A Perspective Towards More Sustainable Production of Biotechnologically Relevant Enzymes Using DESs
Abstract
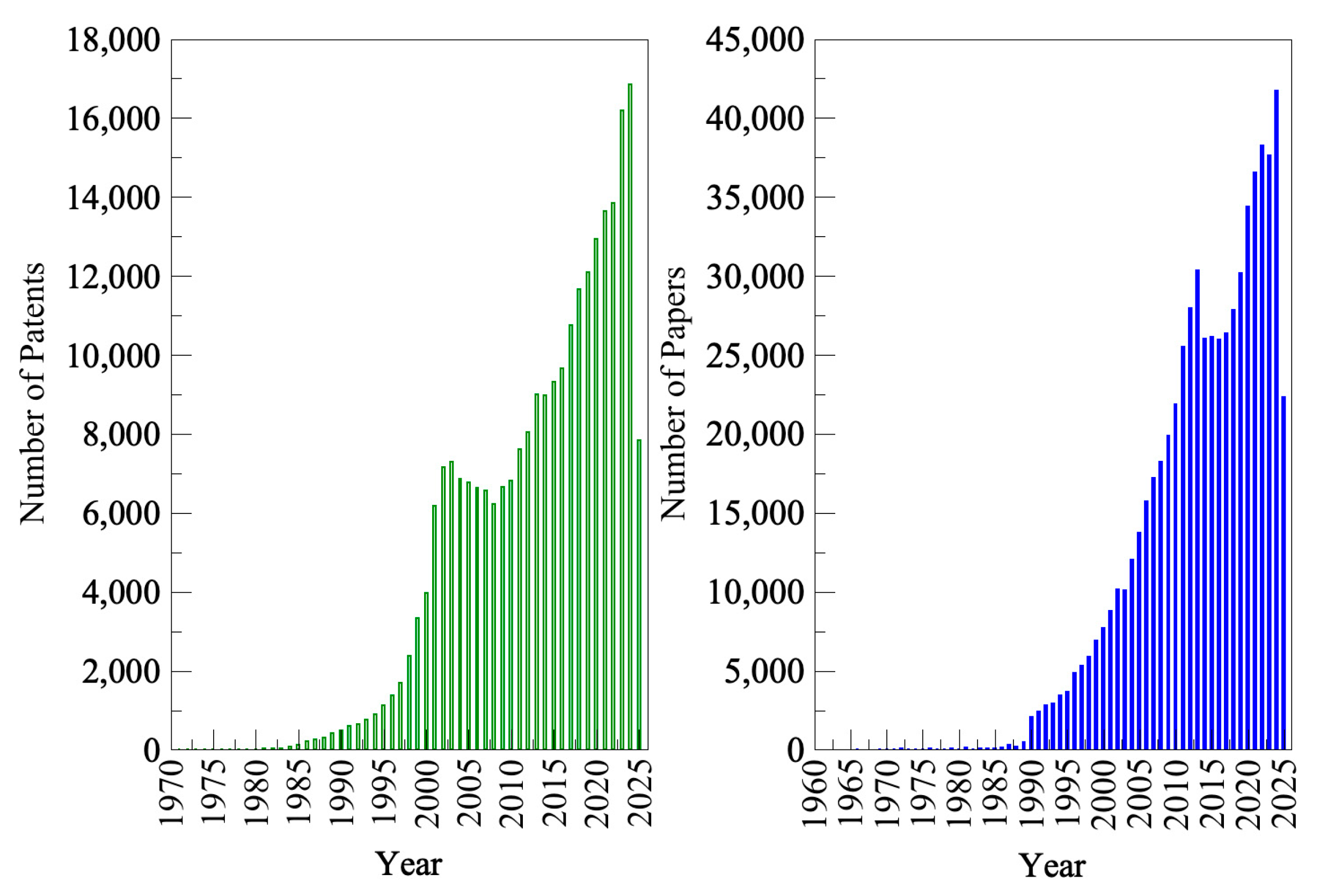
1. Biotechnology Industry
2. Enzymes
2.1. Production of Biocatalysts
2.2. Advances in Genetic Manipulation to Produce New Enzymes
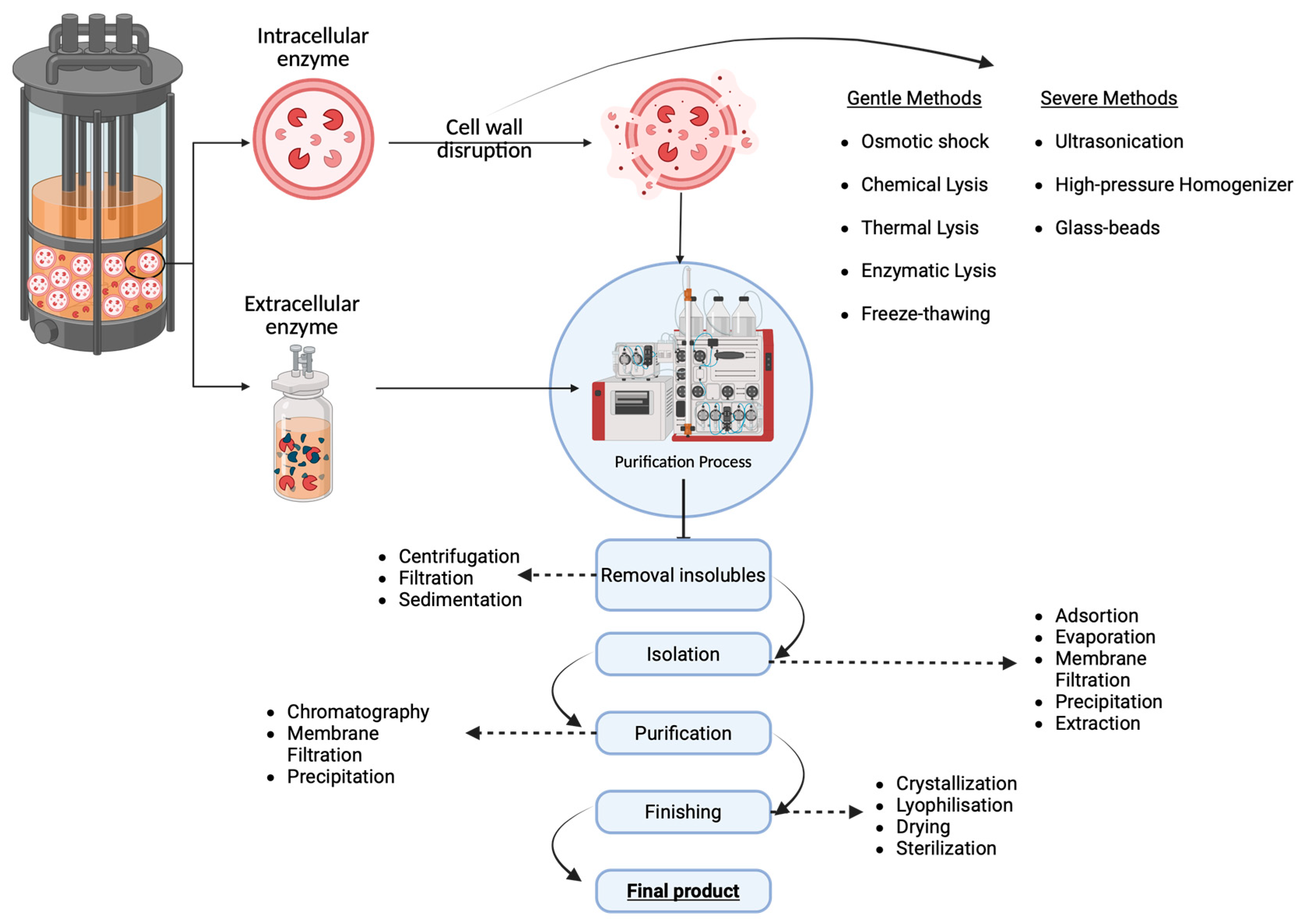
2.3. Production and Purification of Industrial Enzymes
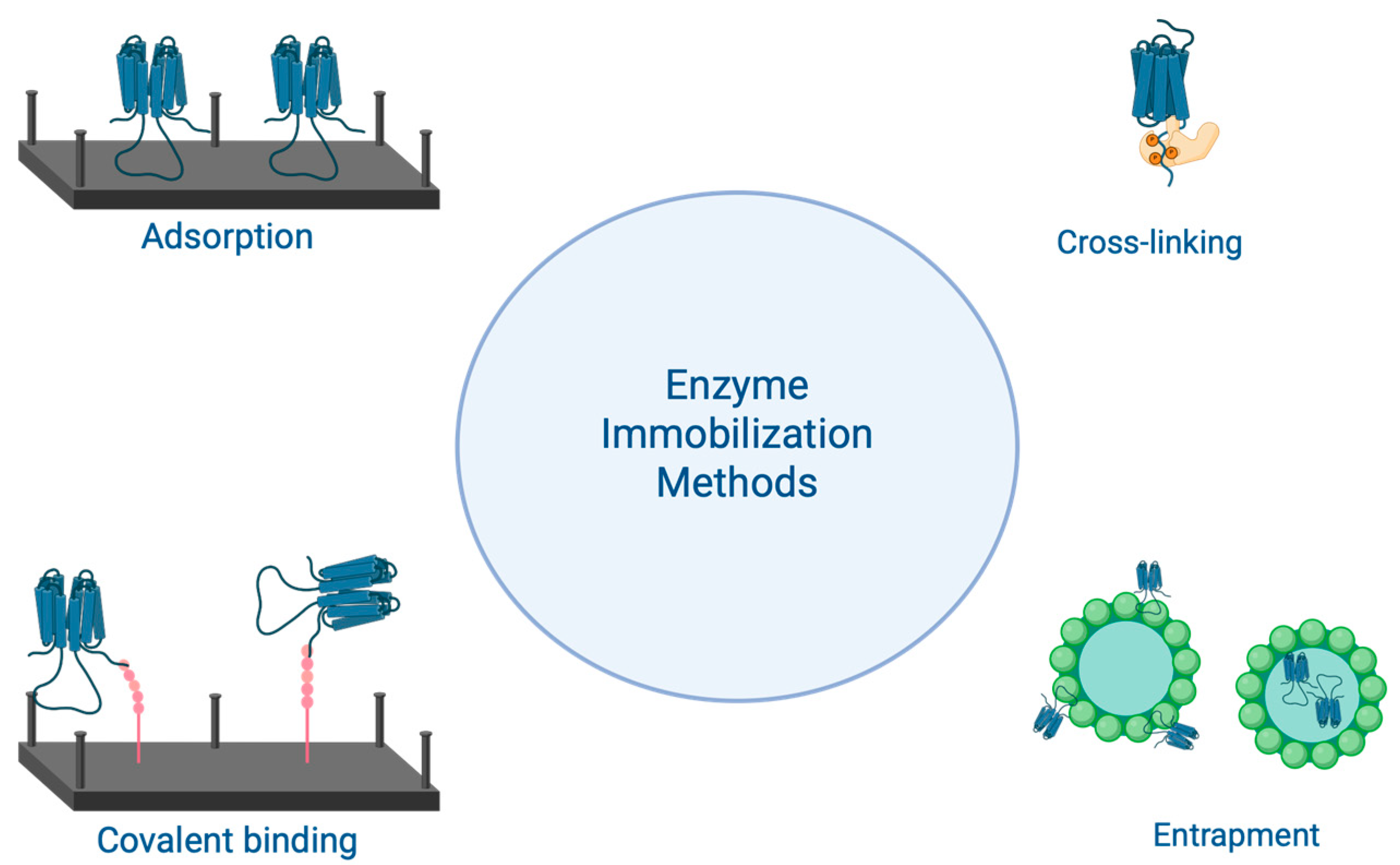
2.4. Stabilization of Commercial Enzymes
3. Deep Eutectic Solvents
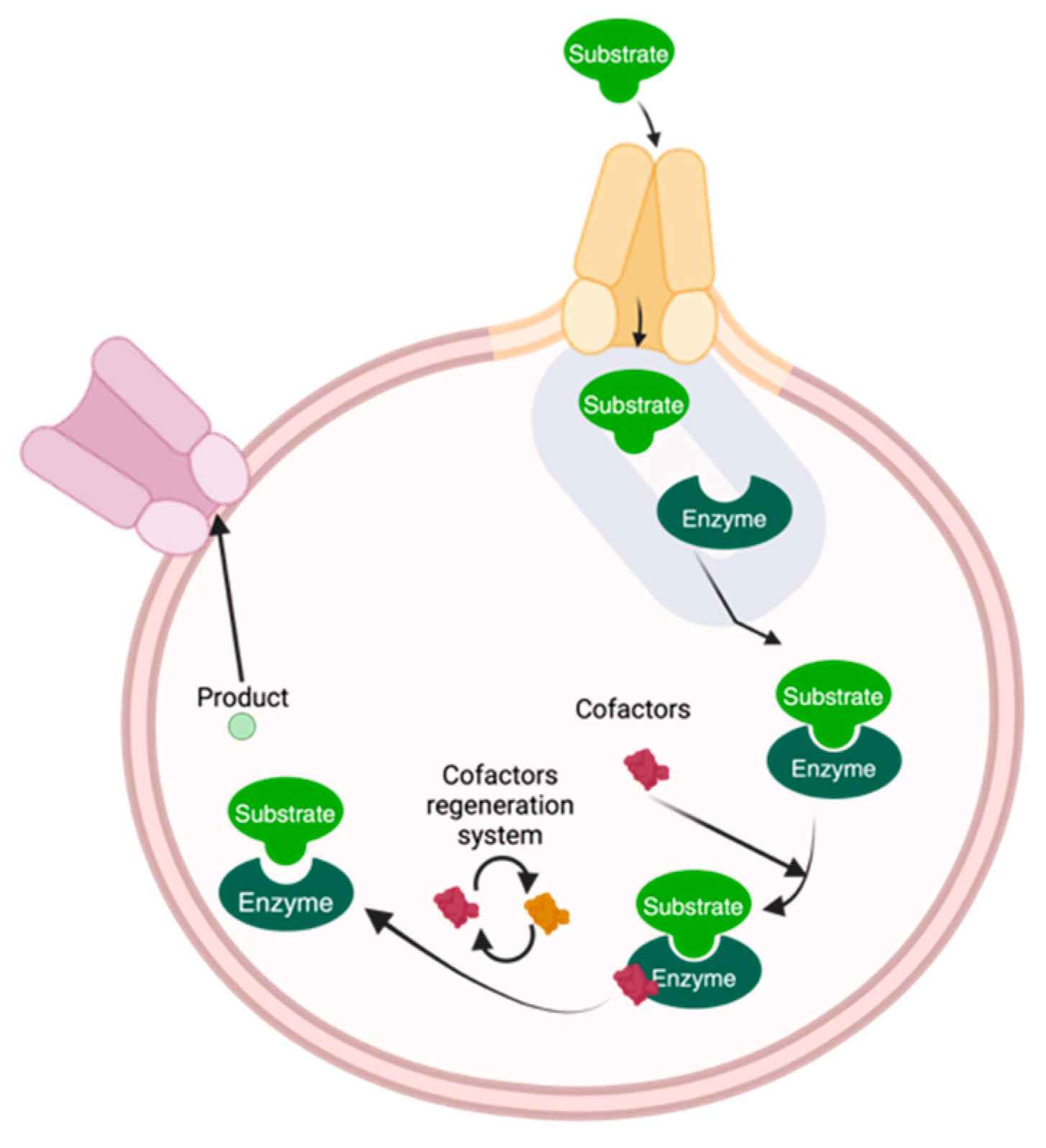
4. DESs and Living Systems
4.1. Studies of Deep Eutectic Solvents and Enzymes
4.2. DESs and Microorganism Lines
