Directed Evolution of AtMP2 Peptide: Unlocking Enhanced Antibacterial Potential from Anabas testudineus
Abstract
1. Introduction
2. Results and Discussion
2.1. Selection of Peptides
2.2. Antimicrobial Activity
2.2.1. Minimum Inhibitory Concentration Determination
2.2.2. Kirby–Bauer Disk Diffusion
2.3. Cytotoxicity Testing—SRB Assay
2.4. Bioinformatics (Protein-Peptide Docking Analysis)
2.4.1. Inhibition and DNA Replication
2.4.2. Suppression of Transcription
2.4.3. Disruption of Protein Translocation and Folding
2.4.4. Inhibition of Protein Homeostasis
2.4.5. Interruption of Cell Division
2.4.6. Limitations and Potential Impact
3. Materials and Methods
3.1. Peptide Selection
3.2. Peptide Synthesis
3.3. Antimicrobial Activity
3.3.1. Kirby–Bauer Disk Diffusion
3.3.2. MIC-Range Growth Inhibition Assay
3.4. Cytotoxicity Testing (SRB Assay)
3.4.1. Treatment Solution Preparation
3.4.2. Cell Preparation
3.4.3. Treatment Exposure
3.4.4. Cell Fixation and Staining
3.5. Protein Docking Simulation
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Larsson, D.G.J.; Flach, C.F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Lathakumari, R.H.; Vajravelu, L.K.; Satheesan, A.; Ravi, S.; Thulukanam, J. Antibiotics and the gut microbiome: Understanding the impact on human health. Med. Microecol. 2024, 20, 100106. [Google Scholar] [CrossRef]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.R.M.; Mitra, S.; Bin Emran, T.; Dhama, K.; Ripon, K.H.; Gajdács, M.; Sahibzada, M.U.K.; et al. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and prospects. J. Infect. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef]
- Lin, F.Y.; Subramaniam, G.; Ambigai, L.; Ong, G.H. Understanding the Use of Antibiotics and Antibiotic Resistance among Science Stream and Non-Science Stream Undergraduate Students in a Malaysian University. J. Liaquat Univ. Med. Health Sci. 2021, 20, 350–357. [Google Scholar]
- Moretta, A.; Scieuzo, C.; Petrone, A.M.; Salvia, R.; Manniello, M.D.; Franco, A.; Lucchetti, D.; Vassallo, A.; Vogel, H.; Sgambato, A.; et al. Antimicrobial Peptides: A New Hope in Biomedical and Pharmaceutical Fields. Front. Cell. Infect. Microbiol. 2021, 11, 668632. [Google Scholar] [CrossRef]
- Nijnik, A.; Hancock, R. Host defence peptides: Antimicrobial and immunomodulatory activity and potential applications for tackling antibiotic-resistant infections. Emerg. Health Threat. J. 2009, 2, 7078. [Google Scholar] [CrossRef]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef]
- Al-Rasheed, A.; Handool, K.O.; Alhelli, A.M.; Garba, B.; Muhialdin, B.J.; Masomian, M.; Hani, H.; Daud, H.H.M. Assessment of Some Immune Components from The Bioactive Crude Extract Derived from The Epidermal Mucus of Climbing Perch Anabas Testudines. Turk. J. Fish Aquat. Sci. 2020, 20, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Najm, A.A.K.; Azfaralariff, A.; Dyari, H.R.E.; Othman, B.A.; Shahid, M.; Khalili, N.; Law, D.; Alwi, S.S.S.; Fazry, S. Anti-breast cancer synthetic peptides derived from the Anabas testudineus skin mucus fractions. Sci. Rep. 2021, 11, 23182. [Google Scholar] [CrossRef]
- Reygaert, W. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018, 4, 482–501. [Google Scholar] [CrossRef]
