, V.; Lauta, F.C.; Modica, J.; Appleton, S.L.; De Franciscis, V.; Catalucci, D. Diagnostic and Therapeutic Aptamers. JACC Basic Transl. Sci. 2024, 9, 260–277. [Google Scholar] [CrossRef] [PubMed]
- Byun, J. Recent Progress and Opportunities for Nucleic Acid Aptamers. Life 2021, 11, 193. [Google Scholar] [CrossRef]
- Ning, Y.; Hu, J.; Lu, F. Aptamers used for biosensors and targeted therapy. Biomed. Pharmacother. 2020, 132, 110902. [Google Scholar] [CrossRef]
- Kelly, L.; Maier, K.E.; Yan, A.; Levy, M. A comparative analysis of cell surface targeting aptamers. Nat. Commun. 2021, 12, 6275. [Google Scholar] [CrossRef]
- Yuan, Y.; Li, Y.; Liu, S.; Gong, P.; Lin, J.; Zhang, X. An overview of aptamer: Design strategy, prominent applications, and potential challenge in plants. J. Plant Physiol. 2024, 296, 154235. [Google Scholar] [CrossRef]
- Napit, R.; Jaysawal, S.K.; Chowdhury, R.; Catague, J.; Melke, H.; Pham, C.V.; Xu, H.; Jia, L.; Lin, J.; Hou, Y.; et al. Aptasensors and Advancement in Molecular Recognition Technology. Adv. Mater. Technol. 2024, 10, 2400504. [Google Scholar] [CrossRef]
- Van den Avont, A.; Sharma-Walia, N. Anti-nucleolin aptamer AS1411, an advancing therapeutic. Front. Mol. Biosci. 2023, 10, 12177691217769. [Google Scholar] [CrossRef]
- Bates, P.J.; Laber, D.A.; Miller, D.M.; Thomas, S.D.; Trent, J.O. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009, 86, 151–164. [Google Scholar] [CrossRef]
- Kitagawa, S.; Matsuda, T.; Washizaki, A.; Murakami, H.; Yamamoto, T.; Yoshioka, Y. Elucidation of the role of nucleolin as a cell surface receptor for nucleic acid-based adjuvants. NPJ Vaccines 2022, 7, 115. [Google Scholar] [CrossRef] [PubMed]
- Safarzadeh Kozani, P.; Safarzadeh Kozani, P.; Rahbarizadeh, F. Chemotherapeutic, Toxin, and Therapeutic Protein Delivery via Nucleolin Aptamer-functionalized Nanoplatforms for Targeted Cancer Therapy. Trends Med. Sci. 2021, 1, 113773. [Google Scholar] [CrossRef]
- Thongchot, S.; Aksonnam, K.; Thuwajit, P.; Yenchitsomanus, P.-T.; Thuwajit, C. Nucleolin-based targeting strategies in cancer treatment: Focus on cancer immunotherapy (Review). Int. J. Mol. Med. 2023, 52, 81. [Google Scholar] [CrossRef] [PubMed]
- Seo, K.; Hwang, K.; Nam, K.M.; Kim, M.J.; Song, Y.-K.; Kim, C.-Y. Nucleolin-Targeting AS1411 Aptamer-Conjugated Nanospheres for Targeted Treatment of Glioblastoma. Pharmaceutics 2024, 16, 566. [Google Scholar] [CrossRef]
- Kozani, P.S.; Kozani, P.S.; Malik, M.T. AS1411-functionalized delivery nanosystems for targeted cancer therapy. Explor. Med. 2021, 2, 146–166. [Google Scholar] [CrossRef]
- Vindigni, G.; Raniolo, S.; Iacovelli, F.; Unida, V.; Stolfi, C.; Desideri, A.; Biocca, S. AS1411 aptamer linked to DNA nanostructures diverts its traffic inside cancer cells and improves its therapeutic efficacy. Pharmaceutics 2021, 13, 1671. [Google Scholar] [CrossRef]
- Lourenço, P.; Miranda, A.; Campello, M.P.C.; Paulo, A.; Louis-Mergny, J.; Cruz, C. Targeting proto-oncogene B-MYB G-quadruplex with a nucleic acid–based fluorescent probe. Int. J. Biol. Macromol. 2024, 266, 131055. [Google Scholar] [CrossRef]
- Dvorkin, S.A.; Karsisiotis, A.I.; Webba da Silva, M. Encoding canonical DNA quadruplex structure. Sci. Adv. 2018, 4, eaat3007. [Google Scholar] [CrossRef]
