Mitochondrial Proteins as Exosomal Cargo: New Breast Cancer Biomarkers & Crucial Players in Carcinogenesis?
Abstract
1. Introduction
2. Results
2.1. Characterization of Exosomes
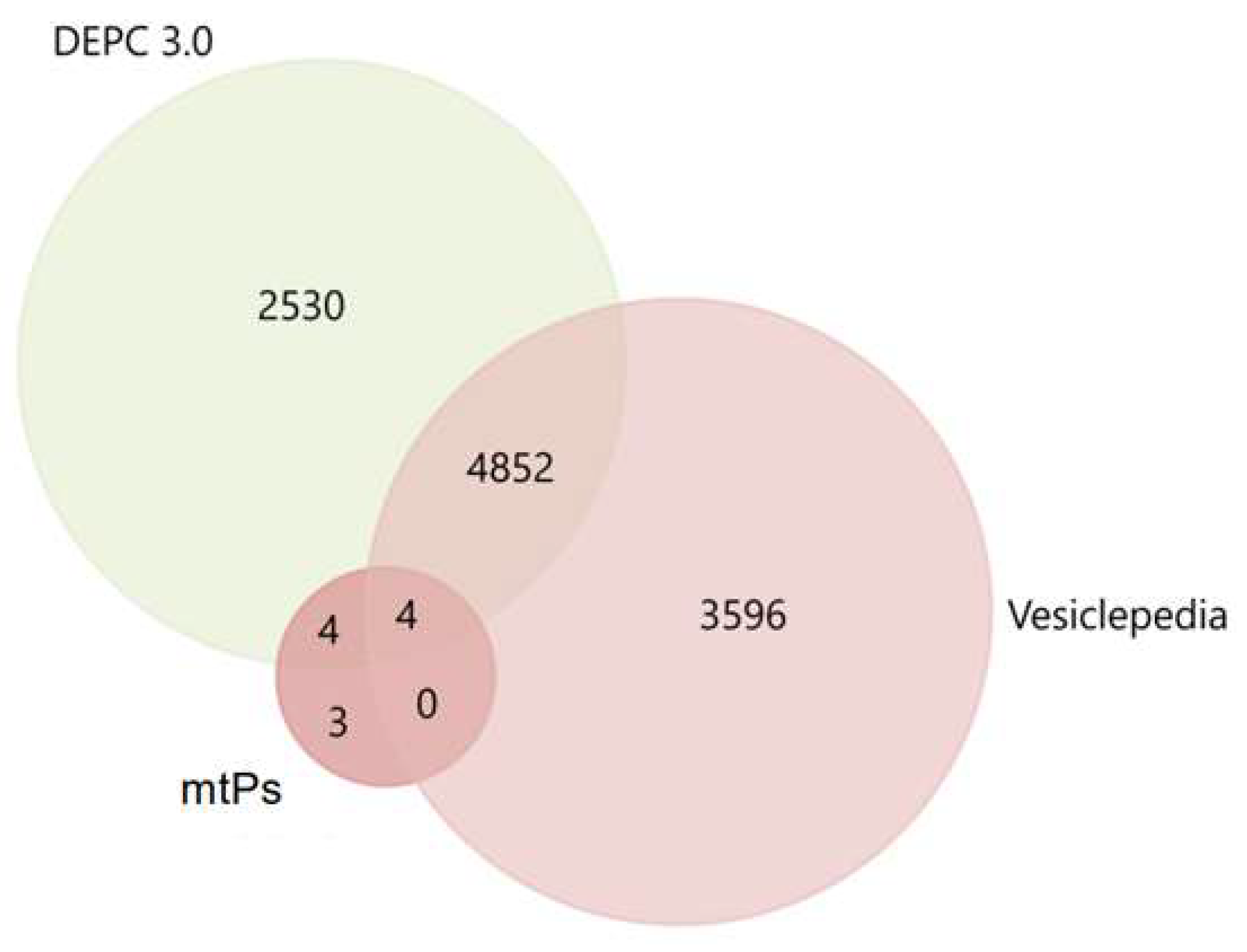
2.2. MtPs in Exosomes Derived from BC Cell Lines
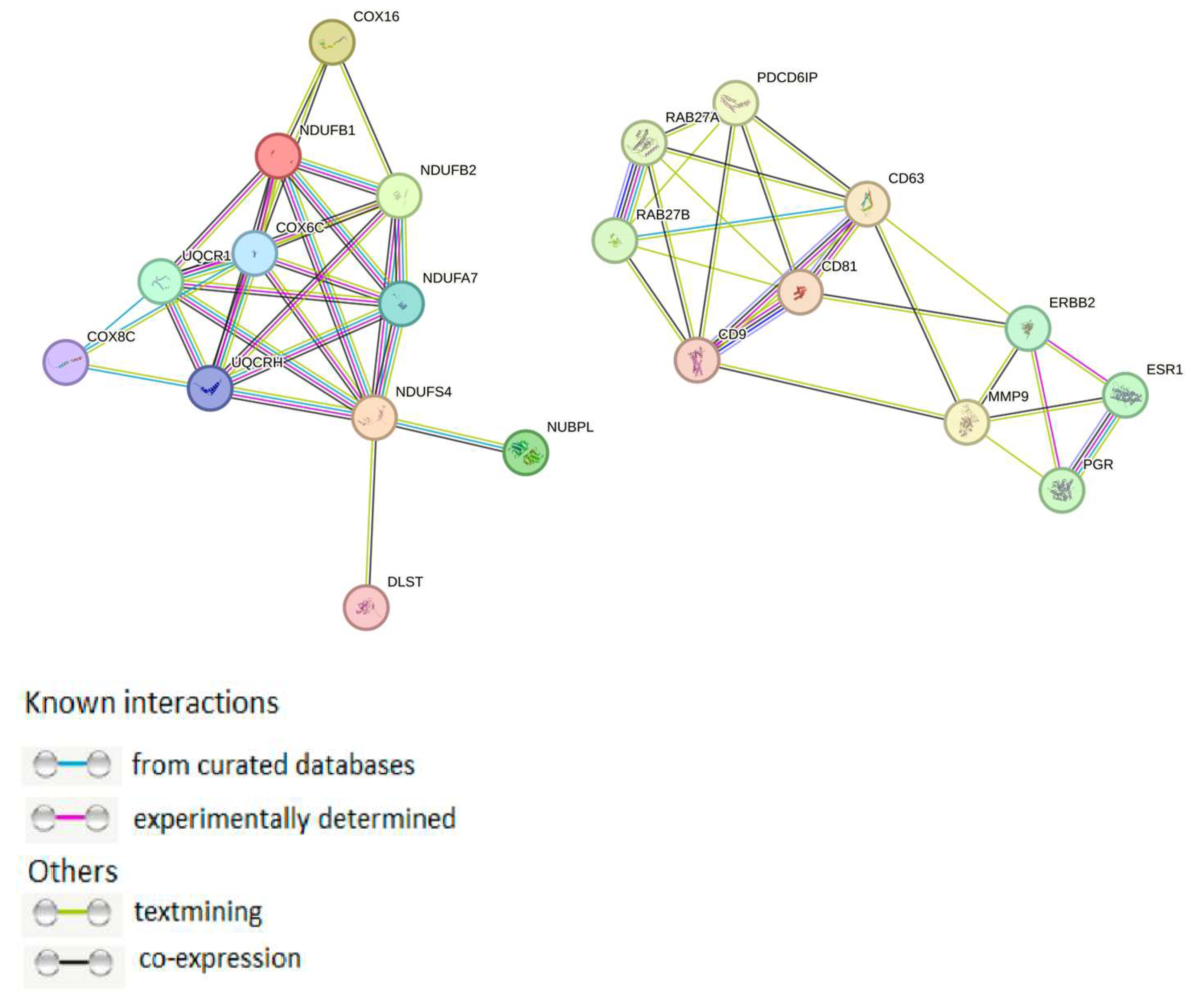
2.3. Exosomal mtPs Are Associated with Cancer Progression
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Exosomes Isolation
4.3. TEM
4.4. Flow Cytometry
4.5. Mass Spectrometry Analysis
4.6. Bioinformatics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BC | breast cancer |
| EMT | epithelial–mesenchymal transition |
| EVs | extracellular vesicles |
| mtDNA | mitochondrial DNA |
| mtPs | mitochondrial proteins |
| OXPHOS | oxidative phosphorylation |
| PPI | protein–protein interactions |
| TEM | transmission electron microscopy |
References
- Andreu, Z.; Hidalgo, M.R.; Masiá, E.; Romera-Giner, S.; Malmierca-Merlo, P.; López-Guerrero, J.A.; García-García, F.; Vicent, M.J. Comparative Profiling of Whole-Cell and Exosome Samples Reveals Protein Signatures That Stratify Breast Cancer Subtypes. Cell. Mol. Life Sci. 2024, 81, 363. [Google Scholar] [CrossRef]
- Jang, S.C.; Crescitelli, R.; Cvjetkovic, A.; Belgrano, V.; Bagge, R.O.; Sundfeldt, K.; Ochiya, T.; Kalluri, R.; Lötvall, J. Mitochondrial Protein Enriched Extracellular Vesicles Discovered in Human Melanoma Tissues Can Be Detected in Patient Plasma. J. Extracell. Vesicles 2019, 8, 1635420. [Google Scholar] [CrossRef]
- Sansone, P.; Savini, C.; Kurelac, I.; Chang, Q.; Amato, L.B.; Strillacci, A.; Stepanova, A.; Iommarini, L.; Mastroleo, C.; Daly, L.; et al. Packaging and Transfer of Mitochondrial DNA via Exosomes Regulate Escape from Dormancy in Hormonal Therapy-Resistant Breast Cancer. Proc. Natl. Acad. Sci. USA 2017, 114, E9066–E9075. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.; Greening, D.W.; Chen, M.; Xu, R.; Ji, H.; Simpson, R.J. Exosomes Derived from Human Primary and Metastatic Colorectal Cancer Cells Contribute to Functional Heterogeneity of Activated Fibroblasts by Reprogramming Their Proteome. Proteomics 2019, 19, 1800148. [Google Scholar] [CrossRef]
