Deciphering the Impact of Mutations on PfDHPS Active Site and Sulfadoxine Binding: Structural Insights from Molecular Dynamics Simulations
Abstract
1. Introduction
2. Results
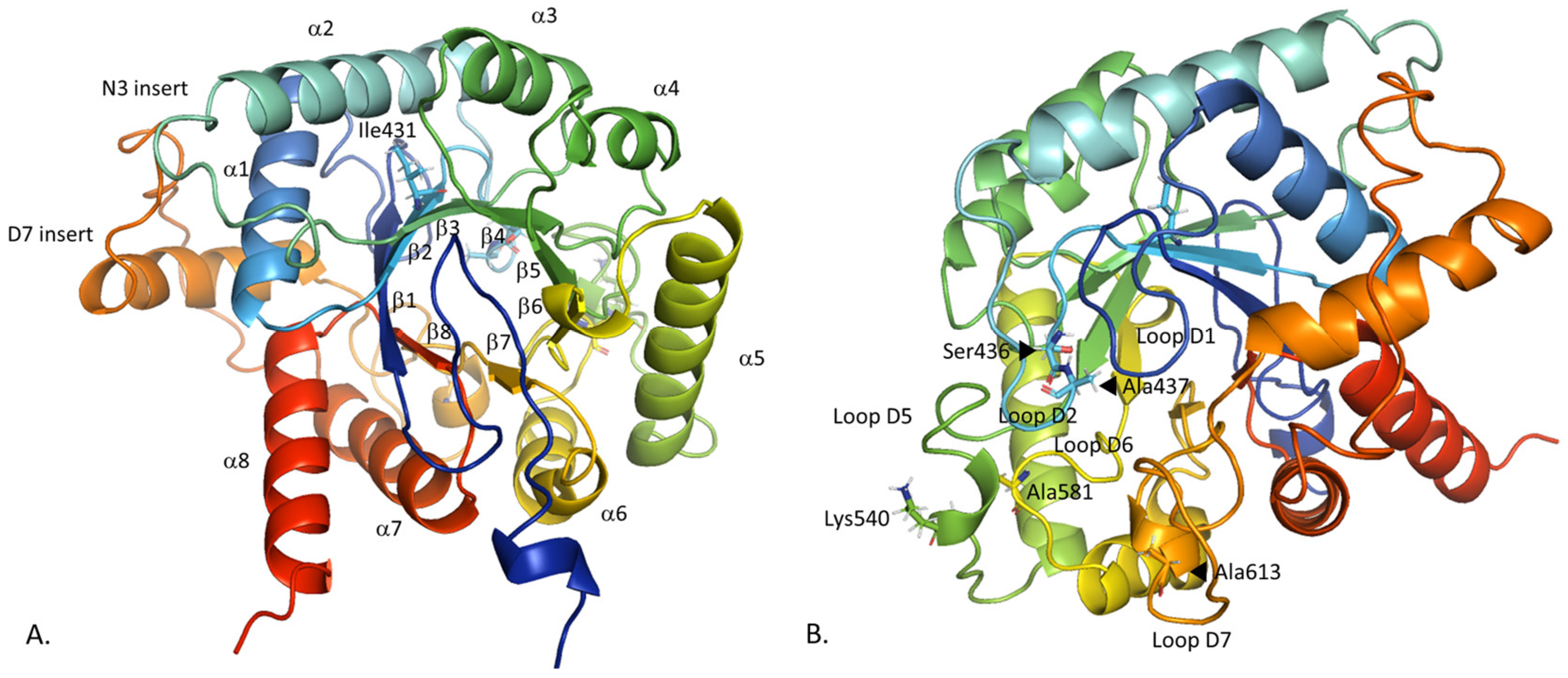
2.1. Comparison of the Constructed PfDHPS Structure with the AlphaFold2 Prediction
2.2. Compared Dynamics Properties of the Wild-Type and Mutated PfDHPS in Complex with pABA