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Maienschein, J. Biotech: The Countercultural Origins of an Industry. BioScience 2007, 57, 373–374. [Google Scholar] [CrossRef]
- Hulse, J.H. Biotechnologies: Past history, present state and future prospects. Trends Food Sci. Technol. 2004, 15, 3–18. [Google Scholar] [CrossRef]
- Amarakoon, I.I.; Hamilton, C.-L.; Mitchell, S.A.; Tennant, P.F.; Roye, M.E. Chapter 28-Biotechnology. In Pharmacognosy; Badal, S., Delgoda, R., Eds.; Academic Press: Boston, MA, USA, 2017; pp. 549–563. ISBN 978-0-12-802104-0. [Google Scholar]
- Fasim, A.; More, V.S.; More, S.S. Large-scale production of enzymes for biotechnology uses. Curr. Opin. Biotechnol. 2021, 69, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Mesbah, N.M. Industrial Biotechnology Based on Enzymes from Extreme Environments. Front. Bioeng. Biotechnol. 2022, 10, 870083. [Google Scholar] [CrossRef]
- Polizzi, K.M.; Bommarius, A.S.; Broering, J.M.; Chaparro-Riggers, J.F. Stability of biocatalysts. Curr. Opin. Chem. Biol. 2007, 11, 220–225. [Google Scholar] [CrossRef]
- Enzymes Market Report 2025–2034|Trends and Forecast. Available online: https://www.thebusinessresearchcompany.com/report/enzyme-global-market-report (accessed on 28 August 2025).