- An, M.; Zhu, J.; Wu, J.; Cuneo, K.C.; Lubman, D.M. Circulating Microvesicles from Pancreatic Cancer Accelerate the Migration and Proliferation of PANC-1 Cells. J. Proteome Res. 2018, 17, 1690–1699. [Google Scholar] [CrossRef] [PubMed]
- Lazar, I.; Clement, E.; Dauvillier, S.; Milhas, D.; Ducoux-Petit, M.; LeGonidec, S.; Moro, C.; Soldan, V.; Dalle, S.; Balor, S.; et al. Adipocyte Exosomes Promote Melanoma Aggressiveness Through Fatty Acid Oxidation: A Novel Mechanism Linking Obesity and Cancer. Cancer Res. 2016, 76, 4051–4057. [Google Scholar] [CrossRef]
- D’Acunzo, P.; Pérez-González, R.; Kim, Y.; Hargash, T.; Miller, C.; Alldred, M.J.; Erdjument-Bromage, H.; Penikalapati, S.C.; Pawlik, M.; Saito, M.; et al. Mitovesicles Are a Novel Population of Extracellular Vesicles of Mitochondrial Origin Altered in Down Syndrome. Sci. Adv. 2021, 7, eabe5085. [Google Scholar] [CrossRef]
- Puhm, F.; Afonyushkin, T.; Resch, U.; Obermayer, G.; Rohde, M.; Penz, T.; Schuster, M.; Wagner, G.; Rendeiro, A.F.; Melki, I.; et al. Mitochondria Are a Subset of Extracellular Vesicles Released by Activated Monocytes and Induce Type I IFN and TNF Responses in Endothelial Cells. Circ. Res. 2019, 125, 43–52. [Google Scholar] [CrossRef]
- Ikeda, G.; Santoso, M.R.; Tada, Y.; Li, A.M.; Vaskova, E.; Jung, J.-H.; O’Brien, C.; Egan, E.; Ye, J.; Yang, P.C. Mitochondria-Rich Extracellular Vesicles From Autologous Stem Cell–Derived Cardiomyocytes Restore Energetics of Ischemic Myocardium. J. Am. Coll. Cardiol. 2021, 77, 1073–1088. [Google Scholar] [CrossRef]
- Gelsomino, L.; Barone, I.; Caruso, A.; Giordano, F.; Brindisi, M.; Morello, G.; Accattatis, F.M.; Panza, S.; Cappello, A.R.; Bonofiglio, D.; et al. Proteomic Profiling of Extracellular Vesicles Released by Leptin-Treated Breast Cancer Cells: A Potential Role in Cancer Metabolism. Int. J. Mol. Sci. 2022, 23, 12941. [Google Scholar] [CrossRef]
- Shafiq, A.; Suwakulsiri, W.; Rai, A.; Chen, M.; Greening, D.W.; Zhu, H.-J.; Xu, R.; Simpson, R.J. Transglutaminase-2, RNA-Binding Proteins and Mitochondrial Proteins Selectively Traffic to MDCK Cell-Derived Microvesicles Following H-Ras-Induced Epithelial-Mesenchymal Transition. Proteomics 2021, 21, 2000221. [Google Scholar] [CrossRef]