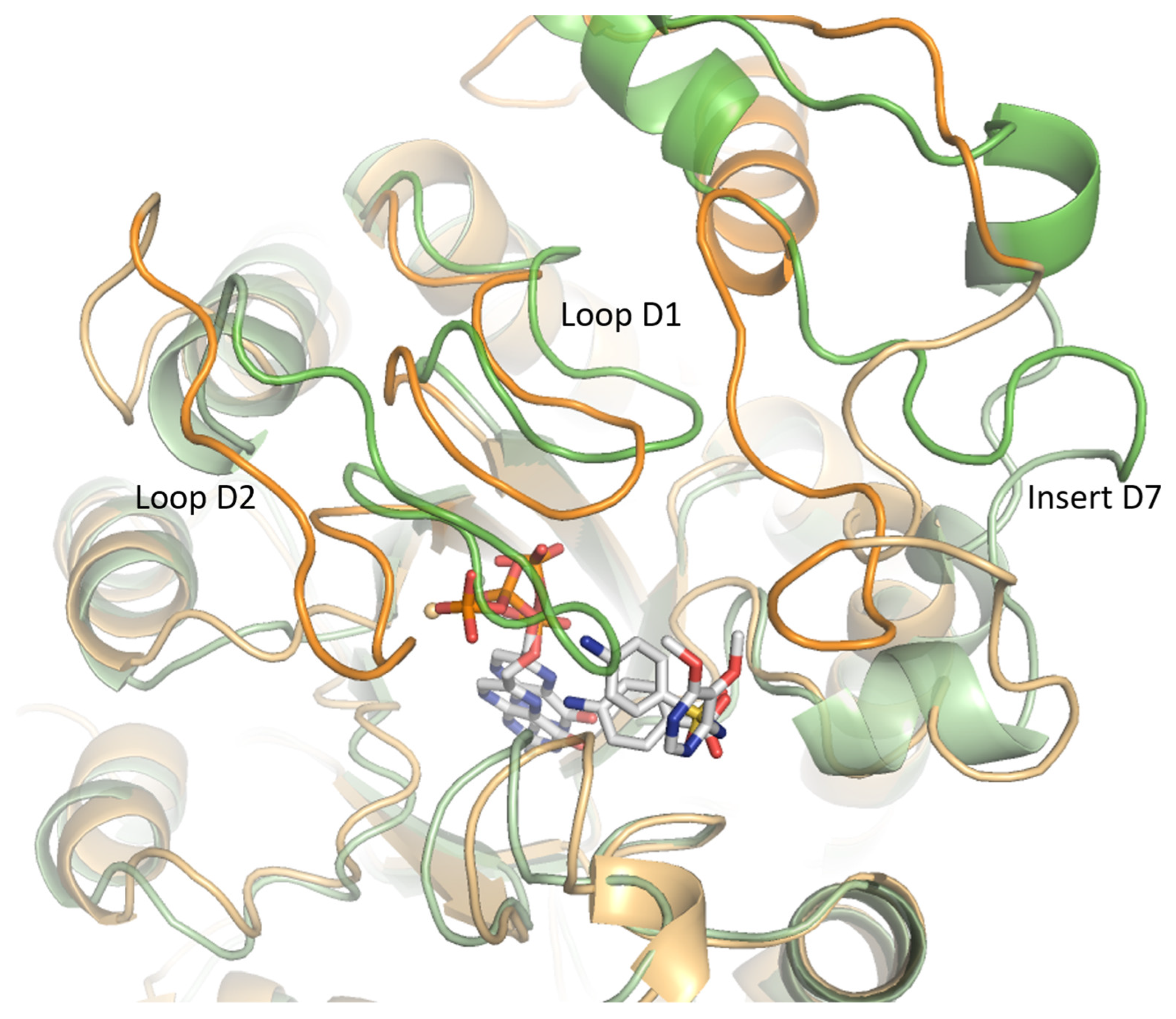
2.2.1. Insert D7 Instability
2.2.2. Loop Flexibility
2.3. Conformational Analysis of PfDHPS Active Site in Complex with pABA
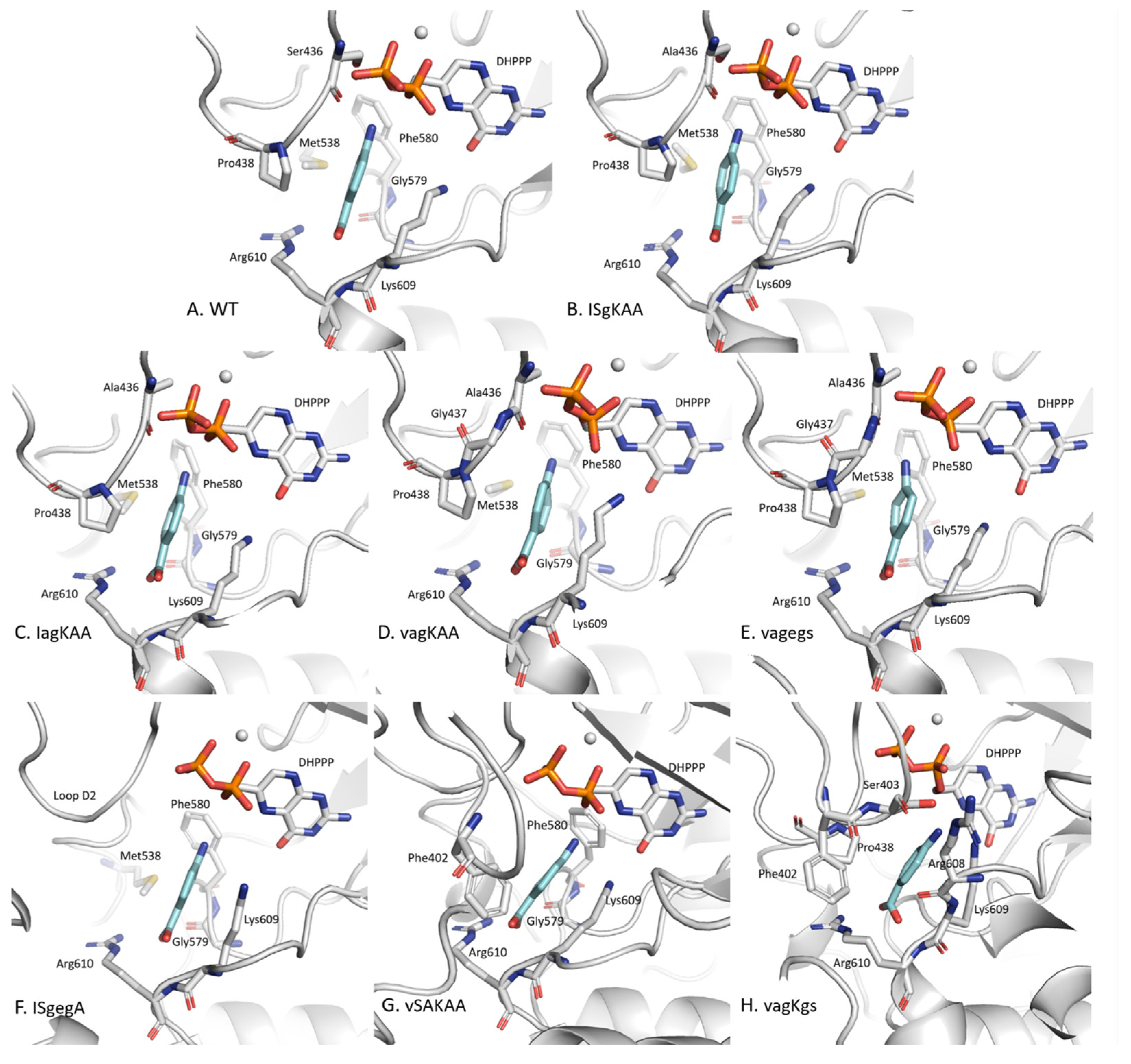
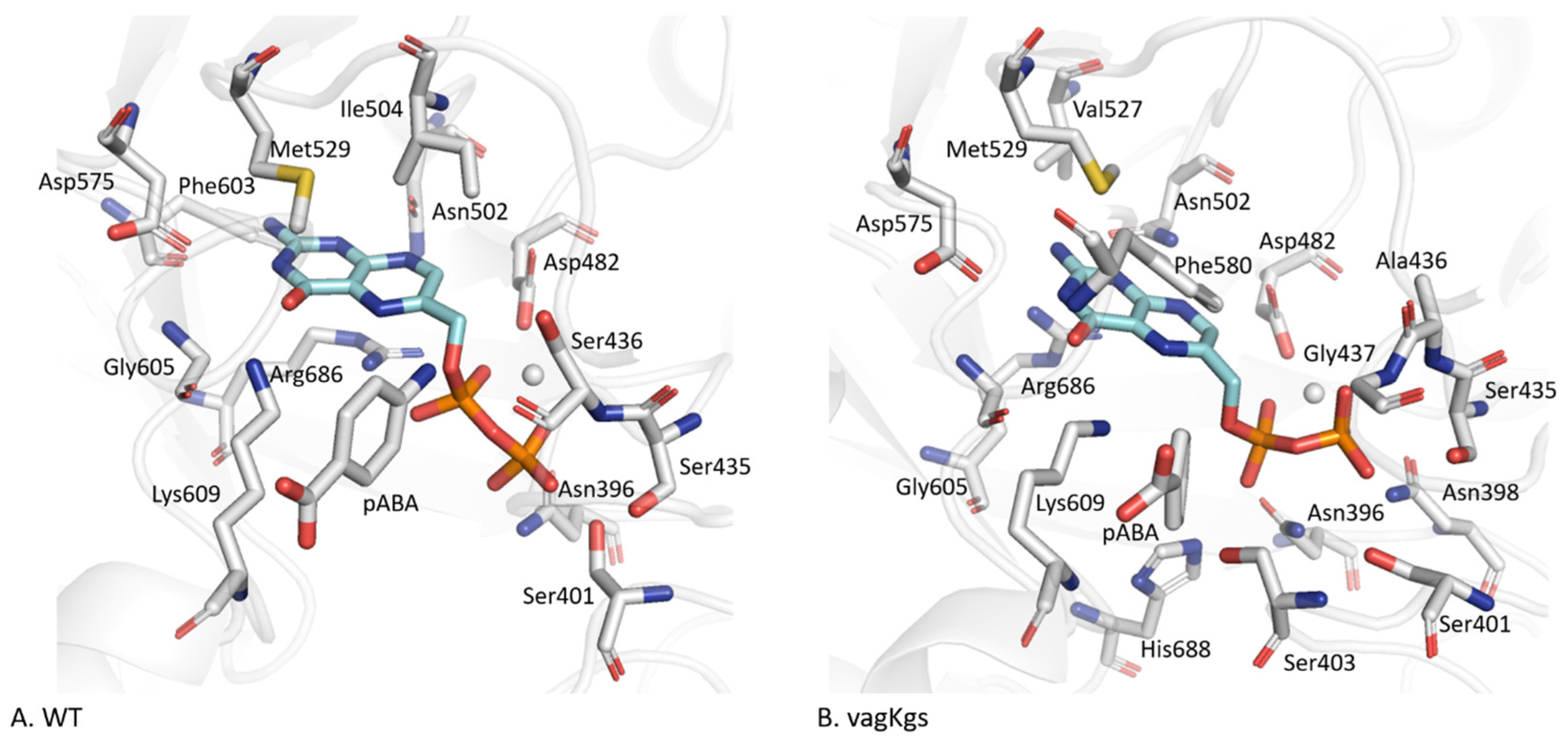
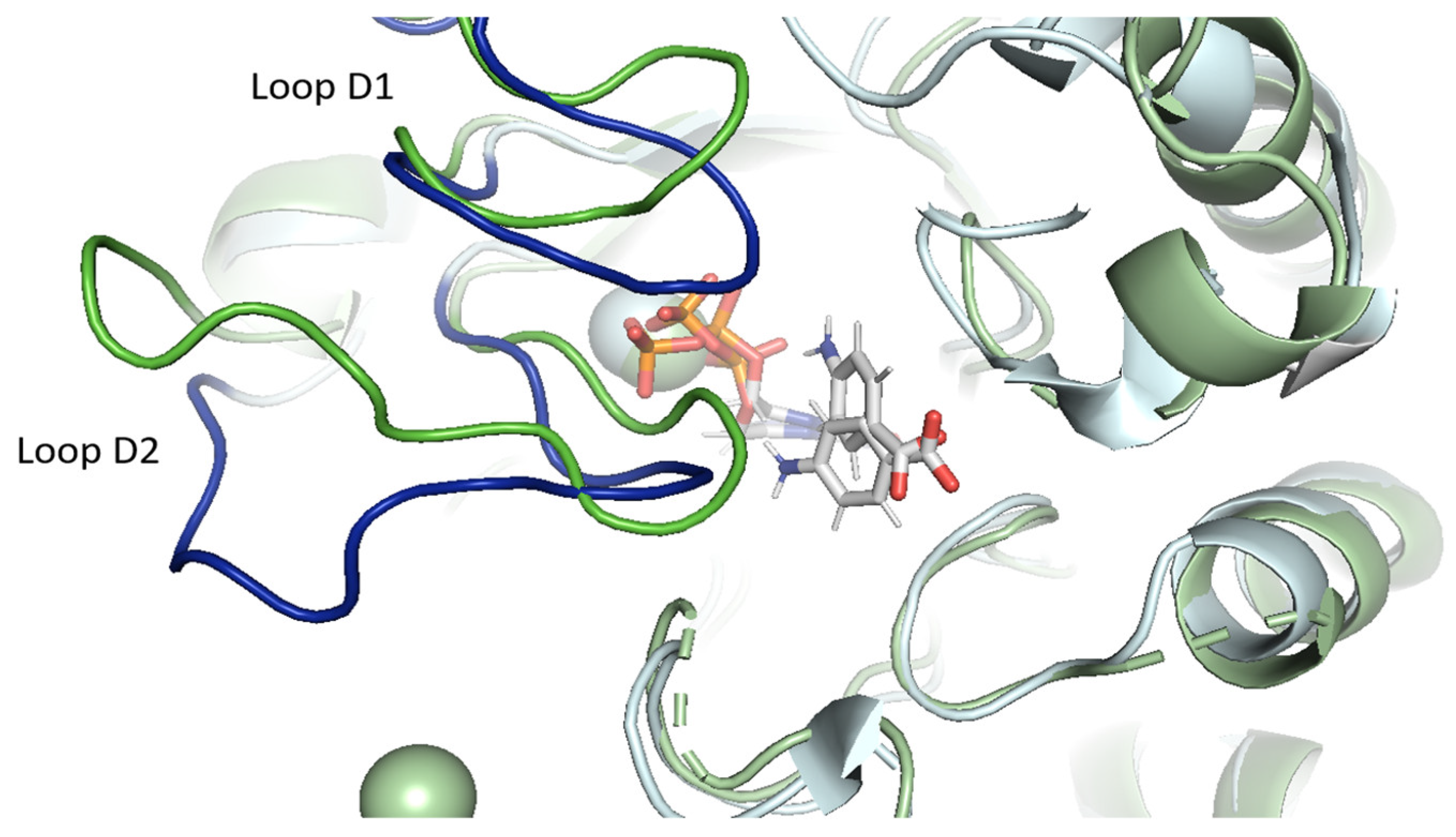
2.3.1. pABA Binding Pocket
2.3.2. The Distinctive Case of the vagKgs System