- Popenda, M.; Miskiewicz, J.; Sarzynska, J.; Zok, T.; Szachniuk, M. Topology-based classification of tetrads and quadruplex structures. Bioinformatics 2020, 36, 1129–1134. [Google Scholar] [CrossRef]
- Jana, J.; Weisz, K. Thermodynamic Stability of G-Quadruplexes: Impact of Sequence and Environment. ChemBioChem 2021, 22, 2848–2856. [Google Scholar] [CrossRef]
- Santos, T.; Salgado, G.F.; Cabrita, E.J.; Cruz, C. G-quadruplexes and their ligands: Biophysical methods to unravel g-quadruplex/ligand interactions. Pharmaceuticals 2021, 14, 769. [Google Scholar] [CrossRef] [PubMed]
- Dhara, M.; Al Hoque, A.; Sen, R.; Dutta, D.; Mukherjee, B.; Paul, B.; Laha, S. Phosphorothioated amino-AS1411 aptamer functionalized stealth nanoliposome accelerates bio-therapeutic threshold of apigenin in neoplastic rat liver: A mechanistic approach. J. Nanobiotechnol. 2023, 21, 28. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Bin Lee, Y.; Lee, J.-H.; Lee, D.H.; Cho, E.J.; Yu, S.J.; Kim, Y.J.; Kim, J.I.; Im, J.H.; Lee, J.H.; et al. Modified AS1411 Aptamer Suppresses Hepatocellular Carcinoma by Up-Regulating Galectin-14. PLoS ONE 2016, 11, e0160822. [Google Scholar] [CrossRef]
- Miranda, A.; Santos, T.; Largy, E.; Cruz, C. Locking up the AS1411 Aptamer with a Flanking Duplex: Towards an Improved Nucleolin-Targeting. Pharmaceuticals 2021, 14, 121. [Google Scholar] [CrossRef]
- Miranda, A.; Santos, T.; Carvalho, J.; Alexandre, D.; Jardim, A.; Caneira, C.; Vaz, V.; Pereira, B.; Godinho, R.; Brito, D.; et al. Aptamer-based approaches to detect nucleolin in prostate cancer. Talanta 2021, 226, 122037. [Google Scholar] [CrossRef]
- Moreira, D.; Leitão, D.; Lopes-Nunes, J.; Santos, T.; Figueiredo, J.; Miranda, A.; Alexandre, D.; Tomaz, C.; Mergny, J.-L.; Cruz, C. G-Quadruplex Aptamer-Ligand Characterization. Molecules 2022, 27, 6781. [Google Scholar] [CrossRef]
- Do, N.Q.; Chung, W.J.; Truong, T.H.A.; Heddi, B.; Phan, A.T. G-quadruplex structure of an anti-proliferative DNA sequence. Nucleic Acids Res. 2017, 45, 7487–7493. [Google Scholar] [CrossRef]
- Santos, T.; Lopes-Nunes, J.; Alexandre, D.; Miranda, A.; Figueiredo, J.; Silva, M.S.; Mergny, J.-L.; Cruz, C. Stabilization of a DNA aptamer by ligand binding. Biochimie 2022, 200, 8–18. [Google Scholar] [CrossRef]
- Miranda, A.; Lopez-Blanco, R.; Lopes-Nunes, J.; Melo, A.M.; Campello, M.P.C.; Paulo, A.; Oliveira, M.C.; Mergny, J.-L.; Oliveira, P.A.; Fernandez-Megia, E.; et al. Gallic Acid–Triethylene Glycol Aptadendrimers Synthesis, Biophysical Characterization and Cellular Evaluation. Pharmaceutics 2022, 14, 2456. [Google Scholar] [CrossRef]
- Lopes-Nunes, J.; Cabral Campello, M.P.; Paulo, A.; Nastruzzi, C.; Oliveira, P.A.; Cruz, C. AT11-guided liposomes for oral cancer cells: From characterization towards in vitro evaluation. J. Drug Deliv. Sci. Technol. 2024, 101, 106214. [Google Scholar] [CrossRef]
- Lopes-Nunes, J.; Oliveira, P.A.; Cruz, C. Enhanced targeted liposomal delivery of imiquimod via aptamer functionalization for head and neck cancer therapy. Colloids Surf. B Biointerfaces 2024, 243, 114121. [Google Scholar] [CrossRef]
- Dailey, M.M.; Miller, M.C.; Bates, P.J.; Lane, A.N.; Trent, J.O. Resolution and characterization of the structural polymorphism of a single quadruplex-forming sequence. Nucleic Acids Res. 2010, 38, 4877–4888. [Google Scholar] [CrossRef] [PubMed]
