Untargeted Lipidomic Reveals Potential Biomarkers in Plasma Samples for the Discrimination of Patients Affected by Parkinson’s Disease
Abstract
1. Introduction
2. Results
2.1. Differentiation Between PD, AD, and Healthy Controls
2.2. Pairwise Comparison
3. Materials and Methods
3.1. Chemicals
3.2. Study Population
3.3. Collection and Handling of Plasma Samples
3.4. Lipid Extraction
3.5. Liquid Chromatography-Mass Spectrometry Analysis
3.6. Data Processing
3.7. Statistical Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ray Dorsey, E.; Elbaz, A.; Nichols, E.; Abd-Allah, F.; Abdelalim, A.; Adsuar, J.C.; Ansha, M.G.; Brayne, C.; Choi, J.Y.J.; Collado-Mateo, D.; et al. Global, Regional, and National Burden of Parkinson’s Disease, 1990–2016: A Systematic Analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2018, 17, 939–953. [Google Scholar] [CrossRef]
- Li, T.; Le, W. Biomarkers for Parkinson’s Disease: How Good Are They? Neurosci. Bull. 2020, 36, 183–194. [Google Scholar] [CrossRef]
- Mhyre, T.R.; Boyd, J.T.; Hamill, R.W.; Maguire-Zeiss, K.A. Parkinson’s Disease. Subcell. Biochem. 2012, 65, 389–455. [Google Scholar] [CrossRef]
- Zella, S.M.A.; Metzdorf, J.; Ciftci, E.; Ostendorf, F.; Muhlack, S.; Gold, R.; Tönges, L. Emerging Immunotherapies for Parkinson Disease. Neurol. Ther. 2019, 8, 29–44. [Google Scholar] [CrossRef]
- Parnetti, L.; Gaetani, L.; Eusebi, P.; Paciotti, S.; Hansson, O.; El-Agnaf, O.; Mollenhauer, B.; Blennow, K.; Calabresi, P. CSF and Blood Biomarkers for Parkinson’s Disease. Lancet Neurol. 2019, 18, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Alves, G.; Pedersen, K.F.; Pedersen, K.F. Epidemiology of Parkinson’s Disease Epidemiology of Parkinson’s Disease. J. Neurol. 2008, 5, 525–535. [Google Scholar]
- Rizzo, G.; Copetti, M.; Arcuti, S.; Martino, D.; Fontana, A.; Logroscino, G. Accuracy of Clinical Diagnosis of Parkinson Disease. Neurology 2016, 86, 566–576. [Google Scholar] [CrossRef] [PubMed]
- Noshin, K.; Boland, M.R.; Hou, B.; Lu, V.; Manning, C.; Shen, L.; Zhang, A. Uncovering Important Diagnostic Features for Alzheimer’s, Parkinson’s and Other Dementias Using. Interpretable Association Mining Methods. HHS Public Access 2025, 30, 631–646. [Google Scholar]