2.3.3. Binding Energies
2.3.4. DHPPP Binding Pocket
2.4. Structural Reorganization of the Active Site in the Case of vagKgs Mutant
2.5. Comparison in Presence of Sulfadoxine
2.5.1. Sulfadoxine Instability
2.5.2. Conformational Variability of Sulfadoxine
2.5.3. Enzyme Stability
2.6. Conformational Analysis of PfDHPS Active Site in Complex with Sulfadoxine
2.6.1. WT PfDHPS
2.6.2. PfDHPS Mutants
3. Discussion
4. Materials and Methods
4.1. Molecular Structures
4.2. System Preparation
4.3. Molecular Dynamics Simulations
4.4. MD Trajectory Analysis
4.5. IGMPlot
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DHFR | dihydrofolate reductase |
| DHP | 7,8-dihydropteroate |
| DHPPP | 6-hydroxymethyl-7,8-dihydropterin pyrophosphate |
| DHPS | dihydropteroate synthase |
| HPPK | 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase |
| MD | molecular dynamics |
| pABA | p-aminobenzoic acid |
| PDB | protein data bank |
| SP | sulfadoxine–pyrimethamine |
| SDX | sulfadoxine |
References
- WHO World Malaria Report 2024. Available online: https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2024 (accessed on 25 July 2025).