- Mba, I.E.; Nweze, E.I. Antimicrobial Peptides Therapy: An Emerging Alternative for Treating Drug-Resistant Bacteria. Yale J. Biol. Med. 2022, 95, 445–463. [Google Scholar]
- Chung, C.R.; Kuo, T.R.; Wu, L.C.; Lee, T.Y.; Horng, J.T. Characterization and identification of antimicrobial peptides with different functional activities. Brief Bioinform. 2020, 21, 1098–1114. [Google Scholar] [CrossRef] [PubMed]
- Hossain, T.J. Methods for screening and evaluation of antimicrobial activity: A review of protocols, advantages, and limitations. Eur. J. Microbiol. Immunol. 2024, 14, 97–115. [Google Scholar] [CrossRef] [PubMed]
- Oyama, L.B.; Olleik, H.; Teixeira, A.C.N.; Guidini, M.M.; Pickup, J.A.; Hui, B.Y.P.; Vidal, N.; Cookson, A.R.; Vallin, H.; Wilkinson, T.; et al. In silico identification of two peptides with antibacterial activity against multidrug-resistant Staphylococcus aureus. NPJ Biofilms Microbiomes 2022, 8, 58. [Google Scholar] [CrossRef]
- Tincho, M.B.; Morris, T.; Meyer, M.; Pretorius, A. Antibacterial Activity of Rationally Designed Antimicrobial Peptides. Int. J. Microbiol. 2020, 2020, 2131535. [Google Scholar] [CrossRef]
- Bahar, A.; Ren, D. Antimicrobial Peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Deber, C.M. Interaction of designed cationic antimicrobial peptides with the outer membrane of Gram-negative bacteria. Sci. Rep. 2024, 14, 1894. [Google Scholar] [CrossRef]
- Malanovic, N.; Lohner, K. Antimicrobial Peptides Targeting Gram-Positive Bacteria. Pharmaceuticals 2016, 9, 59. [Google Scholar] [CrossRef]
- Zhang, Q.-Y.; Yan, Z.-B.; Meng, Y.-M.; Hong, X.-Y.; Shao, G.; Ma, J.-J.; Cheng, X.-R.; Liu, J.; Kang, J.; Fu, C.-Y. Antimicrobial peptides: Mechanism of action, activity and clinical potential. Mil. Med. Res. 2021, 8, 48. [Google Scholar] [CrossRef]
- Theansungnoen, T.; Maijaroen, S.; Jangpromma, N.; Yaraksa, N.; Daduang, S.; Temsiripong, T.; Daduang, J.; Klaynongsruang, S. Cationic Antimicrobial Peptides Derived from Crocodylus siamensis Leukocyte Extract, Revealing Anticancer Activity and Apoptotic Induction on Human Cervical Cancer Cells. Protein J. 2016, 35, 202–211. [Google Scholar] [CrossRef]
- Kamiloglu, S.; Sari, G.; Ozdal, T.; Capanoglu, E. Guidelines for cell viability assays. Food Front. 2020, 1, 332–349. [Google Scholar] [CrossRef]
- Khalef, L.; Lydia, R.; Filicia, K.; Moussa, B. Cell viability and cytotoxicity assays: Biochemical elements and cellular compartments. Cell Biochem. Funct. 2024, 42, e4007. [Google Scholar] [CrossRef]
- Macari, G.; Toti, D.; Pasquadibisceglie, A.; Polticelli, F. DockingApp RF: A State-of-the-Art Novel Scoring Function for Molecular Docking in a User-Friendly Interface to AutoDock Vina. Int. J. Mol. Sci. 2020, 21, 9548. [Google Scholar] [CrossRef] [PubMed]
- Sada, M.; Kimura, H.; Nagasawa, N.; Akagawa, M.; Okayama, K.; Shirai, T.; Sunagawa, S.; Kimura, R.; Saraya, T.; Ishii, H.; et al. Molecular Evolution of the Pseudomonas aeruginosa DNA Gyrase gyrA Gene. Microorganisms 2022, 10, 1660. [Google Scholar] [CrossRef] [PubMed]
- Li, M.-c.; Lu, J.; Lu, Y.; Xiao, T.-y.; Liu, H.-c.; Lin, S.-q.; Xu, D.; Li, G.-l.; Zhao, X.-q.; Liu, Z.-g.; et al. rpoB Mutations and Effects on Rifampin Resistance in Mycobacterium tuberculosis. Infect. Drug Resist. 2021, 14, 4119–4128. [Google Scholar] [CrossRef]