- Wang, J.; Li, X.; Pang, H.; Bu, S.; Zhao, M.; Liu, Y.; Yu, H.; Jiang, Y.; Fan, G. Differential Connectivity Patterns of Mild Cognitive Impairment in Alzheimer’s and Parkinson’s Disease: A Large-Scale Brain Network Study. Acad. Radiol. 2025, in press. [CrossRef] [PubMed]
- Sarallah, R.; Jahani, S.; Soltani Khaboushan, A.; Moaveni, A.K.; Amiri, M.; Majidi Zolbin, M. The Role of CXCL12/CXCR4/CXCR7 Axis in Cognitive Impairment Associated with Neurodegenerative Diseases. Brain Behav. Immun. Health 2025, 43, 100932. [Google Scholar] [CrossRef] [PubMed]
- Deas, E.; Cremades, N.; Angelova, P.R.; Ludtmann, M.H.R.; Yao, Z.; Chen, S.; Horrocks, M.H.; Banushi, B.; Little, D.; Devine, M.J.; et al. Alpha-Synuclein Oligomers Interact with Metal Ions to Induce Oxidative Stress and Neuronal Death in Parkinson’s Disease. Antioxid. Redox Signal 2016, 24, 376–391. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, S.; Wang, D.; Fan, M.; Gao, F.; Sun, W.; Li, Z.; Li, S. The Significance of Uric Acid in the Diagnosis and Treatment of Parkinson Disease. Medicine 2017, 96, e8502. [Google Scholar] [CrossRef] [PubMed]
- Picca, A.; Calvani, R.; Landi, G.; Marini, F.; Biancolillo, A.; Gervasoni, J.; Persichilli, S.; Primiano, A.; Urbani, A.; Bossola, M.; et al. Circulating Amino Acid Signature in Older People with Parkinson’s Disease: A Metabolic Complement to the EXosomes in PArkiNson Disease (EXPAND) Study. Exp. Gerontol. 2019, 128, 110766. [Google Scholar] [CrossRef]
- Yadav, R.S.; Tiwari, N.K. Lipid Integration in Neurodegeneration: An Overview of Alzheimer’s Disease. Mol. Neurobiol. 2014, 50, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Abbott, S.K.; Li, H.; Muñoz, S.S.; Knoch, B.; Batterham, M.; Murphy, K.E.; Halliday, G.M.; Garner, B. Altered Ceramide Acyl Chain Length and Ceramide Synthase Gene Expression in Parkinson’s Disease. Mov. Disord. 2014, 29, 518–526. [Google Scholar] [CrossRef]
- Moll, T.; Marshall, J.N.G.; Soni, N.; Zhang, S.; Cooper-Knock, J.; Shaw, P.J. Membrane Lipid Raft Homeostasis Is Directly Linked to Neurodegeneration. Essays Biochem. 2021, 65, 999–1011. [Google Scholar] [PubMed]