- Kasekarn, W.; Sirawaraporn, R.; Chahomchuen, T.; Cowman, A.F.; Sirawaraporn, W. Molecular Characterization of Bifunctional Hydroxymethyldihydropterin Pyrophosphokinase-Dihydropteroate Synthase from Plasmodium falciparum. Mol. Biochem. Parasitol. 2004, 137, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Chitnumsub, P.; Jaruwat, A.; Talawanich, Y.; Noytanom, K.; Liwnaree, B.; Poen, S.; Yuthavong, Y. The Structure of Plasmodium falciparum Hydroxymethyldihydropterin Pyrophosphokinase-Dihydropteroate Synthase Reveals the Basis of Sulfa Resistance. FEBS J. 2020, 287, 3273–3297. [Google Scholar] [CrossRef]
- Sutherland, C.J.; Fifer, H.; Pearce, R.J.; bin Reza, F.; Nicholas, M.; Haustein, T.; Njimgye-Tekumafor, N.E.; Doherty, J.F.; Gothard, P.; Polley, S.D.; et al. Novel Pfdhps Haplotypes Among Imported Cases of Plasmodium falciparum Malaria in the United Kingdom. Antimicrob. Agents Chemother. 2009, 53, 3405–3410. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chauvin, P.; Menard, S.; Iriart, X.; Nsango, S.E.; Tchioffo, M.T.; Abate, L.; Awono-Ambéné, P.H.; Morlais, I.; Berry, A. Prevalence of Plasmodium falciparum Parasites Resistant to Sulfadoxine/Pyrimethamine in Pregnant Women in Yaoundé, Cameroon: Emergence of Highly Resistant Pfdhfr/Pfdhps Alleles. J. Antimicrob. Chemother. 2015, 70, 2566–2571. [Google Scholar] [CrossRef]
- Guémas, E.; Coppée, R.; Ménard, S.; du Manoir, M.; Nsango, S.; Mvumbi, D.M.; Nakoune, E.; Moukoko, C.E.E.; Akotet, M.K.B.; Mirabeau, T.Y.; et al. Evolution and Spread of Plasmodium falciparum Mutations Associated with Resistance to Sulfadoxine–Pyrimethamine in Central Africa: A Cross-Sectional Study. Lancet Microbe 2023, 4, e983–e993. [Google Scholar] [CrossRef]
- Naidoo, I.; Roper, C. Mapping “Partially Resistant”, “Fully Resistant”, and “super Resistant” Malaria. Trends Parasitol. 2013, 29, 505–515. [Google Scholar] [CrossRef]
- Korsinczky, M.; Fischer, K.; Chen, N.; Baker, J.; Rieckmann, K.; Cheng, Q. Sulfadoxine Resistance in Plasmodium vivax Is Associated with a Specific Amino Acid in Dihydropteroate Synthase at the Putative Sulfadoxine-Binding Site. Antimicrob. Agents Chemother. 2004, 48, 2214–2222. [Google Scholar] [CrossRef]
- de Beer, T.A.P.; Louw, A.I.; Joubert, F. Elucidation of Sulfadoxine Resistance with Structural Models of the Bifunctional Plasmodium falciparum Dihydropterin Pyrophosphokinase-Dihydropteroate Synthase. Bioorg Med. Chem. 2006, 14, 4433–4443. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, C.; Klein, J.; Khartabil, H.; Boisson, J.-C.; Hénon, E. IGMPlot: A Program to Identify, Characterize, and Quantify Molecular Interactions. J. Comput. Chem. 2023, 44, 1750–1766. [Google Scholar] [CrossRef]