- Gupta, R.; Toptygin, D.; Kaiser, C.M. The SecA motor generates mechanical force during protein translocation. Nat. Commun. 2020, 11, 3802. [Google Scholar] [CrossRef]
- Fourie, K.R.; Wilson, H.L. Understanding GroEL and DnaK Stress Response Proteins as Antigens for Bacterial Diseases. Vaccines 2020, 8, 773. [Google Scholar] [CrossRef]
- Nouri, K.; Feng, Y.; Schimmer, A.D. Mitochondrial ClpP serine protease: Biological function and emerging target for cancer therapy. Cell Death Dis. 2020, 11, 841. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Lee, H.; Cho, K.H.; Kim, H.S. Mutations in MetG (methionyl-tRNA synthetase) and TrmD [tRNA (guanine-N1)-methyltransferase] confer meropenem tolerance in Burkholderia thailandensis. J. Antimicrob. Chemother. 2018, 73, 332–338. [Google Scholar] [CrossRef]
- Xiao, J.; Goley, E.D. Redefining the roles of the FtsZ-ring in bacterial cytokinesis. Curr. Opin. Microbiol. 2016, 34, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Roca, J. Transcriptional inhibition by DNA torsional stress. Transcription 2011, 2, 82–85. [Google Scholar] [CrossRef]
- Drlica, K.; Zhao, X.; Malik, M. Quinolones. In Encyclopedia of Microbiology; Elsevier: Amsterdam, The Netherlands, 2009; pp. 707–716. [Google Scholar]
- Dwyer, D.J.; Kohanski, M.A.; Hayete, B.; Collins, J.J. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol. Syst. Biol. 2007, 3, 91. [Google Scholar] [CrossRef]
- Ruan, S.; Tu, C.H.; Bourne, C.R. Friend or Foe: Protein Inhibitors of DNA Gyrase. Biology 2024, 13, 84. [Google Scholar] [CrossRef]
- Vasilyev, N.; Liu, M.M.J.; Epshtein, V.; Shamovsky, I.; Nudler, E. General transcription factor from Escherichia coli with a distinct mechanism of action. Nat. Struct. Mol. Biol. 2024, 31, 141–149. [Google Scholar] [CrossRef]
- Jia, X.; He, X.; Huang, C.; Li, J.; Dong, Z.; Liu, K. Protein translation: Biological processes and therapeutic strategies for human diseases. Signal Transduct. Target Ther. 2024, 9, 44. [Google Scholar] [CrossRef]
- Almanza, A.; Carlesso, A.; Chintha, C.; Creedican, S.; Doultsinos, D.; Leuzzi, B.; Luís, A.; McCarthy, N.; Montibeller, L.; More, S.; et al. Endoplasmic reticulum stress signalling—From basic mechanisms to clinical applications. FEBS J. 2019, 286, 241–278. [Google Scholar] [CrossRef]
- Miller, M.A.; Zachary, Z.J. Mechanisms and Morphology of Cellular Injury, Adaptation, and Death. In Pathologic Basis of Veterinary Disease; Elsevier: Amsterdam, The Netherlands, 2017; p. 2-43.e19. [Google Scholar]
- Ajmal, M.R. Protein Misfolding and Aggregation in Proteinopathies: Causes, Mechanisms, and Cellular Response. Diseases 2023, 11, 30. [Google Scholar] [CrossRef] [PubMed]
- UniProt. SYM_ECOLI Entry P00959. Available online: https://www.uniprot.org/uniprotkb/P00959/entry (accessed on 20 March 2023).
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Waghu, F.H.; Barai, R.S.; Gurung, P.; Idicula-Thomas, S. CAMP R3: A database on sequences, structures and signatures of antimicrobial peptides: Table 1. Nucleic Acids Res. 2016, 44, D1094–D1097. [Google Scholar] [CrossRef]
- Taoyuan (TW): Department of Computer Science and Information Engineering, National Central University [Internet]. AMPfun. Available online: http://fdblab.csie.ncu.edu.tw/AMPfun/index.html (accessed on 20 March 2023).
