Molecular-Genetic Research of Rhodococcus rhodochrous IEGM 1362, an Active (–)-Isopulegol Biotransformer
Abstract
1. Introduction
2. Results and Discussion
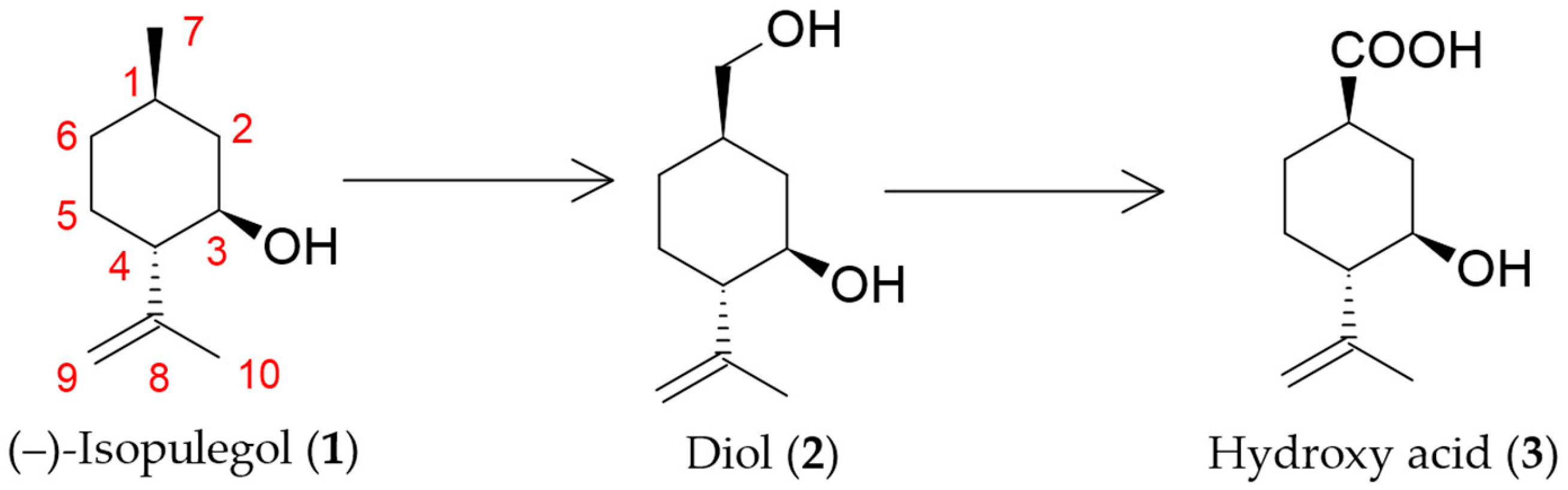
2.1. Determination of Key Enzymes of (–)-Isopulegol Biotransformation
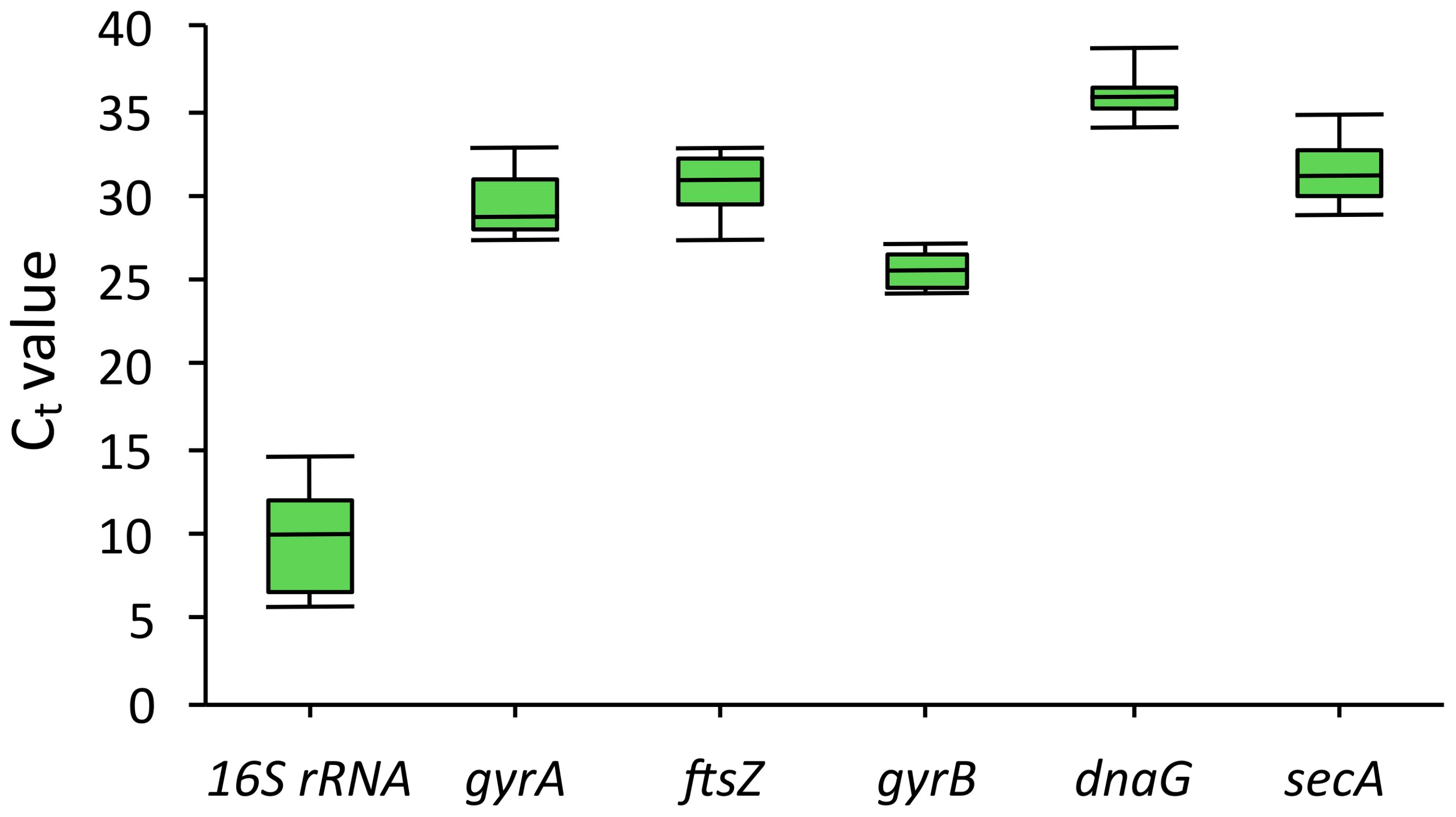
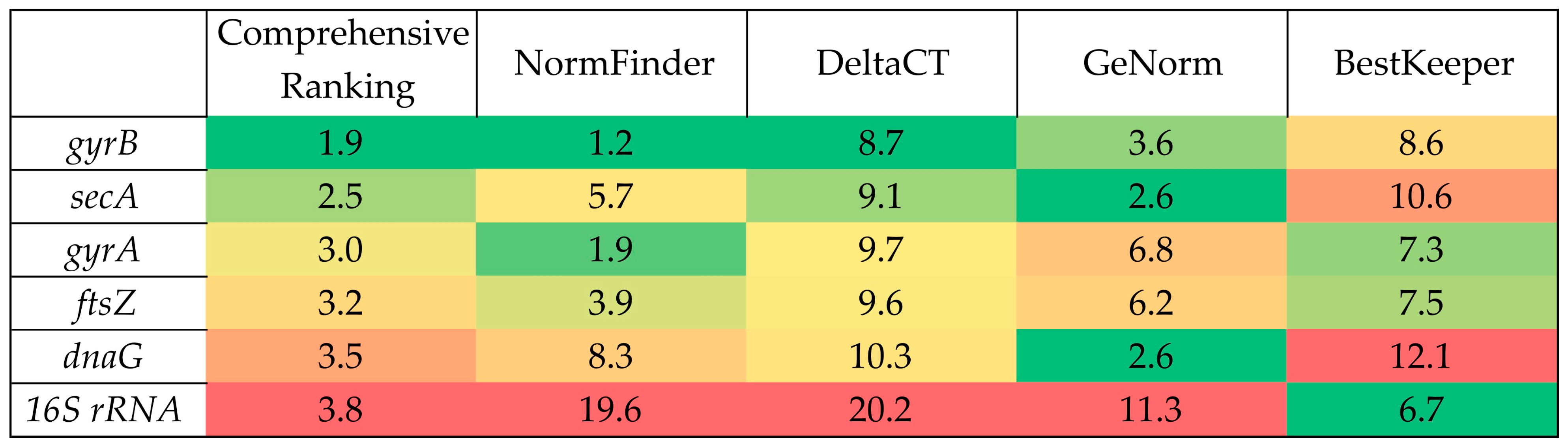
2.2. Selection of Reference Genes
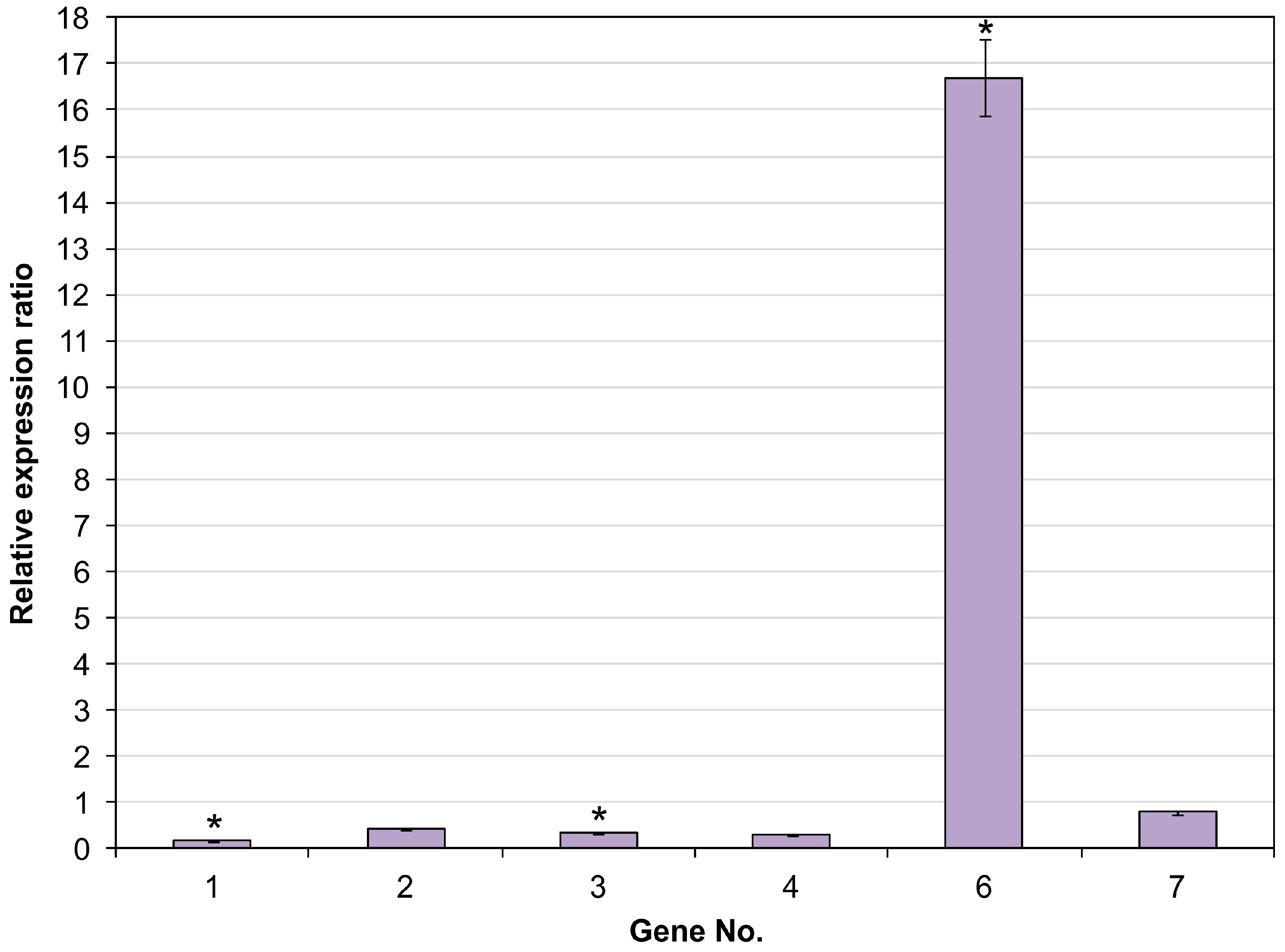
2.3. Expression Analysis of CYP450 Using qRT-PCR
3. Materials and Methods
3.1. Strain and Culture Conditions
3.2. Cell Fractions Obtaining
3.3. Analytical Methods
3.4. RNA Isolation and cDNA Synthesis
3.5. Selection of Reference Genes
3.6. Quantitative Real-Time PCR (qRT-PCR) and Gene Expression Analysis
3.7. Bioinformatics Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CYP450 | Cytochrome P450 |
| qRT-PCR | Quantitative real-time PCR |
| Ct | Cycle threshold |
| TLC | Thin-layer chromatography |
| GC-MS | Gas chromatography–mass spectrometry |
| MPB | Meat–peptone broth |
| LB | Luria–Bertani |
References
- Minh Le, T.; Szakonyi, Z. Enantiomeric Isopulegol as the Chiral Pool in the Total Synthesis of Bioactive Agents. Chem. Rec. 2022, 22, e202100194. [Google Scholar] [CrossRef]
- Ivshina, I.B.; Luchnikova, N.A.; Maltseva, P.Y.; Ilyina, I.V.; Volcho, K.P.; Gatilov, Y.V.; Korchagina, D.V.; Kostrikina, N.A.; Sorokin, V.V.; Mulyukin, A.L. Biotransformation of (–)-Isopulegol by Rhodococcus rhodochrous. Pharmaceuticals 2022, 15, 964. [Google Scholar] [CrossRef] [PubMed]
- Maltseva, P.Y.; Plotnitskaya, N.A.; Krivoruchko, A.V.; Beletskiy, A.V.; Rakitin, A.L.; Mardanov, A.V.; Ivshina, I.B. Bioinformatics Analysis of the Genome of Rhodococcus rhodochrous IEGM 1362, an (–)-Isopulegol Biotransformer. Genes 2024, 15, 992. [Google Scholar] [CrossRef] [PubMed]