- de Souza Vandenberghe, L.P.; Karp, S.G.; Pagnoncelli, M.G.B.; von Linsingen Tavares, M.; Junior, N.L.; Diestra, K.V.; Viesser, J.A.; Soccol, C.R. Classification of enzymes and catalytic properties. In Biomass, Biofuels, Biochemicals: Advances in Enzyme Catalysis and Technologies; Elsevier: Amsterdam, The Netherlands, 2020; pp. 11–30. ISBN 978-0-12-819820-9. [Google Scholar]
- Narváez, A.; Domínguez, E. ENZYMES|Overview. In Encyclopedia of Analytical Science, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2005; pp. 508–523. [Google Scholar] [CrossRef]
- Robinson, P.K. Enzymes: Principles and biotechnological applications. Essays Biochem. 2015, 59, 1–41. [Google Scholar] [CrossRef]
- Meghwanshi, G.K.; Kaur, N.; Verma, S.; Dabi, N.K.; Vashishtha, A.; Charan, P.D.; Purohit, P.; Bhandari, H.S.; Bhojak, N.; Kumar, R. Enzymes for pharmaceutical and therapeutic applications. Biotechnol. Appl. Biochem. 2020, 67, 586–601. [Google Scholar] [CrossRef]
- Cherry, J.R.; Fidantsef, A.L. Directed evolution of industrial enzymes: An update. Curr. Opin. Biotechnol. 2003, 14, 438–443. [Google Scholar] [CrossRef]
- Gogate, P.R.; Pandit, A.B. Application of cavitational reactors for cell disruption for recovery of intracellular enzymes. J. Chem. Technol. Biotechnol. 2008, 83, 1083–1093. [Google Scholar] [CrossRef]
- Gomes, T.A.; Zanette, C.M.; Spier, M.R. An overview of cell disruption methods for intracellular biomolecules recovery. Prep. Biochem. Biotechnol. 2020, 50, 635–654. [Google Scholar] [CrossRef]
- Kirk, O.; Borchert, T.V.; Fuglsang, C.C. Industrial enzyme applications. Curr. Opin. Biotechnol. 2002, 13, 345–351. [Google Scholar] [CrossRef]
- Jaeger, K.-E.; Eggert, T. Enantioselective biocatalysis optimized by directed evolution. Curr. Opin. Biotechnol. 2004, 15, 305–313. [Google Scholar] [CrossRef]
- Gurumallesh, P.; Alagu, K.; Ramakrishnan, B.; Muthusamy, S. A systematic reconsideration on proteases. Int. J. Biol. Macromol. 2019, 128, 254–267. [Google Scholar] [CrossRef]
- Fernández, L.; Breidenstein, E.B.M.; Song, D.; Hancock, R.E.W. Role of intracellular proteases in the antibiotic resistance, motility, and biofilm formation of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2012, 56, 1128–1132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Z.; Zhang, W.X.; Chen, X.L. Mechanisms for induction of microbial extracellular proteases in response to exterior proteins. Appl. Environ. Microbiol. 2020, 86, e01036-20. [Google Scholar] [CrossRef] [PubMed]
- Verma, N.; Kumar, V.; Bansal, M.C. Valorization of Waste Biomass in Fermentative Production of Cellulases: A Review. Waste Biomass Valorization 2021, 12, 613–640. [Google Scholar] [CrossRef]
- Begum, M.F.; Absar, N. Purification and Characterization of Intracellular Cellulase from Aspergillus oryzae ITCC-4857.01. Mycobiology 2009, 37, 121–127. [Google Scholar] [CrossRef]
- Gupta, V.K.; Steindorff, A.S.; de Paula, R.G.; Silva-Rocha, R.; Mach-Aigner, A.R.; Mach, R.L.; Silva, R.N. The Post-genomic Era of Trichoderma reesei: What’s Next? Trends Biotechnol. 2016, 34, 970–982. [Google Scholar] [CrossRef]
- Irfan, M.; Asghar, U.; Nadeem, M.; Nelofer, R.; Syed, Q. Optimization of process parameters for xylanase production by Bacillus sp. in submerged fermentation. J. Radiat. Res. Appl. Sci. 2016, 9, 139–147. [Google Scholar] [CrossRef]
- Marques, N.P.; de Cassia Pereira, J.; Gomes, E.; da Silva, R.; Araújo, A.R.; Ferreira, H.; Rodrigues, A.; Dussán, K.J.; Bocchini, D.A. Cellulases and xylanases production by endophytic fungi by solid state fermentation using lignocellulosic substrates and enzymatic saccharification of pretreated sugarcane bagasse. Ind. Crops Prod. 2018, 122, 66–75. [Google Scholar] [CrossRef]
- Solomon, V.; Teplitsky, A.; Shulami, S.; Zolotnitsky, G.; Shoham, Y.; Shoham, G. Structure-specificity relationships of an intracellular xylanase from Geobacillus stearothermophilus. Acta Crystallogr. D Biol. Crystallogr. 2007, 63, 845–859. [Google Scholar] [CrossRef]
- Knob, A.; Carmona, E.C. Purification and characterization of two extracellular xylanases from Penicillium sclerotiorum: A novel acidophilic xylanase. Appl. Biochem. Biotechnol. 2010, 162, 429–443. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Benjamin, S.; Soccol, C.R.; Nigam, P.; Krieger, N.; Soccol, V.T. The realm of microbial lipases in biotechnology. Biotechnol. Appl. Biochem. 1999, 29, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, F.B.; Shen, W.J. Hormone-sensitive lipase: Control of intracellular tri-(di-)acylglycerol and cholesteryl ester hydrolysis. J. Lipid Res. 2002, 43, 1585–1594. [Google Scholar] [CrossRef] [PubMed]