- Filippov, V.; Song, M.A.; Zhang, K.; Vinters, H.V.; Tung, S.; Kirsch, W.M.; Yang, J.; Duerksen-Hughes, P.J. Increased Ceramide in Brains with Alzheimer’s and Other Neurodegenerative Diseases. J. Alzheimer’s Dis. 2012, 29, 537. [Google Scholar] [CrossRef] [PubMed]
- Huo, Z.; Rana, B.K.; Elman, J.A.; Dong, R.; Engelman, C.D.; Johnson, S.C.; Lyons, M.J.; Franz, C.E.; Kremen, W.S.; Zhao, J. Metabolic Profiling of Cognitive Aging in Midlife. Front. Aging Neurosci. 2020, 12, 555850. [Google Scholar] [CrossRef]
- Nicholls, A.; Theodoridis, G.; Wilson, I.D. Global Metabolic Profiling in Health and Disease. In Global Metabolic Profiling: Clinical Application; Future Science Ltd.: London, UK, 2014; pp. 2–5. [Google Scholar] [CrossRef]
- Han, W.; Sapkota, S.; Camicioli, R.; Dixon, R.A.; Li, L. Profiling Novel Metabolic Biomarkers for Parkinson’s Disease Using In-depth Metabolomic Analysis. Mov. Disord. 2017, 32, 1720–1728. [Google Scholar] [CrossRef]
- Shao, Y.; Le, W. Recent Advances and Perspectives of Metabolomics-Based Investigations in Parkinson’s Disease. Mol. Neurodegener. 2019, 14, 3. [Google Scholar] [CrossRef]
- González-Domínguez, R.; García-Barrera, T.; Gómez-Ariza, J.L. Combination of Metabolomic and Phospholipid-Profiling Approaches for the Study of Alzheimer’s Disease. J. Proteom. 2014, 104, 37–47. [Google Scholar] [CrossRef]
- Peña-Bautista, C.; Álvarez-Sánchez, L.; Roca, M.; García-Vallés, L.; Baquero, M.; Cháfer-Pericás, C. Plasma Lipidomics Approach in Early and Specific Alzheimer’s Disease Diagnosis. J. Clin. Med. 2022, 11, 5030. [Google Scholar] [CrossRef] [PubMed]
- Casanova, R.; Varma, S.; Simpson, B.; Kim, M.; An, Y.; Saldana, S.; Riveros, C.; Moscato, P.; Griswold, M.; Sonntag, D.; et al. Blood Metabolite Markers of Preclinical Alzheimer’s Disease in Two Longitudinally Followed Cohorts of Older Individuals. Alzheimer’s Dement. 2016, 12, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Yu, H.; Xing, S.; Huan, T. Addressing Big Data Challenges in Mass Spectrometry-Based Metabolomics. Chem. Commun. 2022, 58, 9979–9990. [Google Scholar] [CrossRef] [PubMed]