- Greule, A.; Stok, J.E.; De Voss, J.J.; Cryle, M.J. Unrivalled Diversity: The Many Roles and Reactions of Bacterial Cytochromes P450 in Secondary Metabolism. Nat. Prod. Rep. 2018, 35, 757–791. [Google Scholar] [CrossRef] [PubMed]
- Jakubovska, J.; Meškys, R. Characterization of 1,8-Cineole Degradation Encoding Operon from Rhodococcus sp. TMP1. Chemija 2016, 27, 84–91. [Google Scholar]
- Van Der Werf, M.J.; Van Der Ven, C.; Barbirato, F.; Eppink, M.H.M.; De Bont, J.A.M.; Van Berkel, W.J.H. Stereoselective Carveol Dehydrogenase from Rhodococcus erythropolis DCL14. A Novel Nicotinoprotein Belonging to the Short Chain Dehydrogenase/Reductase Superfamily. J. Biol. Chem. 1999, 274, 26296–26304. [Google Scholar] [CrossRef]
- Van Der Werf, M.; Overkamp, K.; de Bont, J. DCL14 Belongs to a Novel Class of Epoxide Hydrolases. J. Bacteriol. 1998, 180, 5052–5057. [Google Scholar] [CrossRef]
- Giang, P.D.; Churchman, L.R.; Buczynski, J.B.; Bell, S.G.; Stok, J.E.; De Voss, J.J. CYP108N14: A Monoterpene Monooxygenase from Rhodococcus globerulus. Arch. Biochem. Biophys. 2024, 752, 109852. [Google Scholar] [CrossRef]
- Tarasova, E.V.; Grishko, V.V.; Ivshina, I.B. Cell Adaptations of Rhodococcus rhodochrous IEGM 66 to Betulin Biotransformation. Process Biochem. 2017, 52, 1–9. [Google Scholar] [CrossRef]
- Cheremnykh, K.M.; Luchnikova, N.A.; Grishko, V.V.; Ivshina, I.B. Bioconversion of Ecotoxic Dehydroabietic Acid Using Rhodococcus Actinobacteria. J. Hazard. Mater. 2018, 346, 103–112. [Google Scholar] [CrossRef]