- Bannwarth, C.; Ehlert, S.; Grimme, S. GFN2-xTB-An Accurate and Broadly Parametrized Self-Consistent Tight-Binding Quantum Chemical Method with Multipole Electrostatics and Density-Dependent Dispersion Contributions. J. Chem. Theory Comput. 2019, 15, 1652–1671. [Google Scholar] [CrossRef]
- Oguike, M.C.; Falade, C.O.; Shu, E.; Enato, I.G.; Watila, I.; Baba, E.S.; Bruce, J.; Webster, J.; Hamade, P.; Meek, S.; et al. Molecular Determinants of Sulfadoxine-Pyrimethamine Resistance in Plasmodium falciparum in Nigeria and the Regional Emergence of Dhps 431V. Int. J. Parasitol. Drugs Drug Resist. 2016, 6, 220–229. [Google Scholar] [CrossRef]
- Boateng, R.A.; Myers-Hansen, J.L.; Dolling, N.N.O.; Mensah, B.A.; Brodsky, E.; Mazumder, M.; Ghansah, A. Global Analysis of Plasmodium falciparum Dihydropteroate Synthase Variants Associated with Sulfadoxine Resistance Reveals Variant Distribution and Mechanisms of Resistance: A Computational-Based Study. Molecules 2022, 28, 145. [Google Scholar] [CrossRef]
- Birkholtz, L.-M.; Wrenger, C.; Joubert, F.; Wells, G.A.; Walter, R.D.; Louw, A.I. Parasite-Specific Inserts in the Bifunctional S-Adenosylmethionine Decarboxylase/Ornithine Decarboxylase of Plasmodium falciparum Modulate Catalytic Activities and Domain Interactions. Biochem. J. 2004, 377, 439–448. [Google Scholar] [CrossRef]
- Yogavel, M.; Nettleship, J.E.; Sharma, A.; Harlos, K.; Jamwal, A.; Chaturvedi, R.; Sharma, M.; Jain, V.; Chhibber-Goel, J.; Sharma, A. Structure of 6-Hydroxymethyl-7,8-Dihydropterin Pyrophosphokinase-Dihydropteroate Synthase from Plasmodium vivax Sheds Light on Drug Resistance. J. Biol. Chem. 2018, 293, 14962–14972. [Google Scholar] [CrossRef]
- DeLano, W.L. Pymol: An Open-Source Molecular Graphics Tool. CCP4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Case, D.A.; Aktulga, H.M.; Belfon, K.; Cerutti, D.S.; Cisneros, G.A.; Cruzeiro, V.W.D.; Forouzesh, N.; Giese, T.J.; Götz, A.W.; Gohlke, H.; et al. AmberTools. J. Chem. Inf. Model. 2023, 63, 6183–6191. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and Testing of a General Amber Force Field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Antechamber: An Accessory Software Package for Molecular Mechanical Calculations. J. Am. Chem. Soc. 2001, 222, 2001. [Google Scholar]
- Mark, P.; Nilsson, L. Structure and Dynamics of the TIP3P, SPC, and SPC/E Water Models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA Methods to Estimate Ligand-Binding Affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Ester, M.; Kriegel, H.-P.; Sander, J.; Xu, X. A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. In Proceedings of the Second International Conference on Knowledge Discovery and Data Mining, Portland, OR, USA, 2–4 August 1996; AAAI Press: Portland, OR, USA, 1996; pp. 226–231. [Google Scholar]