- Han, X. Lipidomics: Comprehensive Mass Spectrometry of Lipids; John Wiley & Sons: Hoboken, NJ, USA, 2016; ISBN 9781118893128. [Google Scholar]
- Lv, J.; Zhang, L.; Yan, F.; Wang, X. Clinical Lipidomics: A New Way to Diagnose Human Diseases. Clin. Transl. Med. 2018, 7, e12. [Google Scholar] [CrossRef]
- Pizarro, C.; Arenzana-Rámila, I.; Pérez-Del-Notario, N.; Pérez-Matute, P.; González-Sáiz, J.M. Plasma Lipidomic Profiling Method Based on Ultrasound Extraction and Liquid Chromatography Mass Spectrometry. Anal. Chem. 2013, 85, 12085–12092. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; De Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the Gap between Raw Spectra and Functional Insights. Nucleic Acids Res 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Cruciani-Guglielmacci, C.; López, M.; Campana, M.; le Stunff, H. Brain Ceramide Metabolism in the Control of Energy Balance. Front. Physiol. 2017, 8, 787. [Google Scholar] [CrossRef]
- Wang, G.; Bieberich, E. Sphingolipids in Neurodegeneration (with Focus on Ceramide and S1P). Adv. Biol. Regul. 2018, 70, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Castro, B.M.; Prieto, M.; Silva, L.C. Ceramide: A Simple Sphingolipid with Unique Biophysical Properties. Prog. Lipid Res. 2014, 54, 53–67. [Google Scholar] [CrossRef]
- Mapstone, M.; Cheema, A.K.; Fiandaca, M.S.; Zhong, X.; Mhyre, T.R.; Macarthur, L.H.; Hall, W.J.; Fisher, S.G.; Peterson, D.R.; Haley, J.M.; et al. Plasma Phospholipids Identify Antecedent Memory Impairment in Older Adults. Nat. Med. 2014, 20, 415. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Nevado-Holgado, A.; Whiley, L.; Snowden, S.G.; Soininen, H.; Kloszewska, I.; Mecocci, P.; Tsolaki, M.; Vellas, B.; Thambisetty, M.; et al. Association between Plasma Ceramides and Phosphatidylcholines and Hippocampal Brain Volume in Late Onset Alzheimer’s Disease. J. Alzheimer’s Dis. 2017, 60, 809. [Google Scholar] [CrossRef]
- Patel, D.; Witt, S.N. Ethanolamine and Phosphatidylethanolamine: Partners in Health and Disease. Oxid. Med. Cell Longev. 2017, 2017, 4829180. [Google Scholar] [CrossRef]