- Zhou, Z.; Qu, C.; Zhou, P.; Zhou, Q.; Li, D.; Wu, X.; Yang, L. Extracellular Vesicles Activated Cancer-Associated Fibroblasts Promote Lung Cancer Metastasis through Mitophagy and mtDNA Transfer. J. Exp. Clin. Cancer Res. 2024, 43, 158. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zang, T.; Wang, Y. Human Mitochondrial Genome Compression Using Machine Learning Techniques. Hum. Genom. 2019, 13 (Suppl. S1), 49. [Google Scholar] [CrossRef] [PubMed]
- Todkar, K.; Chikhi, L.; Desjardins, V.; El-Mortada, F.; Pépin, G.; Germain, M. Selective Packaging of Mitochondrial Proteins into Extracellular Vesicles Prevents the Release of Mitochondrial DAMPs. Nat. Commun. 2021, 12, 1971. [Google Scholar] [CrossRef]
- Shefer, A.; Yalovaya, A.; Tamkovich, S. Exosomes in Breast Cancer: Involvement in Tumor Dissemination and Prospects for Liquid Biopsy. Int. J. Mol. Sci. 2022, 23, 8845. [Google Scholar] [CrossRef]
- Ellinger, J.; Poss, M.; Brüggemann, M.; Gromes, A.; Schmidt, D.; Ellinger, N.; Tolkach, Y.; Dietrich, D.; Kristiansen, G.; Müller, S.C. Systematic Expression Analysis of Mitochondrial Complex I Identifies NDUFS1 as a Biomarker in Clear-Cell Renal-Cell Carcinoma. Clin. Genitourin. Cancer 2017, 15, e551–e562. [Google Scholar] [CrossRef]
- Nikas, J.B.; Boylan, K.L.; Skubitz, A.P.; Low, W.C. Mathematical Prognostic Biomarker Models for Treatment Response and Survival in Epithelial Ovarian Cancer. Cancer Inform. 2011, 10, 233–247. [Google Scholar] [CrossRef]
- Marquez, J.; Kratchmarova, I.; Akimov, V.; Unda, F.; Ibarretxe, G.; Clerigué, A.S.; Osinalde, N.; Badiola, I. NADH Dehydrogenase Complex I Is Overexpressed in Incipient Metastatic Murine Colon Cancer Cells. Oncol. Rep. 2019, 41, 742–752. [Google Scholar] [CrossRef]
- Cheng, T.; Jiang, B.; Xu, M.; Yuan, C.; Tai, M.; Wu, H.; Lu, B.; Sun, P.; Jiang, X.; Zhang, X. NDUFS4 Promotes Tumor Progression and Predicts Prognosis in Gastric Cancer. Carcinogenesis 2022, 43, 980–987. [Google Scholar] [CrossRef]
- Liang, J.; Vitale, T.; Zhang, X.; Jackson, T.D.; Yu, D.; Jedrychowski, M.; Gygi, S.P.; Widlund, H.R.; Wucherpfennig, K.W.; Puigserver, P. Selective Deficiency of Mitochondrial Respiratory Complex I Subunits Ndufs4/6 Causes Tumor Immunogenicity. Nat. Cancer 2025, 6, 323–337. [Google Scholar] [CrossRef]