- Esnault, C.; Magat, T.; El Aabidine, A.Z.; Garcia-Oliver, E.; Cucchiarini, A.; Bouchouika, S.; Lleres, D.; Goerke, L.; Luo, Y.; Verga, D.; et al. G4access identifies G-quadruplexes and their associations with open chromatin and imprinting control regions. Nat. Genet. 2023, 55, 1359–1369. [Google Scholar] [CrossRef]
- Luo, Y.; Granzhan, A.; Verga, D.; Mergny, J. FRET-MC: A fluorescence melting competition assay for studying G4 structures in vitro. Biopolymers 2021, 112, e93. [Google Scholar] [CrossRef]
- Luo, Y.; Verga, D.; Mergny, J.-L. Iso-FRET: An isothermal competition assay to analyze quadruplex formation in vitro. Nucleic Acids Res. 2022, 1, 13–14. [Google Scholar] [CrossRef]
- Needleman, S.B.; Wunsch, C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970, 48, 443–453. [Google Scholar] [CrossRef]
- Largy, E.; Mergny, J.-L. Shape matters: Size-exclusion HPLC for the study of nucleic acid structural polymorphism. Nucleic Acids Res. 2014, 42, e149. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Arumugam, S.; Clarke Miller, M.; Maliekal, J.; Bates, P.J.; Trent, J.O.; Lane, A.N. Solution structure of the RBD1,2 domains from human nucleolin. J. Biomol. NMR 2010, 47, 79–83. [Google Scholar] [CrossRef]
- Berman, H.M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Carvalho, J.; Santos, T.; Carrilho, R.; Sousa, F.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. Ligand screening to pre-miRNA 149 G-quadruplex investigated by molecular dynamics. J. Biomol. Struct. Dyn. 2020, 38, 2276–2286. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, J.; Lopes-Nunes, J.; Vialet, B.; Rosado, T.; Gallardo, E.; Vale, J.; Eloy, C.; Ferreira, S.; Palmeira-de-Oliveira, R.; Cabral Campello, M.P.; et al. Nanoaggregate-forming lipid-conjugated AS1411 aptamer as a promising tum.or-targeted delivery system of anticancer agents in vitro. Nanomedicine 2021, 36, 102429. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Luo, H.; Gubu, A.; Yu, S.; Zhang, H.; Dai, H.; Zhang, Y.; Zhang, B.; Ma, Y.; Lu, A.; et al. Chemically modified aptamers for improving binding affinity to the target proteins via enhanced non-covalent bonding. Front. Cell Dev. Biol. 2023, 11, 1091809. [Google Scholar] [CrossRef] [PubMed]
- Ni, S.; Yao, H.; Wang, L.; Lu, J.; Jiang, F.; Lu, A.; Zhang, G. Chemical Modifications of Nucleic Acid Aptamers for Therapeutic Purposes. Int. J. Mol. Sci. 2017, 18, 1683. [Google Scholar] [CrossRef]
- Do, N.Q.; Lim, K.W.; Teo, M.H.; Heddi, B.; Phan, A.T. Stacking of G-quadruplexes: NMR structure of a G-rich oligonucleotide with potential anti-HIV and anticancer activity†. Nucleic Acids Res. 2011, 39, 9448–9457. [Google Scholar] [CrossRef]
- Carvalho, J.; Queiroz, J.A.; Cruz, C. Circular dichroism of G-Quadruplex: A laboratory experiment for the study of topology and ligand binding. J. Chem. Educ. 2017, 94, 1547–1551. [Google Scholar] [CrossRef]
- Šket, P.; Plavec, J. Tetramolecular DNA quadruplexes in solution: Insights into structural diversity and cation movement. J. Am. Chem. Soc. 2010, 132, 12724–12732. [Google Scholar] [CrossRef]
- Mergny, J.L.; Li, J.; Lacroix, L.; Amrane, S.; Chaires, J.B. Thermal difference spectra: A specific signature for nucleic acid structures. Nucleic Acids Res. 2005, 33, e138. [Google Scholar] [CrossRef]