- Ye, L.; Zhu, X.; Wu, T.; Wang, W.; Zhao, D.; Bi, C.; Zhang, X. Optimizing the Localization of Astaxanthin Enzymes for Improved Productivity. Biotechnol. Biofuels 2018, 11, 278. [Google Scholar] [CrossRef]
- Rocha, D.J.P.G.; Castro, T.L.P.; Aguiar, E.R.G.R.; Pacheco, L.G.C. Gene Expression Analysis in Bacteria by RT-QPCR. Methods Mol. Biol. 2020, 2065, 119–137. [Google Scholar]
- Delorenzo, D.M.; Moon, T.S. Selection of Stable Reference Genes for RT-qPCR in Rhodococcus opacus PD630. Sci. Rep. 2018, 8, 6019. [Google Scholar] [CrossRef]
- Di Canito, A.; Zampolli, J.; Orro, A.; D’Ursi, P.; Milanesi, L.; Sello, G.; Steinbüchel, A.; Di Gennaro, P. Genome-Based Analysis for the Identification of Genes Involved in o-Xylene Degradation in Rhodococcus opacus R7. BMC Genom. 2018, 19, 587. [Google Scholar] [CrossRef]
- Li, Y.; Ma, Q.; Zhang, J.; Meng, N.; Su, J.; Wang, J. Transcriptomic Profiling Reveals the Molecular Responses of Rhodococcus aetherivorans DMU1 to Skatole Stress. Ecotoxicol. Environ. Saf. 2023, 249, 114464. [Google Scholar] [CrossRef]
- Iminova, L.; Delegan, Y.; Frantsuzova, E.; Bogun, A.; Zvonarev, A.; Suzina, N.; Anbumani, S.; Solyanikova, I. Physiological and Biochemical Characterization and Genome Analysis of Rhodococcus qingshengii Strain 7B Capable of Crude Oil Degradation and Plant Stimulation. Biotechnol. Rep. 2022, 35, e00741. [Google Scholar] [CrossRef] [PubMed]
- Zampolli, J.; Orro, A.; Manconi, A.; Ami, D.; Natalello, A.; Di Gennaro, P. Transcriptomic Analysis of Rhodococcus opacus R7 Grown on Polyethylene by RNA-Seq. Sci. Rep. 2021, 11, 21311. [Google Scholar] [CrossRef] [PubMed]