- Vu, T.; Datta, P.K. Regulation of EMT in Colorectal Cancer: A Culprit in Metastasis. Cancers 2017, 9, 171. [Google Scholar] [CrossRef]
- Zhang, X.; Hong, B.; Li, H.; Sun, Z.; Zhao, J.; Li, M.; Wei, D.; Wang, Y.; Zhang, N. Disulfidptosis and Ferroptosis Related Genes Define the Immune Microenvironment and NUBPL Serves as a Potential Biomarker for Predicting Prognosis and Immunotherapy Response in Bladder Cancer. Heliyon 2024, 10, e37638. [Google Scholar] [CrossRef]
- Olsen, T.K.; Panagopoulos, I.; Gorunova, L.; Micci, F.; Andersen, K.; Andersen, H.K.; Meling, T.R.; Due-Tønnessen, B.; Scheie, D.; Heim, S.; et al. Novel Fusion Genes and Chimeric Transcripts in Ependymal Tumors. Genes Chromosom. Cancer 2016, 55, 944–953. [Google Scholar] [CrossRef] [PubMed]
- Sotgia, F.; Lisanti, M.P. Mitochondrial Biomarkers Predict Tumor Progression and Poor Overall Survival in Gastric Cancers: Companion Diagnostics for Personalized Medicine. Oncotarget 2017, 8, 67117–67128. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.X.; Sun, W.; Wang, S.H.; Liu, P.J.; Wang, Y.C. Differential Expression and Clinical Significance of COX6C in Human Diseases. Am. J. Transl. Res. 2021, 13, 1–10. [Google Scholar]
- Wang, F.L.; Wang, Y.; Wong, W.K.; Liu, Y.; Addivinola, F.J.; Liang, P.; Chen, L.B.; Kantoff, P.; Pardee, A. Two Differentially Expressed Genes in Normal Human Prostate Tissue and in Carcinoma. Cancer Res. 1996, 56, 3634–3637. [Google Scholar]
- Li, L.H.; Li, Y.J.; Huang, Y.G.; Ouyang, Y.; Zhu, Y.; Wang, Y.Z.; Guo, X.D.; Yuan, Y.; Gong, K.M. Long Non-Coding RNA MIF-AS1 Promotes Gastric Cancer Cell Proliferation and Reduces Apoptosis to Upregulate NDUFA4. Cancer Sci. 2018, 109, 3714–3725. [Google Scholar] [CrossRef]
- Kurose, K.; Mine, N.; Doi, D.; Ota, Y.; Yoneyama, K.; Konishi, H.; Araki, T.; Emi, M. Novel Gene Fusion of COX6C at 8q22-23 to HMGIC at 12q15 in a Uterine Leiomyoma. Genes Chromosom. Cancer 2000, 27, 303–307. [Google Scholar] [CrossRef]
- Swierniak, M.; Pfeifer, A.; Stokowy, T.; Rusinek, D.; Chekan, M.; Lange, D.; Krajewska, J.; Oczko-Wojciechowska, M.; Czarniecka, A.; Jarzab, M.; et al. Somatic Mutation Profiling of Follicular Thyroid Cancer by Next Generation Sequencing. Mol. Cell. Endocrinol. 2016, 433, 130–137. [Google Scholar] [CrossRef]