- Chandra, P.; Enespa; Singh, R.; Arora, P.K. Microbial lipases and their industrial applications: A comprehensive review. Microb. Cell Factories 2020, 19, 169. [Google Scholar] [CrossRef]
- Farooq, M.A.; Ali, S.; Hassan, A.; Tahir, H.M.; Mumtaz, S.; Mumtaz, S. Biosynthesis and industrial applications of α-amylase: A review. Arch. Microbiol. 2021, 203, 1281–1292. [Google Scholar] [CrossRef]
- Xu, Q.; Cao, Y.; Li, X.; Liu, L.; Qin, S.; Wang, Y.; Cao, Y.; Xu, H.; Qiao, D. Purification and characterization of a novel intracellular α-amylase with a wide variety of substrates hydrolysis and transglycosylation activity from Paenibacillus sp. SSG-1. Protein Expr. Purif. 2018, 144, 62–70. [Google Scholar] [CrossRef]
- Krishnan, A.; Alias, Z.; Convey, P.; González-Aravena, M.; Smykla, J.; Rizman-Idid, M.; Alias, S.A. Temperature and pH Profiling of Extracellular Amylase from Antarctic and Arctic Soil Microfungi. Fermentation 2022, 8, 601. [Google Scholar] [CrossRef]
- Buddhiwant, P.; Bhavsar, K.; Kumar, V.R.; Khire, J.M. Phytase production by solid-state fermentation of groundnut oil cake by Aspergillus niger: A bioprocess optimization study for animal feedstock applications. Prep. Biochem. Biotechnol. 2015, 46, 531–538. [Google Scholar] [CrossRef]
- Sumengen, M.; Dincer, S.; Kaya, A. Production and Characterization of Phytase from Lactobacillus plantarum. Food Biotechnol. 2013, 27, 105–118. [Google Scholar] [CrossRef]
- Steiner, K.; Schwab, H. Recent advances in rational approaches for enzyme engineering. Comput. Struct. Biotechnol. J. 2012, 2, e201209010. [Google Scholar] [CrossRef]
- Reetz, M.T.; Sun, Z.; Qu, G. Introduction to Directed Evolution and Rational Design as Protein Engineering Techniques. In Enzyme Engineering; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2023; pp. 1–28. ISBN 978-3-527-83689-5. [Google Scholar]
- Rybarczyk, A.; Sultan, T.; Hussain, N.; Azam, H.M.H.; Rafique, S.; Zdarta, J.; Jesionowski, T. Fusion of enzymatic proteins: Enhancing biological activities and facilitating biological modifications. Adv. Colloid Interface Sci. 2025, 340, 103473. [Google Scholar] [CrossRef] [PubMed]
- Schüürmann, J.; Quehl, P.; Festel, G.; Jose, J. Bacterial whole-cell biocatalysts by surface display of enzymes: Toward industrial application. Appl. Microbiol. Biotechnol. 2014, 98, 8031–8046. [Google Scholar] [CrossRef] [PubMed]
- Drienovská, I.; Roelfes, G. Expanding the enzyme universe with genetically encoded unnatural amino acids. Nat. Catal. 2020, 3, 193–202. [Google Scholar] [CrossRef]
- McKelvey, S.M.; Murphy, R.A. Biotechnological Use of Fungal Enzymes. In Fungi; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2017; pp. 201–225. ISBN 978-1-119-37431-2. [Google Scholar]
- van Beilen, J.B.; Li, Z. Enzyme technology: An overview. Curr. Opin. Biotechnol. 2002, 13, 338–344. [Google Scholar] [CrossRef]
- Li, S.; Yang, X.; Yang, S.; Zhu, M.; Wang, X. Technology prospecting on enzymes: Application, marketing and engineering. Comput. Struct. Biotechnol. J. 2012, 2, e201209017. [Google Scholar] [CrossRef]
- Liu, X.; Kokare, C. Microbial Enzymes of Use in Industry. In Biotechnology of Microbial Enzymes: Production, Biocatalysis and Industrial Applications; Elsevier Inc.: Amsterdam, The Netherlands, 2017; pp. 267–298. ISBN 978-0-12-803746-1. [Google Scholar]
- Suenaga, H. Targeted metagenomics unveils the molecular basis for adaptive evolution of enzymes to their environment. Front. Microbiol. 2015, 6, 1018. [Google Scholar] [CrossRef]
- Li, L.; Li, W.; Gong, J.; Xu, Y.; Wu, Z.; Jiang, Z.; Cheng, Y.S.; Li, Q.; Ni, H. An effective computational-screening strategy for simultaneously improving both catalytic activity and thermostability of α-l-rhamnosidase. Biotechnol. Bioeng. 2021, 118, 3409–3419. [Google Scholar] [CrossRef]
- Wu, H.; Chen, Q.; Zhang, W.; Mu, W. Overview of strategies for developing high thermostability industrial enzymes: Discovery, mechanism, modification and challenges. Crit. Rev. Food Sci. Nutr. 2023, 63, 2057–2073. [Google Scholar] [CrossRef]
- de Freitas-Júnior, A.C.V.; da Costa, H.M.S.; Marcuschi, M.; Icimoto, M.Y.; Machado, M.F.M.; Machado, M.F.M.; Ferreira, J.C.; de Oliveira, V.M.S.B.B.; Buarque, D.S.; Bezerra, R.S. Substrate specificity, physicochemical and kinetic properties of a trypsin from the giant Amazonian fish pirarucu (Arapaima gigas). Biocatal. Agric. Biotechnol. 2021, 35, 102073. [Google Scholar] [CrossRef]
- Li, Y.; Yang, Y.; Zhang, M.; Yue, X.; Guo, R.T.; Huang, Z.; Chen, F. Mutagenesis of a Single Site Inverts the Stereopreference of Imine Reductase. ACS Catal. 2025, 15, 2192–2199. [Google Scholar] [CrossRef]
- Sharma, A.; Gupta, G.; Ahmad, T.; Mansoor, S.; Kaur, B. Enzyme Engineering: Current Trends and Future Perspectives. Food Rev. Int. 2021, 37, 121–154. [Google Scholar] [CrossRef]