- Hudzicki, J. Kirby-Bauer disk diffusion susceptibility test protocol. Am. Soc. Microbiol. 2009, 15, 1–23. [Google Scholar]
- Koca, Ö. Microbiological Characteristics of Bacillus subtilis Species and their Relationship with Hospital Infections. In Bacterial, Viral, Fungal, and Parasitic Coinfections; IntechOpen: London, UK, 2024. [Google Scholar]
- Lee, Z.Z.; Abraham, R.; O’dEa, M.; Harb, A.; Hunt, K.; Lee, T.; Abraham, S.; Jordan, D. Validation of Selective Agars for Detection and Quantification of Escherichia coli Strains Resistant to Critically Important Antimicrobials. Microbiol. Spectr. 2021, 9, e0066421. [Google Scholar] [CrossRef] [PubMed]
- Kourmouli, A.; Valenti, M.; van Rijn, E.; Beaumont, H.J.E.; Kalantzi, O.-I.; Schmidt-Ott, A.; Biskos, G. Can disc diffusion susceptibility tests assess the antimicrobial activity of engineered nanoparticles? J. Nanopart. Res. 2018, 20, 62. [Google Scholar] [CrossRef]
- Mercer, D.K.; Torres, M.D.T.; Duay, S.S.; Lovie, E.; Simpson, L.; von Köckritz-Blickwede, M.; de la Fuente-Nunez, C.; O’NEil, D.A.; Angeles-Boza, A.M. Antimicrobial Susceptibility Testing of Antimicrobial Peptides to Better Predict Efficacy. Front. Cell. Infect. Microbiol. 2020, 10, 326. [Google Scholar] [CrossRef]
- Kowalska-Krochmal, B.; Dudek-Wicher, R. The Minimum Inhibitory Concentration of Antibiotics: Methods, Interpretation, Clinical Relevance. Pathogens 2021, 10, 165. [Google Scholar] [CrossRef]
- Werner, A. How to Do Serial Dilutions (Including calculations) [Internet]. Available online: https://www.integra-biosciences.com/global/en/blog/article/how-do-serial-dilutions-including-calculations (accessed on 20 November 2023).
- Orellana, E.; Kasinski, A. Sulforhodamine B (SRB) Assay in Cell Culture to Investigate Cell Proliferation. Bio Protoc. 2016, 6, e1984. [Google Scholar] [CrossRef]
- Alsedfy, M.Y.; Ebnalwaled, A.A.; Moustafa, M.; Said, A.H. Investigating the binding affinity, molecular dynamics, and ADMET properties of curcumin-IONPs as a mucoadhesive bioavailable oral treatment for iron deficiency anemia. Sci. Rep. 2024, 14, 22027. [Google Scholar] [CrossRef] [PubMed]









| No | AMPs Sequence | AMPFun | APD3 | ||||
|---|---|---|---|---|---|---|---|
| Antimicrobial Activity | Anticancer Activity | Gram+ | Gram− | Total Hydrophobic Ratio % | Total Net Charge | ||
| 1 | TGIATSGLFTFTLHTGSLAPAT | YES 0.9607 | YES 0.6884 | YES 0.6333 | YES 0.7283 | 41% | 0.25 |
| 2 | TGWATSGLATFTLHTGSLAPAT | YES 0.965 | YES 0.6102 | YES 0.6 | YES 0.7363 | 41% | 0.25 |
| 3 | TGTATSGLATFTLHTGSLAPAT | YES 0.9767 | YES 0.5691 | YES 0.575 | YES 0.6961 | 36% | 0.25 |
| 4 | TGTATSGLATFTLHTGSLAPAT | YES 0.9607 | YES 0.5441 | YES 0.5583 | YES 0.6913 | 36% | 0.25 |
| 5 | TGLATSGLATFTLHTGSLAPAT | YES 0.965 | YES 0.5231 | YES 0.6167 | YES 0.7078 | 41% | 0.25 |
| 6 | TGIATSGLATFTLHTGSLAIAT | YES 0.9719 | YES 0.5218 | YES 0.7417 | YES 0.7798 | 45% | 0.25 |
| 7 | TGMATSGLATFTLHTGSLAPAT | YES 0.956 | YES 0.5191 | YES 0.5583 | YES 0.6837 | 41% | 0.25 |
| 8 | TGIATSGLATFTLHTISLAPAT | YES 0.9526 | YES 0.5151 | YES 0.65 | YES 0.7754 | 45% | 0.25 |
| 9 | TGIATSGLATFTLHTGSLAPIT | YES 0.9638 | YES 0.5113 | YES 0.6333 | YES 0.792 | 41% | 0.25 |
| 10 | TGIATSGLATFTLHTGSLIPAT | YES 0.9583 | YES 0.5113 | YES 0.6583 | YES 0.7509 | 41% | 0.15 |