- Sazykin, I.; Makarenko, M.; Khmelevtsova, L.; Seliverstova, E.; Rakin, A.; Sazykina, M. Cyclohexane, Naphthalene, and Diesel Fuel Increase Oxidative Stress, CYP153, SodA, and RecA Gene Expression in Rhodococcus erythropolis. Microbiologyopen 2019, 8, e00855. [Google Scholar] [CrossRef] [PubMed]
- da Silveira, B.P.; Gressler, L.T.; Cargnelutti, J.F.; Bordin, A.I.; de Vargas, A.C. GAPDH, RhbC, and VapA Gene Expression in Rhodococcus equi Cultured under Different Iron Concentrations. Microb. Pathog. 2020, 139, 103885. [Google Scholar] [CrossRef]
- Tian, K.; Meng, Q.; Li, S.; Chang, M.; Meng, F.; Yu, Y.; Li, H.; Qiu, Q.; Shao, J.; Huo, H. Mechanism of 17β-Estradiol Degradation by Rhodococcus equi via the 4,5-Seco Pathway and Its Key Genes. Environ. Pollut. 2022, 312, 120021. [Google Scholar] [CrossRef]
- Tian, K.; Meng, F.; Meng, Q.; Gao, Y.; Zhang, L.; Wang, L.; Wang, Y.; Li, X.; Huo, H. The Analysis of Estrogen-Degrading and Functional Metabolism Genes in Rhodococcus equi DSSKP-R-001. Int. J. Genom. 2020, 2020, 9369182. [Google Scholar] [CrossRef] [PubMed]
- Rocha, D.J.P.; Santos, C.S.; Pacheco, L.G.C. Bacterial Reference Genes for Gene Expression Studies by RT-QPCR: Survey and Analysis. Antonie Van Leeuwenhoek 2015, 108, 685–693. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Plotnitskaya, N.A.; Maltseva, P.Y.; Ivshina, I.B. Genome Annotation and Catalytic Profile of Rhodococcus rhodochrous IEGM 107, Mono- and Diterpenoid Biotransformer. Genes 2025, 16, 739. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Wang, J.; Zhang, B. RefFinder: A Web-Based Tool for Comprehensively Analyzing and Identifying Reference Genes. Funct. Integr. Genom. 2023, 23, 125. [Google Scholar] [CrossRef]