- Xu, Y.; Xiao, Y.J.; Zhu, K.; Baudhuin, L.M.; Lu, J.; Hong, G.; Kim, K.S.; Cristina, K.L.; Song, L.; Williams, F.S.; et al. Unfolding the Pathophysiological Role of Bioactive Lysophospholipids. Curr. Drug Targets Immune Endocr. Metab. Disord. 2003, 3, 23–32. [Google Scholar] [CrossRef]
- Tian, Q.; Mitchell, B.A.; Zampino, M.; Ferrucci, L. Longitudinal Associations between Blood Lysophosphatidylcholines and Skeletal Muscle Mitochondrial Function. Geroscience 2022, 44, 2213–2221. [Google Scholar] [CrossRef] [PubMed]
- Grzelczyk, A.; Gendaszewska-Darmach, E. Novel Bioactive Glycerol-Based Lysophospholipids: New Data-New Insight into Their Function. Biochimie 2013, 95, 667–679. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, I.; Lim, Y.S.; Ng, S.Y.; Ling, S.C. Deciphering Lipid Dysregulation in ALS: From Mechanisms to Translational Medicine. Transl. Neurodegener. 2022, 11, 48. [Google Scholar] [CrossRef] [PubMed]
- Miletić Vukajlović, J.; Drakulić, D.; Pejić, S.; Ilić, T.V.; Stefanović, A.; Petković, M.; Schiller, J. Increased Plasma Phosphatidylcholine/Lysophosphatidylcholine Ratios in Patients with Parkinson’s Disease. Rapid Commun. Mass. Spectrom. 2020, 34, e8595. [Google Scholar] [CrossRef] [PubMed]
- López de Frutos, L.; Almeida, F.; Murillo-Saich, J.; Conceição, V.A.; Guma, M.; Queheberger, O.; Giraldo, P.; Miltenberger-Miltenyi, G. Serum Phospholipid Profile Changes in Gaucher Disease and Parkinson’s Disease. Int. J. Mol. Sci. 2022, 23, 10387. [Google Scholar] [CrossRef]
- Ngo, S.T. Lipids: Key Players in Central Nervous System Cell Physiology and Pathology. Semin. Cell Dev. Biol. 2021, 112, 59–60. [Google Scholar] [CrossRef]
- Fahy, E.; Subramaniam, S.; Brown, H.A.; Glass, C.K.; Merrill, A.H.; Murphy, R.C.; Raetz, C.R.H.; Russell, D.W.; Seyama, Y.; Shaw, W.; et al. A Comprehensive Classification System for Lipids. J. Lipid Res. 2005, 46, 839–861. [Google Scholar] [CrossRef]