| 11 | TGIATSGLATFTLHTGSLAFAT | YES 0.9719 | YES 0.5001 | YES 0.725 | YES 0.7497 | 45% | 0.25 |
| 12 | TGIATSGLATITLHTGSLAPAT | YES 0.9538 | YES 0.4991 | YES 0.65 | YES 0.7514 | 41% | 0.25 |
| 13 | TGIATSGLATFTLHTGWLAPAT | YES 0.9626 | YES 0.4953 | YES 0.6583 | YES 0.7774 | 45% | 0.25 |
| 14 | TGIATSGLATFTLHTLSLAPAT | YES 0.9526 | YES 0.4953 | YES 0.65 | YES 0.762 | 45% | 0.25 |
| 15 | TGIATSGLATFTLHTMSLAPAT | YES 0.9396 | YES 0.4953 | YES 0.625 | YES 0.7301 | 45% | 0.25 |
| 16 | TGIATSGLATFTLHTTSLAPAT | YES 0.9626 | YES 0.4953 | YES 0.6583 | YES 0.7288 | 41% | 0.25 |
| 17 | TGIATSGLATFTLHTVSLAPAT | YES 0.9479 | YES 0.4953 | YES 0.65 | YES 0.7538 | 45% | 0.25 |
| 18 | TGIATSGLATFTLHTGSLAPAI | YES 0.983 | YES 0.4952 | YES 0.6833 | YES 0.7967 | 45% | 0.25 |
| 19 | TGIITSGLATFTLHTGSLAPAT | YES 0.9504 | YES 0.4918 | YES 0.5833 | YES 0.7409 | 41% | 0.25 |
| 20 | TGIATSGLATFTLHTGSLACAT | YES 0.9719 | YES 0.4911 | YES 0.7417 | YES 0.768 | 45% | 0.25 |
| Protein | ZDOCK Server | HPEPDOCK Server | Docking Score & Bond Type | Docking Score & Bond Type | ||
|---|---|---|---|---|---|---|
| AtMP2-1 (TGTATSGLATFTLHTGSLAPAT) | AtMP2-2 (TGWATSGLATFTLHTGSLAPAT) | AtMP2-1 (TGTATSGLATFTLHTGSLAPAT) | AtMP2-2 (TGWATSGLATFTLHTGSLAPAT) | AtMP2-1 Retrieved on (25 September 2024) | AtMP2-2 Retrieved on (25 September 2024) | |
| GyrA |  Link Retrieved on (25 September 2024) |  Link Retrieved on (25 September 2024) |  Link Retrieved on (25 September 2024) |  Link Retrieved on (26 September 2024) | −199.731 Single | −209.673 Single |
| GyrB |  Link Retrieved on (25 September 2024) |  Link Retrieved on (25 September 2024) |  Link Retrieved on (25 September 2024) |  Link Retrieved on (26 September 2024) | −215.102 Single | −204.34 Single |
| RpoB |  Link Retrieved on (25 September 2024) |  Link Retrieved on (25 September 2024) |  Link Retrieved on (25 September 2024) |  Link Retrieved on (26 September 2024) | −184.743 Single | −182.555 Single |
| SecA |  Link Retrieved on (25 September 2024) |  Link Retrieved on (25 September 2024) |  Link Retrieved on (25 September 2024) |  Link Retrieved on (26 September 2024) | −216.083 Single | −230.044 Single |
| GroEL |  Link Retrieved on (25 September 2024) |  Link Retrieved on (25 September 2024) |  Link Retrieved on (25 September 2024) |  Link Retrieved on (26 September 2024) | −184.075 Single | −186.028 Single |
| ParE |  Link Retrieved on (25 September 2024) |  Link Retrieved on (25 September 2024) |  Link Retrieved on (25 September 2024) |  Link Retrieved on (26 September 2024) | −201.482 Single | −195.991 Single |
| DnaK |  Link Retrieved on (25 September 2024) |  Link Retrieved on (25 September 2024) |  Link Retrieved on (26 September 2024) |  Link Retrieved on (26 September 2024) | −211.956 Single | −211.956 Single |
| ClpP |  Link Retrieved on (25 September 2024) |  Link Retrieved on (25 September 2024) |  Link Retrieved on (26 September 2024) |  Link Retrieved on (26 September 2024) | −235.844 Single | −507.438 Single |
| MetG |  Link Retrieved on (25 September 2024) |  Link Retrieved on (25 September 2024) | 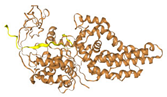 Link Retrieved on (26 September 2024) |  Link Retrieved on (26 September 2024) | −238.677 Single | −235.14 Single |