- Madeira, F.; Madhusoodanan, N.; Lee, J.; Eusebi, A.; Niewielska, A.; Tivey, A.R.N.; Lopez, R.; Butcher, S. The EMBL-EBI Job Dispatcher Sequence Analysis Tools Framework in 2024. Nucleic Acids Res. 2024, 52, 521–525. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis Version 12 for Adaptive and Green Computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]


| Gene No. | Enzyme | Locus Tag | Primers (Forward; Reverse) | Amplicon Size, bp | PCR Efficiency, % | R2 |
|---|---|---|---|---|---|---|
| 1 | Putative cytochrome P450 hydroxylase | NOT90_25380 | TACAGCCCCGAACTCGACTA; TGTCCGGTATCGATGAAGCG | 311 | 91.5 | 0.973 |
| 2 | Putative cytochrome P450 hydroxylase | NOT90_21400 | GTTCATCGAGGGCCTGAACA; CTCGAAACGCAGTGTCTCCT | 373 | 93 | 0.999 |
| 3 | Cytochrome P450 | NOT90_15505 | CGACGACATCTTCTCGGTGT; TGTGGTGGGAGAAGTGCATC | 389 | 94.4 | 0.998 |
| 4 | Putative cytochrome P450 | NOT90_24995 | TCGTTCGCTTTGCACTACCT; CGAATGGCTTGTATGCGTGG | 370 | 99 | 0.999 |
| 5 | Cytochrome P450 | NOT90_03190 | CACGTCGACCATCACGATCT; TTGGTGGACGATCGCTTTGA | 448 | 100.2 | 0.999 |
| 6 | Putative cytochrome P450 hydroxylase | NOT90_19635 | GCTCTGACGCAGGAGTTCTT; ACGCAGTGTGTAGTCCTGTG | 444 | 98.7 | 0.996 |
| 7 | Cytochrome P450 monooxygenase | NOT90_21345 | ATCGGTTCACCCAGAACCTG; GAGGACAGGATCACGAAGCC | 403 | 100 | 0.966 |
| 8 | Cytochrome P450 | NOT90_02535 | CCGATCTCGTAGCCCAGTTC; TTCGCTGATCTCGATTCCCG | 554 | 96.9 | 0.997 |
| 9 | Putative cytochrome P450 hydroxylase | NOT90_16895 | CATCTCCCACGGCCTGTATC; TGTACTCGACACGAAGGTGC | 461 | 102.9 | 0.993 |
| Protein Name | Organism | Length, AA | Identity, % | Entry |
|---|---|---|---|---|
| Cytochrome P450 | Rhodococcus sp. CX | 440 | 94.3 | A0A931HIH7 |
| Cytochrome P450-terp | Rhodococcus sp. T7 | 440 | 92.3 | A0A6N7CCE3 |