- Kuznetsova, E.; Proudfoot, M.; Sanders, S.A.; Reinking, J.; Savchenko, A.; Arrowsmith, C.H.; Edwards, A.M.; Yakunin, A.F. Enzyme genomics: Application of general enzymatic screens to discover new enzymes. FEMS Microbiol. Rev. 2005, 29, 263–279. [Google Scholar] [CrossRef]
- Adam, G.C.; Sorensen, E.J.; Cravatt, B.F. Proteomic profiling of mechanistically distinct enzyme classes using a common chemotype. Nat. Biotechnol. 2002, 20, 805–809. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, T.; de Zafra, C.; Kim, A.; Gelzleichter, T.R. Chapter 1-Overview of Biopharmaceuticals and Comparison with Small-molecule Drug Development. In Nonclinical Development of Novel Biologics, Biosimilars, Vaccines and Specialty Biologics; Plitnick, L.M., Herzyk, D.J., Eds.; Academic Press: San Diego, CA, USA, 2013; pp. 3–33. ISBN 978-0-12-394810-6. [Google Scholar]
- Singh, P.; Kumar, S. Microbial enzyme in food biotechnology. In Enzymes in Food Biotechnology: Production, Applications, and Future Prospects; Elsevier: Amsterdam, The Netherlands, 2018; pp. 19–28. ISBN 978-0-12-813280-7. [Google Scholar]
- Sanchez, S.; Demain, A.L. Metabolic regulation of fermentation processes. Enzyme Microb. Technol. 2002, 31, 895–906. [Google Scholar] [CrossRef]
- Bussamara, R.; Fuentefria, A.M.; de Oliveira, E.S.; Broetto, L.; Simcikova, M.; Valente, P.; Schrank, A.; Vainstein, M.H. Isolation of a lipase-secreting yeast for enzyme production in a pilot-plant scale batch fermentation. Bioresour. Technol. 2010, 101, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.C.; Inwood, S.; Gong, T.; Sharma, A.; Yu, L.Y.; Zhu, P. Fed-batch high-cell-density fermentation strategies for Pichia pastoris growth and production. Crit. Rev. Biotechnol. 2019, 39, 258–271. [Google Scholar] [CrossRef]
- Li, T.; Chen, X.B.; Chen, J.C.; Wu, Q.; Chen, G.Q. Open and continuous fermentation: Products, conditions and bioprocess economy. Biotechnol. J. 2014, 9, 1503–1511. [Google Scholar] [CrossRef]
- Farinas, C.S. Developments in solid-state fermentation for the production of biomass-degrading enzymes for the bioenergy sector. Renew. Sustain. Energy Rev. 2015, 52, 179–188. [Google Scholar] [CrossRef]
- Ouedraogo, J.-P.; Tsang, A. Production of Native and Recombinant Enzymes by Fungi for Industrial Applications. In Encyclopedia of Mycology; Zaragoza, Ó., Casadevall, A., Eds.; Elsevier: Oxford, UK, 2021; pp. 222–232. ISBN 978-0-323-85180-0. [Google Scholar]
- Novelli, P.K.; Barros, M.M.; Fleuri, L.F. Novel inexpensive fungi proteases: Production by solid state fermentation and characterization. Food Chem. 2016, 198, 119–124. [Google Scholar] [CrossRef]
- Mascarin, G.M.; Golo, P.S.; de Ribeiro-Silva, C.S.; Muniz, E.R.; de Franco, A.O.; Kobori, N.N.; Fernandes, É.K.K. Advances in submerged liquid fermentation and formulation of entomopathogenic fungi. Appl. Microbiol. Biotechnol. 2024, 108, 1–20. [Google Scholar] [CrossRef]
- Hölker, U.; Höfer, M.; Lenz, J. Biotechnological advantages of laboratory-scale solid-state fermentation with fungi. Appl. Microbiol. Biotechnol. 2004, 64, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Linke, D.; Berger, R.G. Foaming of proteins: New prospects for enzyme purification processes. J. Biotechnol. 2011, 152, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Schwaminger, S.P.; Zimmermann, I.; Berensmeier, S. Current research approaches in downstream processing of pharmaceutically relevant proteins. Curr. Opin. Biotechnol. 2022, 77, 102768. [Google Scholar] [CrossRef] [PubMed]
- Gomis-Fons, J.; Löfgren, A.; Andersson, N.; Nilsson, B.; Berghard, L.; Wood, S. Integration of a complete downstream process for the automated lab-scale production of a recombinant protein. J. Biotechnol. 2019, 301, 45–51. [Google Scholar] [CrossRef]
- Mohapatra, S.; Thatoi, H.N. Purification and Characterization of Extracellular enzyme from Aspergillus fumigatus and Its Application on a pennisetum sp for enhanced glucose production. Can. J. Biotechnol. 2017, 1, 273. [Google Scholar] [CrossRef]
- de Carvalho, J.C.; Medeiros, A.B.P.; Letti, L.A.J.; Kirnev, P.C.S.; Soccol, C.R. Cell Disruption and Isolation of Intracellular Products. In Current Developments in Biotechnology and Bioengineering: Production, Isolation and Purification of Industrial Products; Elsevier Inc.: Amsterdam, The Netherlands, 2016; pp. 807–822. ISBN 978-0-444-63662-1. [Google Scholar]
- Nishikawa, A.H.; Bailon, P. Affinity purification methods: Nonspecific adsorption of proteins due to ionic groups in cyanogen bromide treated agarose. Arch. Biochem. Biophys. 1975, 168, 576–584. [Google Scholar] [CrossRef]