- Bie, L.; Wang, Y.; Jiang, F.; Xiao, Z.; Zhang, L.; Wang, J. Insights into the binding mode of AS1411 aptamer to nucleolin. Front. Mol. Biosci. 2022, 9, 1025313. [Google Scholar] [CrossRef]
- Santos, T.; Miranda, A.; Campello, M.P.; Paulo, A.; Salgado, G.; Cabrita, E.J.; Cruz, C. Recognition of nucleolin through interaction with RNA G-quadruplex. Biochem. Pharmacol. 2021, 189, 114208. [Google Scholar] [CrossRef]
- Lopes-Nunes, J.; Carvalho, J.; Figueiredo, J.; Ramos, C.I.; Lourenço, L.M.; Tomé, J.P.; Neves, M.G.; Mergny, J.-L.; Queiroz, J.A.; Salgado, G.F.; et al. Phthalocyanines for G-quadruplex aptamers binding. Bioorg. Chem. 2020, 100, 103920. [Google Scholar] [CrossRef] [PubMed]
- Santos, T.; Silva, M.; Imbert, L.; Campello, M.P.C.; Paulo, A.; Amrane, S.; Salgado, G.F.; Cruz, C.; Cabrita, E.J. A Structural Perspective into the Interaction of an Oncogenesis-Relevant pre-miRNA G-Quadruplex Ligand Carrier with the Protein Nucleolin. Chem. Eur. J. 2023, 29, e202301181. [Google Scholar] [CrossRef] [PubMed]
- Santos, T.; Salgado, G.F.; Cabrita, E.J.; Cruz, C. Nucleolin: A binding partner of G-quadruplex structures. Trends Cell Biol. 2022, 32, 561–564. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, J.; Paiva, A.; Campello, M.P.C.; Paulo, A.; Mergny, J.-L.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. Aptamer-based Targeted Delivery of a G-quadruplex Ligand in Cervical Cancer Cells. Sci. Rep. 2019, 9, 7945. [Google Scholar] [CrossRef]
- Figueiredo, J.; Lopes-Nunes, J.; Carvalho, J.; Antunes, F.; Ribeiro, M.; Campello, M.P.C.; Paulo, A.; Paiva, A.; Salgado, G.F.; Queiroz, J.A.; et al. AS1411 derivatives as carriers of G-quadruplex ligands for cervical cancer cells. Int. J. Pharm. 2019, 568, 118511. [Google Scholar] [CrossRef]
- Bates, P.J.; Reyes-Reyes, E.M.; Malik, M.T.; Murphy, E.M.; O’Toole, M.G.; Trent, J.O. G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent: Uses and mechanisms. Biochim. Biophys. Acta General Subj. 2017, 1861, 1414–1428. [Google Scholar] [CrossRef]
- Reyes-Reyes, E.M.; Teng, Y.; Bates, P.J. A new paradigm for aptamer therapeutic AS1411 action: Uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res. 2010, 70, 8617–8629. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lourenço, P.; Moreira, D.; Miranda, A.; Lopes-Nunes, J.; Maocha, I.; Santos, T.; Ferreira, P.L.; Sousa, F.; Paiva, A.; Cruz, C. Exploring the G-Quadruplex Formation of AS1411 Derivatives. Molecules 2025, 30, 1673. https://doi.org/10.3390/molecules30081673
Lourenço P, Moreira D, Miranda A, Lopes-Nunes J, Maocha I, Santos T, Ferreira PL, Sousa F, Paiva A, Cruz C. Exploring the G-Quadruplex Formation of AS1411 Derivatives. Molecules. 2025; 30(8):1673. https://doi.org/10.3390/molecules30081673
Chicago/Turabian StyleLourenço, Pedro, David Moreira, André Miranda, Jéssica Lopes-Nunes, Izamara Maocha, Tiago Santos, Pedro L. Ferreira, Fani Sousa, Artur Paiva, and Carla Cruz. 2025. "Exploring the G-Quadruplex Formation of AS1411 Derivatives" Molecules 30, no. 8: 1673. https://doi.org/10.3390/molecules30081673
APA StyleLourenço, P., Moreira, D., Miranda, A., Lopes-Nunes, J., Maocha, I., Santos, T., Ferreira, P. L., Sousa, F., Paiva, A., & Cruz, C. (2025). Exploring the G-Quadruplex Formation of AS1411 Derivatives. Molecules, 30(8), 1673. https://doi.org/10.3390/molecules30081673