- Schmitt, F.; Hussain, G.; Dupuis, L.; Loeffler, J.P.; Henriques, A. A Plural Role for Lipids in Motor Neuron Diseases: Energy, Signaling and Structure. Front. Cell Neurosci. 2014, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Morrow, A.; Panyard, D.J.; Deming, Y.K.; Jonaitis, E.; Dong, R.; Vasiljevic, E.; Betthauser, T.J.; Kollmorgen, G.; Suridjan, I.; Bayfield, A.; et al. Cerebrospinal Fluid Sphingomyelins in Alzheimer’s Disease, Neurodegeneration, and Neuroinflammation. J. Alzheimer’s Dis. 2022, 90, 667. [Google Scholar] [CrossRef] [PubMed]
- Baloni, P.; Arnold, M.; Buitrago, L.; Nho, K.; Moreno, H.; Huynh, K.; Brauner, B.; Louie, G.; Kueider-Paisley, A.; Suhre, K.; et al. Multi-Omic Analyses Characterize the Ceramide/Sphingomyelin Pathway as a Therapeutic Target in Alzheimer’s Disease. Commun. Biol. 2022, 5, 1074. [Google Scholar] [CrossRef]
- Custodia, A.; Aramburu-Núñez, M.; Correa-Paz, C.; Posado-Fernández, A.; Gómez-Larrauri, A.; Castillo, J.; Gómez-Muñoz, A.; Sobrino, T.; Ouro, A. Ceramide Metabolism and Parkinson’s Disease—Therapeutic Targets. Biomolecules 2021, 11, 945. [Google Scholar] [CrossRef] [PubMed]
- Alaamery, M.; Albesher, N.; Aljawini, N.; Alsuwailm, M.; Massadeh, S.; Wheeler, M.A.; Chao, C.C.; Quintana, F.J. Role of Sphingolipid Metabolism in Neurodegeneration. J. Neurochem. 2021, 158, 25–35. [Google Scholar] [CrossRef]
- Estes, R.E.; Lin, B.; Khera, A.; Davis, M.Y. Lipid Metabolism Influence on Neurodegenerative Disease Progression: Is the Vehicle as Important as the Cargo? Front. Mol. Neurosci. 2021, 14, 788695. [Google Scholar] [CrossRef] [PubMed]
- Guedes, L.C.; Chan, R.B.; Gomes, M.A.; Conceição, V.A.; Machado, R.B.; Soares, T.; Xu, Y.; Gaspar, P.; Carriço, J.A.; Alcalay, R.N.; et al. Serum Lipid Alterations in GBA-Associated Parkinson’s Disease. Park. Relat. Disord. 2017, 44, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Plotegher, N.; Bubacco, L.; Greggio, E.; Civiero, L. Ceramides in Parkinson’s Disease: From Recent Evidence to New Hypotheses. Front. Neurosci. 2019, 13, 330. [Google Scholar] [CrossRef] [PubMed]