- Liu, W.S.; Liu, Y.D.; Fu, Q.; Zhang, W.J.; Xu, L.; Chang, Y.; Xu, J.J. Prognostic Significance of Ubiquinol-Cytochrome c Reductase Hinge Protein Expression in Patients with Clear Cell Renal Cell Carcinoma. Am. J. Cancer Res. 2016, 6, 797–805. [Google Scholar]
- Anderson, N.M.; Qin, X.; Finan, J.M.; Lam, A.; Athoe, J.; Missiaen, R.; Skuli, N.; Kennedy, A.; Saini, A.S.; Tao, T.; et al. Metabolic Enzyme DLST Promotes Tumor Aggression and Reveals a Vulnerability to OXPHOS Inhibition in High-Risk Neuroblastoma. Cancer Res. 2021, 81, 4417–4430. [Google Scholar] [CrossRef]
- Shen, N.; Korm, S.; Karantanos, T.; Li, D.; Zhang, X.; Ritou, E.; Xu, H.; Xu, P.; Xu, Y.; Xu, C.; et al. DLST-Dependence Dictates Metabolic Heterogeneity in TCA-Cycle Usage among Triple-Negative Breast Cancer. Commun. Biol. 2021, 4, 1289. [Google Scholar] [CrossRef] [PubMed]
- Shinde, A.; Jung, H.; Lee, H.; Libring, S.; Saleh, A.M.; Solorio, L.; Lee, S.-H.; Hwang, S.-Y.; Park, E.S.; Lee, K.-M.; et al. TNF-α Differentially Modulates Subunit Levels of Respiratory Electron Transport Complexes of ER/PR +ve/−ve Breast Cancer Cells to Regulate Mitochondrial Complex Activity and Tumorigenic Potential. Cancer Metab. 2021, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, N.; Sun, D.; Sun, H.; Tong, D.; Liu, D.; Pang, B.; Li, S.; Wei, J.; Dai, J.; et al. NUBPL, a Novel Metastasis-Related Gene, Promotes Colorectal Carcinoma Cell Motility by Inducing Epithelial-Mesenchymal Transition. Cancer Sci. 2017, 108, 1169–1176. [Google Scholar] [CrossRef] [PubMed]
- Michail, A.; Gkikas, D.; Stellas, D.; Kaltezioti, V.; Politis, P.K. Prox1 Suppresses the Proliferation of Breast Cancer Cells via Direct Inhibition of c-Myc Gene Expression. Cells 2023, 12, 1869. [Google Scholar] [CrossRef]
- Luo, Y.; Medina Bengtsson, L.; Wang, X.; Grawé, J.; Maecke, B.; Rydén, L.; Wennerberg, J.; Kjellström, L.; Kjellman, C. UQCRH Downregulation Promotes Warburg Effect in Renal Cell Carcinoma Cells. Sci. Rep. 2020, 10, 15021. [Google Scholar] [CrossRef]
- Wang, M.; Wei, R.; Li, G.; Bi, H.L.; Jia, Z.; Zhang, M.; Pang, M.; Li, X.; Ma, L.; Tang, Y. SUMOylation of SYNJ2BP-COX16 Promotes Breast Cancer Progression through DRP1-Mediated Mitochondrial Fission. Cancer Lett. 2022, 547, 215871. [Google Scholar] [CrossRef]