| FtsZ |  Link Retrieved on (25 September 2024) |  Link Retrieved on (25 September 2024) |  Link Retrieved on (26 September 2024) |  Link Retrieved on (26 September 2024) | −173.444 Single | −186.951 Single |
| Gene/ Protein | Role in Bacterial Cell | Cell Cycle/ Process | Putative Mechanism (Based on Docking Predictions and Literature) | Reference |
|---|---|---|---|---|
| GyrA | Involved in DNA replication; mutations can lead to resistance | DNA Replication | Disrupts DNA replication, leading to bacterial cell death | [25] |
| RpoB | Part of RNA polymerase; mutations can affect transcription and antibiotic resistance | Transcription | Interferes with transcription, reducing bacterial protein production | [26] |
| SecA | Involved in protein translocation across membranes | Protein Transport | Disrupts protein transport, impairing cell functionality | [27] |
| GyrB | Assists in DNA replication; plays a role in cell division | DNA Replication and Cell Division | Disrupts DNA replication, affecting cell division and survival | [25] |
| GroEL | Chaperone protein involved in protein folding; essential for cell survival | Protein Folding | Inhibits protein folding, leading to the accumulation of misfolded proteins | [28] |
| ParE | Involved in DNA replication and repair; contributes to bacterial growth | DNA Replication and Repair | Interferes with DNA repair mechanisms, reducing bacterial viability | [26] |
| DnaK | Chaperone protein aids in stress response and protein refolding | Stress Response and Protein Refolding | Disrupts stress response, preventing refolding of damaged proteins | [28] |
| ClpP | Protease that degrades misfolded proteins, crucial for bacterial survival | Protein Quality Control | Inhibits protease activity, leading to the accumulation of damaged proteins | [29] |
| MetG | Involved in protein synthesis, specifically tRNA synthetase activity | Protein Synthesis | Impairs protein synthesis, reducing bacterial growth | [30] |
| FtsZ | Key player in bacterial cell division, forming the Z-ring at the site of division | Cell Division | Disrupts cell division by preventing the proper formation of the Z-ring | [31] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, L.T.; Ang, A.; Najm, A.; Adnan, A.M.; Nordin, A.M.; Mahmood, I.; Dunkhorol, S.; Fazry, S.; Law, D. Directed Evolution of AtMP2 Peptide: Unlocking Enhanced Antibacterial Potential from Anabas testudineus. Molecules 2025, 30, 4590. https://doi.org/10.3390/molecules30234590
Lee LT, Ang A, Najm A, Adnan AM, Nordin AM, Mahmood I, Dunkhorol S, Fazry S, Law D. Directed Evolution of AtMP2 Peptide: Unlocking Enhanced Antibacterial Potential from Anabas testudineus. Molecules. 2025; 30(23):4590. https://doi.org/10.3390/molecules30234590
Chicago/Turabian StyleLee, Li Ting, Arnold Ang, Ahmed Najm, Adura Mohd Adnan, Akram Mohd Nordin, Ibrahim Mahmood, Sarantuya Dunkhorol, Shazrul Fazry, and Douglas Law. 2025. "Directed Evolution of AtMP2 Peptide: Unlocking Enhanced Antibacterial Potential from Anabas testudineus" Molecules 30, no. 23: 4590. https://doi.org/10.3390/molecules30234590
APA StyleLee, L. T., Ang, A., Najm, A., Adnan, A. M., Nordin, A. M., Mahmood, I., Dunkhorol, S., Fazry, S., & Law, D. (2025). Directed Evolution of AtMP2 Peptide: Unlocking Enhanced Antibacterial Potential from Anabas testudineus. Molecules, 30(23), 4590. https://doi.org/10.3390/molecules30234590