| With Insert D7 | Without Insert D7 | |
|---|---|---|
| Average ± Std. Dev. (Å) | Average ± Std. Dev. (Å) | |
| WT | 6.20 ± 0.69 | 2.73 ± 0.23 |
| ISgKAA | 4.85 ± 1.04 | 2.55 ± 0.38 |
| IagKAA | 4.91 ± 0.93 | 2.94 ± 0.50 |
| vSAKAA | 4.08 ± 0.73 | 2.87 ± 0.51 |
| vagKAA | 4.94 ± 1.63 | 2.69 ± 0.41 |
| vagKgs | 3.58 ± 0.63 | 2.59 ± 0.33 |
| ISgegA | 4.56 ± 1.85 | 2.80 ± 0.51 |
| vagegs | 4.28 ± 1.16 | 2.92 ± 0.57 |
| Average (kcal/mol) | Std. Dev. (kcal/mol) | |
|---|---|---|
| WT | −19.72 | 2.85 |
| ISgKAA | −18.42 | 3.42 |
| IagKAA | −20.43 | 2.79 |
| vSAKAA | −18.41 | 3.96 |
| vagKAA | −19.79 | 3.86 |
| vagKgs | −23.60 | 3.69 |
| ISgegA | −16.51 | 4.89 |
| vagegs | −20.33 | 2.74 |
| Average (Å) | Std. Dev. (Å) | |
|---|---|---|
| WT | 3.64 | 0.32 |
| ISgKAA | 3.83 | 0.44 |
| IagKAA | 3.60 | 0.30 |
| vSAKAA | 3.82 | 0.77 |
| vagKAA | 3.45 | 0.28 |
| vagKgs | 3.43 | 0.25 |
| ISgegA | 3.84 | 0.47 |
| vagegs | 3.45 | 0.24 |
| Average ± Std. Dev. (Å) | |
|---|---|
| WT | 2.69 ± 0.36 |
| ISgKAA | 3.42 ± 0.38 |
| IagKAA | 3.18 ± 0.38 |
| vSAKAA | 2.96 ± 0.31 |
| vagKAA | 2.68 ± 0.36 |
| vagKgs | 2.79 ± 0.39 |
| ISgegA | 3.72 ± 0.60 |
| vagegs | 3.36 ± 0.59 |
| Average (kcal/mol) | Std. Dev. (kcal/mol) | |
|---|---|---|
| WT | −19.06 | 3.56 |
| ISgKAA | −16.67 | 5.58 |
| IagKAA | −20.24 | 4.45 |
| vagKAA | −17.94 | 5.80 |
| vagKgs | −18.92 | 4.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guémas, E.; Ménard, S.; Jeanne, N.; Landa, G.; Berry, A.; Brut, M. Deciphering the Impact of Mutations on PfDHPS Active Site and Sulfadoxine Binding: Structural Insights from Molecular Dynamics Simulations. Molecules 2025, 30, 4118. https://doi.org/10.3390/molecules30204118
Guémas E, Ménard S, Jeanne N, Landa G, Berry A, Brut M. Deciphering the Impact of Mutations on PfDHPS Active Site and Sulfadoxine Binding: Structural Insights from Molecular Dynamics Simulations. Molecules. 2025; 30(20):4118. https://doi.org/10.3390/molecules30204118
Chicago/Turabian StyleGuémas, Emilie, Sandie Ménard, Nicolas Jeanne, Georges Landa, Antoine Berry, and Marie Brut. 2025. "Deciphering the Impact of Mutations on PfDHPS Active Site and Sulfadoxine Binding: Structural Insights from Molecular Dynamics Simulations" Molecules 30, no. 20: 4118. https://doi.org/10.3390/molecules30204118
APA StyleGuémas, E., Ménard, S., Jeanne, N., Landa, G., Berry, A., & Brut, M. (2025). Deciphering the Impact of Mutations on PfDHPS Active Site and Sulfadoxine Binding: Structural Insights from Molecular Dynamics Simulations. Molecules, 30(20), 4118. https://doi.org/10.3390/molecules30204118