- Li, X.; Liu, Y.; Sun, Y. Development of poly(methacrylate)-grafted Sepharose FF for cation-exchange chromatography of proteins. J. Chromatogr. A 2020, 1634, 461669. [Google Scholar] [CrossRef]
- Takahashi, H.; Li, B.; Sasaki, T.; Miyazaki, C.; Kajino, T.; Inagaki, S. Immobilized enzymes in ordered mesoporous silica materials and improvement of their stability and catalytic activity in an organic solvent. Microporous Mesoporous Mater. 2001, 44, 755–762. [Google Scholar] [CrossRef]
- Rudolph, F.B.; Cooper, B.F.; Greenhut, J. Enzyme purification by high-performance ion-exchange liquid chromatography. In Progress in HPLC; CRC Press: Florida, FL, USA, 2020; pp. 133–147. [Google Scholar] [CrossRef]
- dos Santos, R.; Carvalho, A.L.; Roque, A.C.A. Renaissance of protein crystallization and precipitation in biopharmaceuticals purification. Biotechnol. Adv. 2017, 35, 41–50. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Berenguer-Murcia, Á.; Carballares, D.; Morellon-Sterling, R.; Fernandez-Lafuente, R. Stabilization of enzymes via immobilization: Multipoint covalent attachment and other stabilization strategies. Biotechnol. Adv. 2021, 52, 107821. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.M.; Han, S.S.; Kim, H.S. Industrial applications of enzyme biocatalysis: Current status and future aspects. Biotechnol. Adv. 2015, 33, 1443–1454. [Google Scholar] [CrossRef] [PubMed]
- Papamichael, E.M.; Stergiou, P.-Y. Chapter 12-Enzyme immobilization strategies and bioprocessing applications. In Biomass, Biofuels, Biochemicals; Singh, S.P., Pandey, A., Singhania, R.R., Larroche, C., Li, Z., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 217–241. ISBN 978-0-12-819820-9. [Google Scholar]
- Brady, D.; Jordaan, J. Advances in enzyme immobilisation. Biotechnol. Lett. 2009, 31, 1639–1650. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.H.; Kim, M. An Overview of Techniques in Enzyme Immobilization. Appl. Sci. Converg. Technol. 2017, 26, 157–163. [Google Scholar] [CrossRef]
- Remonatto, D.; Izidoro, B.F.; Mazziero, V.T.; Catarino, B.P.; do Nascimento, J.F.C.; Cerri, M.O.; Andrade, G.S.S.; de Paula, A.V. 3D printing and enzyme immobilization: An overview of current trends. Bioprinting 2023, 33, e00289. [Google Scholar] [CrossRef]
- Dey, G.; Nagpal, V.; Banerjee, R. Immobilization of α-amylase from Bacillus circulans GRS 313 on coconut fiber. Appl. Biochem. Biotechnol. 2002, 102, 303–313. [Google Scholar] [CrossRef]
- Rosales-Hernández, M.; Kispert, L.; Torres-Ramírez, E.; Ramírez-Rosales, D.; Zamorano-Ulloa, R.; Trujillo-Ferrara, J. Electron paramagnetic resonance analyses of biotransformation reactions with cytochrome P-450 immobilized on mesoporous molecular sieves. Biotechnol. Lett. 2007, 29, 919–924. [Google Scholar] [CrossRef]
- Mitchell, S.; Pérez-Ramírez, J. Mesoporous zeolites as enzyme carriers: Synthesis, characterization, and application in biocatalysis. Catal. Today 2011, 168, 28–37. [Google Scholar] [CrossRef]
- Fu, J.; Reinhold, J.; Woodbury, N.W. Peptide-modified surfaces for enzyme immobilization. PLoS ONE 2011, 6, e18692. [Google Scholar] [CrossRef]
- Ispas, C.; Sokolov, I.; Andreescu, S. Enzyme-functionalized mesoporous silica for bioanalytical applications. Anal. Bioanal. Chem. 2009, 393, 543–554. [Google Scholar] [CrossRef]
- Sardar, M.; Roy, I.; Gupta, M.N. Simultaneous purification and immobilization of Aspergillus niger xylanase on the reversibly soluble polymer EudragitTM L-100. Enzyme Microb. Technol. 2000, 27, 672–679. [Google Scholar] [CrossRef]
- Shi, Q.H.; Tian, Y.; Dong, X.Y.; Bai, S.; Sun, Y. Chitosan-coated silica beads as immobilized metal affinity support for protein adsorption. Biochem. Eng. J. 2003, 16, 317–322. [Google Scholar] [CrossRef]
- Sardar, M.; Gupta, M.N. Immobilization of tomato pectinase on Con A-Seralose 4B by bioaffinity layering. Enzyme Microb. Technol. 2005, 37, 355–359. [Google Scholar] [CrossRef]
- Ho, L.F.; Li, S.Y.; Lin, S.C.; Hsu, W.H. Integrated enzyme purification and immobilization processes with immobilized metal affinity adsorbents. Process Biochem. 2004, 39, 1573–1581. [Google Scholar] [CrossRef]
- Zhang, C.; Xing, X.H. Enzyme Bioreactors. In Comprehensive Biotechnology, 2nd ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2011; Volume 2, pp. 319–329. ISBN 978-0-08-088504-9. [Google Scholar]
- Dai, D.; Xia, L. Effect of lipase immobilization on resolution of (R, S)-2-octanol in nonaqueous media using modified ultrastable-Y molecular sieve as support. Appl. Biochem. Biotechnol. 2006, 134, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Erdemir, S.; Yilmaz, M. Catalytic effect of calix[n]arene based sol-gel encapsulated or covalent immobilized lipases on enantioselective hydrolysis of (R/S)-naproxen methyl ester. J. Incl. Phenom. Macrocycl. Chem. 2012, 72, 189–196. [Google Scholar] [CrossRef]