- Katsel, P.; Li, C.; Haroutunian, V. Gene Expression Alterations in the Sphingolipid Metabolism Pathways during Progression of Dementia and Alzheimer’s Disease: A Shift toward Ceramide Accumulation at the Earliest Recognizable Stages of Alzheimer’s Disease? Neurochem. Res. 2007, 32, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Speziale, R.; Montesano, C.; De Leonibus, M.L.; Bonelli, F.; Fezzardi, P.; Beconi, M.G.; Monteagudo, E.; Elbaum, D.; Orsatti, L. Determination of Acetyl Coenzyme A in Human Whole Blood by Ultra-Performance Liquid Chromatography-Mass Spectrometry. J. Chromatogr. B 2018, 1083, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Currais, A.; Huang, L.; Goldberg, J.; Petrascheck, M.; Ates, G.; Pinto-Duarte, A.; Shokhirev, M.N.; Schubert, D.; Maher, P. Elevating Acetyl-CoA Levels Reduces Aspects of Brain Aging. Elife 2019, 8, e47866. [Google Scholar] [CrossRef]
- Hu, L.; Huang, Y.-J.; Wei, Y.-D.; Li, T.; Ke, W.; Chen, G.-H.; Dong, M.-X. Plasma Metabolomics Profiles Indicate Sex Differences of Lipid Metabolism in Patients with Parkinson’s Disease. Sci. Rep. 2024, 14, 31262. [Google Scholar] [CrossRef] [PubMed]
- Santiago, J.A.; Potashkin, J.A. Shared Dysregulated Pathways Lead to Parkinson’s Disease and Diabetes. Trends Mol. Med. 2013, 19, 176–186. [Google Scholar] [CrossRef]
- Wang, M.C.; Li, C.I.; Liu, C.S.; Lin, C.H.; Yang, S.Y.; Li, T.C.; Lin, C.C. Effect of Blood Lipid Variability on Mortality in Patients with Type 2 Diabetes: A Large Single-Center Cohort Study. Cardiovasc. Diabetol. 2021, 20, 228. [Google Scholar] [CrossRef] [PubMed]

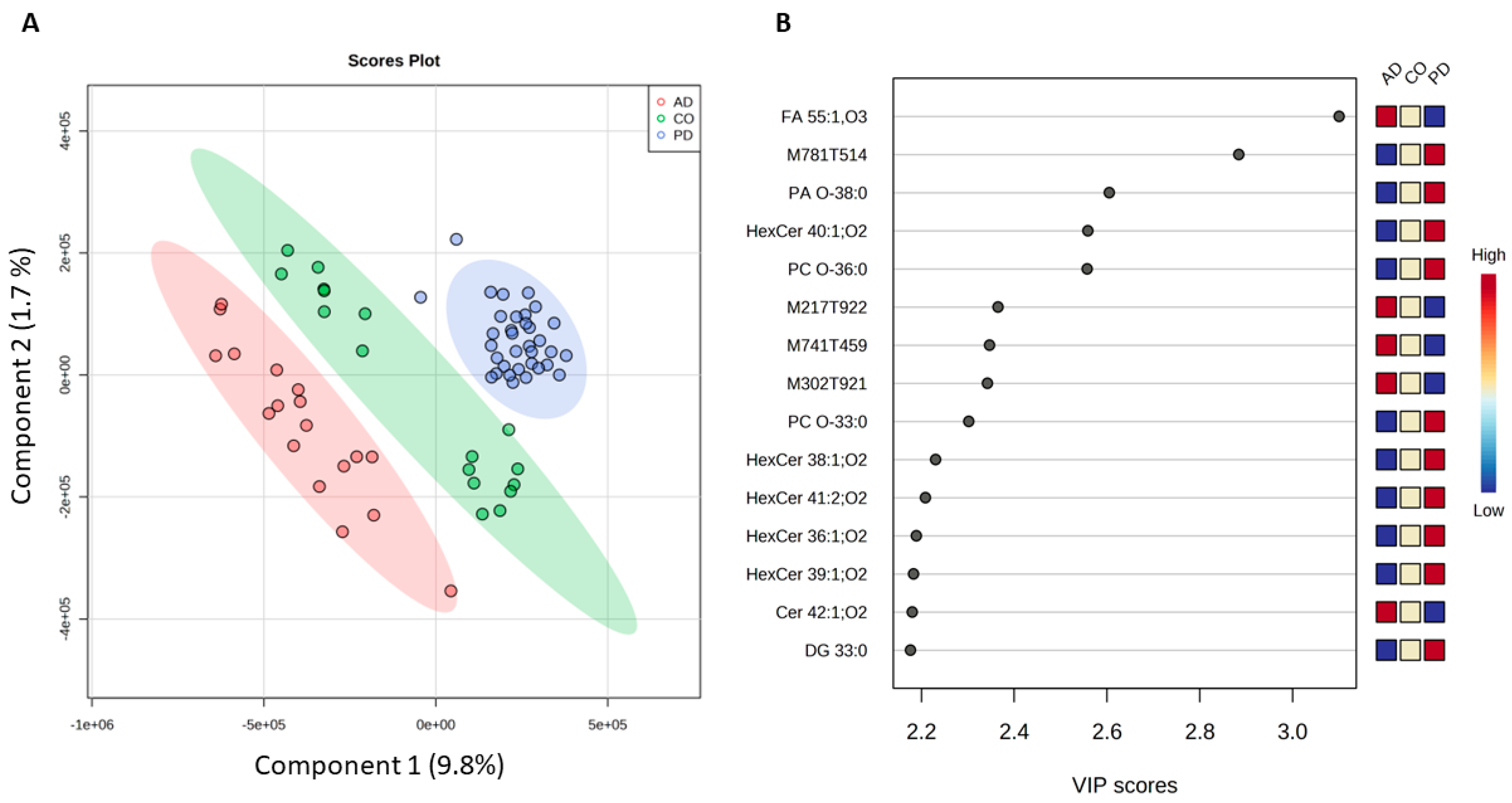
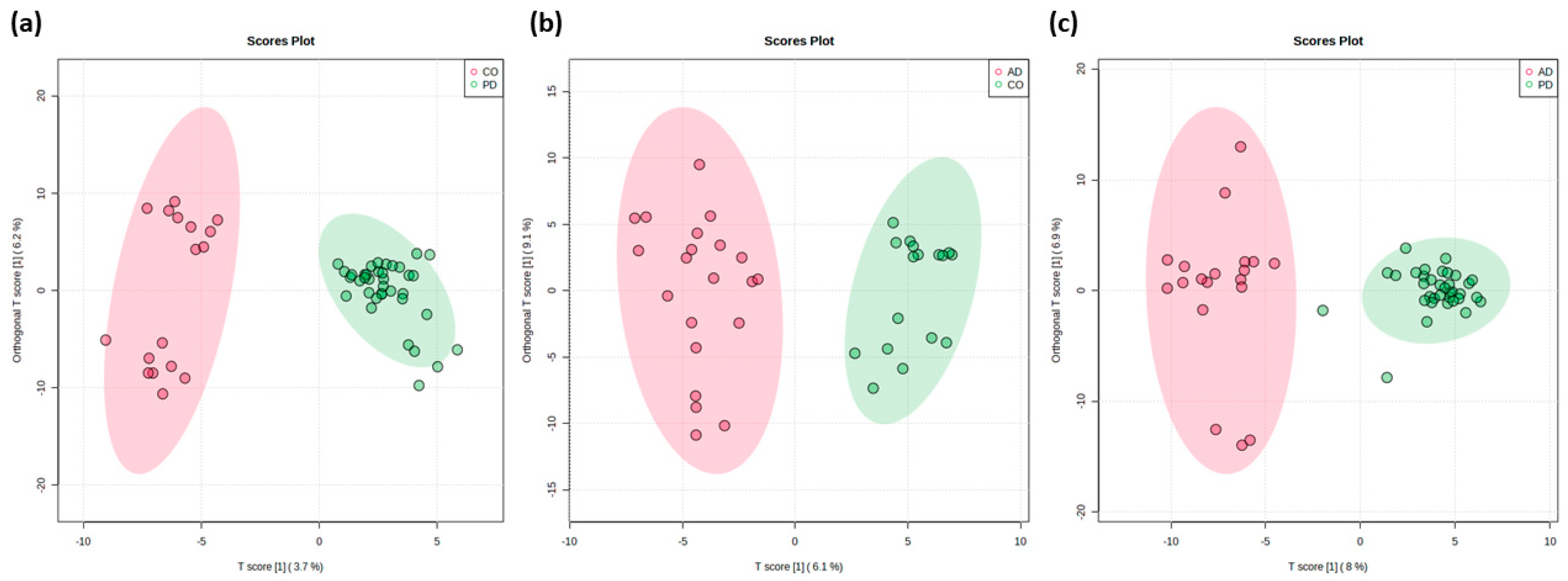

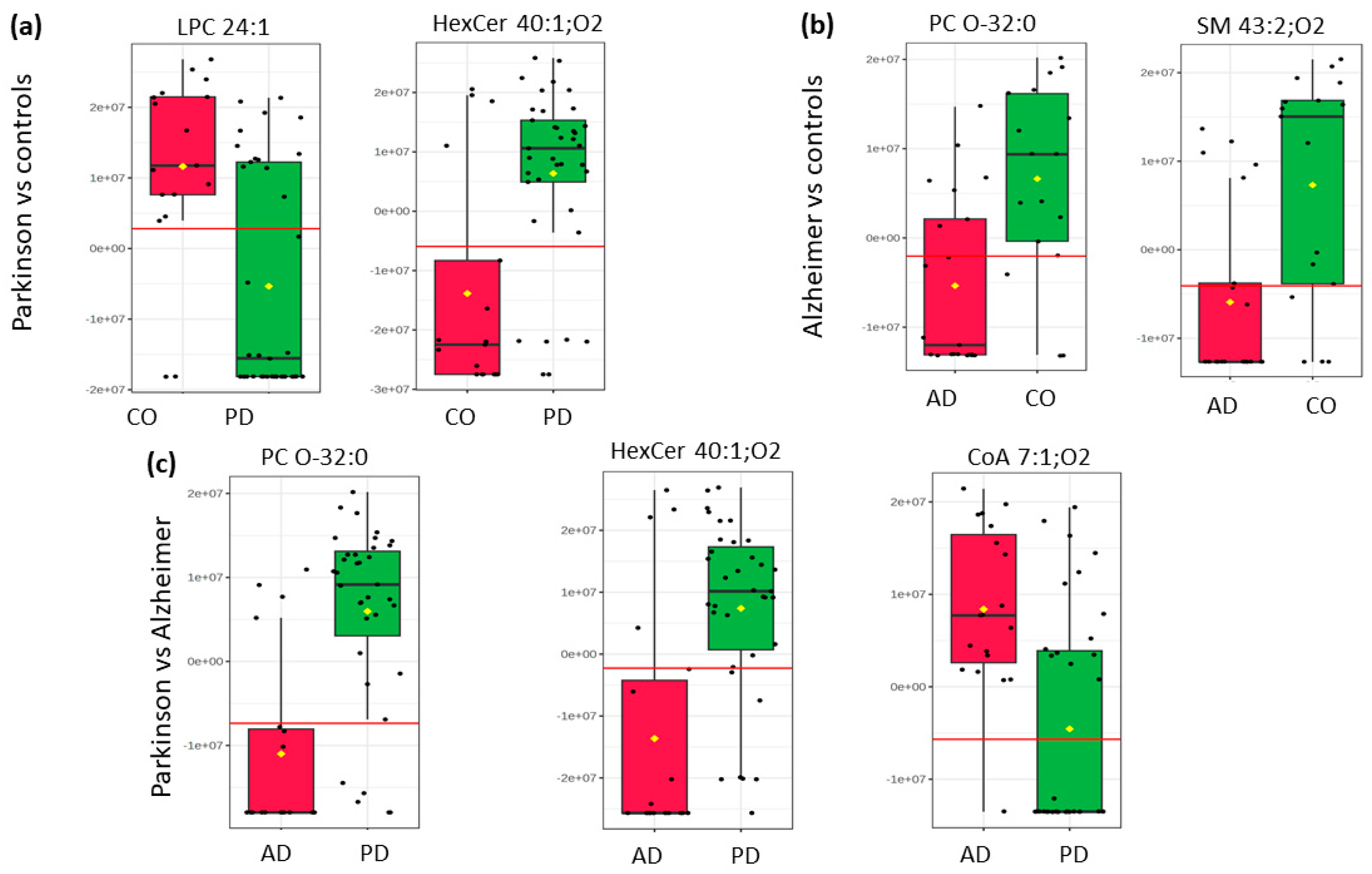
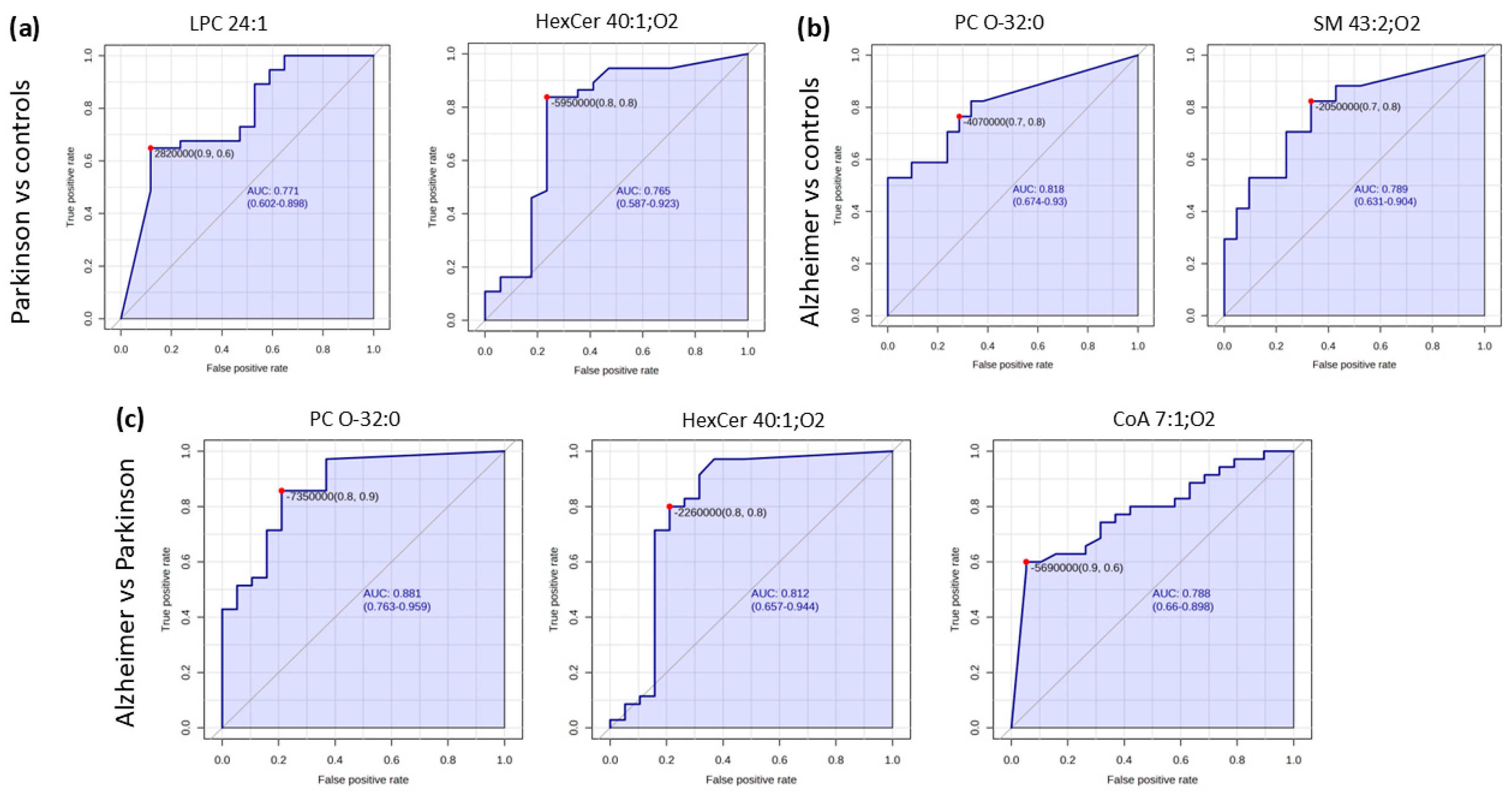
| m/z | ±/m/z | Name | Ion | Category | t Stat | p Value | log2(FC) | VIP Score |
|---|---|---|---|---|---|---|---|---|
| PD vs. AD | ||||||||
| 782.6632 | 0.0117 | HexCer 40:1;O2 | [M − H]− | Sphingolipids | −4.57 | 0.00003 | −1.4582 | 2.43 |
| 764.5892 | 0.0081 | PC O-32:0 | [M + Formate]− | Glycerophospholipids | −5.46 | 0.00000 | −1.7798 | 2.21 |
| 312.7256 | 0.0103 | CoA 7:1;O4 | [M − 3H]3− | Fatty Acyls | 4.20 | 0.00011 | 1.258 | 2.24 |
| CO vs. AD | ||||||||
| 811.6900 | 0.0202 | SM 43:2;O2 | [M − CH3]− | Phosphosphingolipids | −3.65 | 0.00083 | −1.3194 | 2.00 |