| Cytochrome P450 | R. oxybenzonivorans | 440 | 91.1 | A0A2S2C1W6 |
| Cytochrome P450-terp | Nocardia cerradoensis | 440 | 85.6 | A0A231GXP1 |
| Putative cytochrome P450 | Gordonia aichiensis NBRC 108223 | 443 | 77.9 | L7KKR0 |
| Cytochrome P450 | G. asplenii | 442 | 77.2 | A0A848KZE1 |
| Cytochrome P450 | G. desulfuricans | 443 | 77 | A0A7K3LNL8 |
| Cytochrome P450-terp | G. insulae | 441 | 76.8 | A0A3G8JNI1 |
| Cytochrome P450 | N. jinanensis | 443 | 76.5 | A0A917RPB9 |
| Cytochrome P450 | Amycolatopsis acidiphila | 441 | 71.2 | A0A558AL67 |
| Linalool 8-monooxygenase | Candidatus Protofrankia datiscae | 441 | 70.6 | F8AXG5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maltseva, P.Y.; Plotnitskaya, N.A.; Ivshina, I.B. Molecular-Genetic Research of Rhodococcus rhodochrous IEGM 1362, an Active (–)-Isopulegol Biotransformer. Molecules 2025, 30, 3976. https://doi.org/10.3390/molecules30193976
Maltseva PY, Plotnitskaya NA, Ivshina IB. Molecular-Genetic Research of Rhodococcus rhodochrous IEGM 1362, an Active (–)-Isopulegol Biotransformer. Molecules. 2025; 30(19):3976. https://doi.org/10.3390/molecules30193976
Chicago/Turabian StyleMaltseva, Polina Y., Natalia A. Plotnitskaya, and Irina B. Ivshina. 2025. "Molecular-Genetic Research of Rhodococcus rhodochrous IEGM 1362, an Active (–)-Isopulegol Biotransformer" Molecules 30, no. 19: 3976. https://doi.org/10.3390/molecules30193976
APA StyleMaltseva, P. Y., Plotnitskaya, N. A., & Ivshina, I. B. (2025). Molecular-Genetic Research of Rhodococcus rhodochrous IEGM 1362, an Active (–)-Isopulegol Biotransformer. Molecules, 30(19), 3976. https://doi.org/10.3390/molecules30193976