- Wang, C.; Lv, J.; Xue, C.; Li, H.; Li, T.; Xu, P.; Li, J.; Wang, Q.; Zhao, W.; Duan, Y.; et al. Novel Role of COX6c in the Regulation of Oxidative Phosphorylation and Diseases. Cell Death Discov. 2022, 8, 336. [Google Scholar] [CrossRef]
- Tutanov, O.; Orlova, E.; Proskura, K.; Grigor’eva, A.; Yunusova, N.; Tsentalovich, Y.; Alexandrova, A.; Tamkovich, S. Proteomic Analysis of Blood Exosomes from Healthy Females and Breast Cancer Patients Reveals an Association between Different Exosomal Bioactivity on Non-Tumorigenic Epithelial Cell and Breast Cancer Cell Migration In Vitro. Biomolecules 2020, 10, 495. [Google Scholar] [CrossRef]
- Aebersold, R.; Mann, M. Mass Spectrometry-Based Proteomics. Nature 2003, 422, 198–207. [Google Scholar] [CrossRef]
- Domon, B.; Aebersold, R. Mass Spectrometry and Protein Analysis. Science 2006, 312, 212–217. [Google Scholar] [CrossRef]
- Suckau, D.; Resemann, A.; Schuerenberg, M.; Hufnagel, P.; Franzen, J.; Holle, A. A Novel MALDI LIFT-TOF/TOF Mass Spectrometer for Proteomics. Anal. Bioanal. Chem. 2003, 376, 952–965. [Google Scholar] [CrossRef]
- Yang, Q.; Zhang, Y.; Cui, H.; Chen, L.; Zhao, Y.; Lin, Y.; Zhang, M.; Xie, L. dbDEPC 3.0: The Database of Differentially Expressed Proteins in Human Cancer with Multi-Level Annotation and Drug Indication. Database 2018, 2018, bay015. [Google Scholar] [CrossRef]

| BC Subtype | Cell Line | MFI 1 CD9+ CD81+ |
|---|---|---|
| Non-tumorogenic breast cell lines | HBL-100 | 1750 |
| MCF-10A | 4050 | |
| Luminal A BC cell lines | MCF-7 | 3170 |
| ZR-75-1 | 2120 | |
| T-47D | 4120 | |
| Triple-positive BC cell line | BT-474 | 2870 |
| HER2+ BC cell lines | SK-BR-3 | 2330 |
| Triple-negative BC cell lines | BT-549 | 4800 |
| MDA-MB-231 | 4075 | |
| HCC-1937 | 1630 | |
| Negative control | 520 |
| BC Subtype | Cell Line | Gene | Uniprot ID | Protein Name | Mitochondria Component |
|---|---|---|---|---|---|
| HER2+ | SKBR-3 | NDUFB1 | O75438 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 1 | Complex I |
| Triplepositive | BT-474 | NDUFB1 | O75438 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 1 | Complex I |
| NDUFS4 | O43181 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 4 | Complex I | ||