| 764.5892 | 0.0081 | PC O-32:0 | [M + Formate]− | Glycerophosphocholines | −3.54 | 0.00114 | −1.3693 | 1.91 |
| CO vs. PD | ||||||||
| 782.6633 | 0.0117 | HexCer 40:1;O2 | [M − H]− | Sphingolipids | −4.25 | 0.00009 | −1.3118 | 2.63 |
| 301.7632 | 0.0494 | LPC 24:1 | [M − 2H]2− | Ceramides | 3.85 | 0.00032 | 1.1784 | 2.54 |
| Compound | AUC | Sensitivity | Specificity |
|---|---|---|---|
| PD vs. AD | |||
| HexCer 40:1;O2 | 0.806 | 80.00% | 78.00% |
| PC O-32:0 | 0.874 | 77.10% | 78.90% |
| CoA 7:1;O4 | 0.782 | 60.00% | 94.70% |
| CO vs. AD | |||
| SM 43:2;O2 | 0.787 | 70.00% | 76.20% |
| PC O-32:0 | 0.808 | 70.00% | 76.20% |
| CO vs. PD | |||
| Hex Cer 40:1;O2 | 0.765 | 78.40% | 76.50% |
| LPC 24:1 | 0.768 | 62.20% | 88.20% |
| Gender | N· | Age Distribution | Mean Age | |
|---|---|---|---|---|
| Controls | Male | 9 | 60–80 | 69 ± 1 |
| Female | 16 | 43–76 | 59 ± 1 | |
| PD | Male | 29 | 46–82 | 67 ± 1 |
| Female | 16 | 59–85 | 67 ± 1 | |
| AD | Male | 6 | 68–81 | 76 ± 1 |
| Female | 21 | 60–85 | 73 ± 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tkachenko, K.; González-Sáiz, J.M.; Pizarro, C. Untargeted Lipidomic Reveals Potential Biomarkers in Plasma Samples for the Discrimination of Patients Affected by Parkinson’s Disease. Molecules 2025, 30, 850. https://doi.org/10.3390/molecules30040850
Tkachenko K, González-Sáiz JM, Pizarro C. Untargeted Lipidomic Reveals Potential Biomarkers in Plasma Samples for the Discrimination of Patients Affected by Parkinson’s Disease. Molecules. 2025; 30(4):850. https://doi.org/10.3390/molecules30040850
Chicago/Turabian StyleTkachenko, Kateryna, Jose María González-Sáiz, and Consuelo Pizarro. 2025. "Untargeted Lipidomic Reveals Potential Biomarkers in Plasma Samples for the Discrimination of Patients Affected by Parkinson’s Disease" Molecules 30, no. 4: 850. https://doi.org/10.3390/molecules30040850
APA StyleTkachenko, K., González-Sáiz, J. M., & Pizarro, C. (2025). Untargeted Lipidomic Reveals Potential Biomarkers in Plasma Samples for the Discrimination of Patients Affected by Parkinson’s Disease. Molecules, 30(4), 850. https://doi.org/10.3390/molecules30040850