| COX16 | Q9P0S2 | Cytochrome c oxidase assembly protein COX16 homolog | Complex IV | ||
| Luminal A | ZR-75-1 | NDUFB2 | O95178 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 2 | Complex I |
| MCF-7 | NUBPL | Q8TB37 | Iron-sulfur cluster transfer protein NUBPL | Complex I | |
| T-47D | UQCR10 | Q9UDW1 | Cytochrome b-c1 complex subunit 9 | Complex III | |
| Triplenegative | BT-549 | NDUFA7 | O95182 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7 | Complex I |
| COX6C | P09669 | Cytochrome c oxidase subunit 6C | Complex IV | ||
| HCC-1937 | UQCRH | P07919 | Cytochrome b-c1 complex subunit 6 | Complex III | |
| MDA-MD-463 | COX8C | Q7Z4L0 | Cytochrome c oxidase subunit 8C | Complex IV | |
| MDA-MB-231 | DLST | P36957 | Dihy-drolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehy-drogenase complex | Tricarboxylic acid cycle component |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shefer, A.; Yanshole, L.; Grygor’eva, A.; Yunusova, N.; Kondakova, I.; Spirina, L.; Shevela, A.; Chernyshova, A.; Romanov, A.; Tamkovich, S. Mitochondrial Proteins as Exosomal Cargo: New Breast Cancer Biomarkers & Crucial Players in Carcinogenesis? Molecules 2025, 30, 4112. https://doi.org/10.3390/molecules30204112
Shefer A, Yanshole L, Grygor’eva A, Yunusova N, Kondakova I, Spirina L, Shevela A, Chernyshova A, Romanov A, Tamkovich S. Mitochondrial Proteins as Exosomal Cargo: New Breast Cancer Biomarkers & Crucial Players in Carcinogenesis? Molecules. 2025; 30(20):4112. https://doi.org/10.3390/molecules30204112
Chicago/Turabian StyleShefer, Aleksei, Lyudmila Yanshole, Alina Grygor’eva, Natalia Yunusova, Irina Kondakova, Liudmila Spirina, Andrey Shevela, Alyona Chernyshova, Alexander Romanov, and Svetlana Tamkovich. 2025. "Mitochondrial Proteins as Exosomal Cargo: New Breast Cancer Biomarkers & Crucial Players in Carcinogenesis?" Molecules 30, no. 20: 4112. https://doi.org/10.3390/molecules30204112
APA StyleShefer, A., Yanshole, L., Grygor’eva, A., Yunusova, N., Kondakova, I., Spirina, L., Shevela, A., Chernyshova, A., Romanov, A., & Tamkovich, S. (2025). Mitochondrial Proteins as Exosomal Cargo: New Breast Cancer Biomarkers & Crucial Players in Carcinogenesis? Molecules, 30(20), 4112. https://doi.org/10.3390/molecules30204112