- Betigeri, S.S.; Neau, S.H. Immobilization of lipase using hydrophilic polymers in the form of hydrogel beads. Biomaterials 2002, 23, 3627–3636. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.C.; Kuan, I.C.; Hong, J.R.; Tsai, B.H.; Lee, S.L.; Yu, C.Y. Activity enhancement and stabilization of lipase from Pseudomonas cepacia in polyallylamine-mediated biomimetic silica. Biotechnol. Lett. 2011, 33, 525–529. [Google Scholar] [CrossRef]
- Cabadaj, P.; Illeová, V.; Červeňanský, I.; Rupčíková, V.; Krajčovič, T.; Bučko, M.; Polakovič, M. Investigation of process stability of a whole-cell biocatalyst with Baeyer–Villiger monooxygenase activity in continuous bioreactors. Environ. Technol. Innov. 2023, 30, 103083. [Google Scholar] [CrossRef]
- Wachtmeister, J.; Rother, D. Recent advances in whole cell biocatalysis techniques bridging from investigative to industrial scale. Curr. Opin. Biotechnol. 2016, 42, 169–177. [Google Scholar] [CrossRef]
- de Carvalho, C.C.C.R. Whole cell biocatalysts: Essential workers from Nature to the industry. Microb. Biotechnol. 2017, 10, 250–263. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, H.; Zhu, Y.; Zhang, Y.; Li, X.; Wang, F. Saccharomyces cerevisiae cell surface display technology: Strategies for improvement and applications. Front. Bioeng. Biotechnol. 2022, 10, 1056804. [Google Scholar] [CrossRef]
- Wei, P.; Pan, X.; Chen, C.-Y.; Li, H.-Y.; Yan, X.; Li, C.; Chu, Y.-H.; Yan, B. Emerging impacts of ionic liquids on eco-environmental safety and human health. Chem. Soc. Rev. 2021, 50, 13609–13627. [Google Scholar] [CrossRef] [PubMed]
- Taklimi, S.M.; Divsalar, A.; Ghalandari, B.; Ding, X.; Di Gioia, M.L.; Omar, K.A.; Saboury, A.A. Effects of deep eutectic solvents on the activity and stability of enzymes. J. Mol. Liq. 2023, 377, 121562. [Google Scholar] [CrossRef]
- Smith, E.L.; Abbott, A.P.; Ryder, K.S. Deep Eutectic Solvents (DESs) and Their Applications. Chem. Rev. 2014, 114, 11060–11082. [Google Scholar] [CrossRef] [PubMed]
- Vanda, H.; Dai, Y.; Wilson, E.G.; Verpoorte, R.; Choi, Y.H. Green solvents from ionic liquids and deep eutectic solvents to natural deep eutectic solvents. Comptes Rendus Chim. 2018, 21, 628–638. [Google Scholar] [CrossRef]
- Juneidi, I.; Hayyan, M.; Hashim, M.A. Intensification of biotransformations using deep eutectic solvents: Overview and outlook. Process Biochem. 2018, 66, 33–60. [Google Scholar] [CrossRef]
- Perna, F.M.; Vitale, P.; Capriati, V. Deep eutectic solvents and their applications as green solvents. Curr. Opin. Green Sustain. Chem. 2020, 21, 27–33. [Google Scholar] [CrossRef]
- Zhekenov, T.; Toksanbayev, N.; Kazakbayeva, Z.; Shah, D.; Mjalli, F.S. Formation of type III Deep Eutectic Solvents and effect of water on their intermolecular interactions. Fluid Phase Equilibria 2017, 441, 43–48. [Google Scholar] [CrossRef]
- Agieienko, V.; Buchner, R. Densities, Viscosities, and Electrical Conductivities of Pure Anhydrous Reline and Its Mixtures with Water in the Temperature Range (293.15 to 338.15) K. J. Chem. Eng. Data 2019, 64, 4763–4774. [Google Scholar] [CrossRef]
- Choi, Y.H.; van Spronsen, J.; Dai, Y.; Verberne, M.; Hollmann, F.; Arends, I.W.C.E.; Witkamp, G.J.; Verpoorte, R. Are natural deep eutectic solvents the missing link in understanding cellular metabolism and physiology? Plant Physiol. 2011, 156, 1701–1705. [Google Scholar] [CrossRef]
- Gao, Y.; Fan, M.; Cheng, X.; Liu, X.; Yang, H.; Ma, W.; Guo, M.; Li, L. Deep eutectic solvent: Synthesis, classification, properties and application in macromolecular substances. Int. J. Biol. Macromol. 2024, 278, 134593. [Google Scholar] [CrossRef]
- Buzatu, A.R.; Todea, A.; Pop, R.; Dreavă, D.M.; Paul, C.; Bîtcan, I.; Motoc, M.; Peter, F.; Boeriu, C.G. Designed Reactive Natural Deep Eutectic Solvents for Lipase-Catalyzed Esterification. Molecules 2025, 30, 778. [Google Scholar] [CrossRef]
- El Achkar, T.; Fourmentin, S.; Greige-Gerges, H. Deep eutectic solvents: An overview on their interactions with water and biochemical compounds. J. Mol. Liq. 2019, 288, 111028. [Google Scholar] [CrossRef]
- Pätzold, M.; Siebenhaller, S.; Kara, S.; Liese, A.; Syldatk, C.; Holtmann, D. Deep Eutectic Solvents as Efficient Solvents in Biocatalysis. Trends Biotechnol. 2019, 37, 943–959. [Google Scholar] [CrossRef] [PubMed]
- Arnodo, D.; Maffeis, E.; Marra, F.; Nejrotti, S.; Prandi, C. Combination of Enzymes and Deep Eutectic Solvents as Powerful Toolbox for Organic Synthesis. Molecules 2023, 28, 516. [Google Scholar] [CrossRef] [PubMed]
- Meneses, L.; Gajardo-Parra, N.F.; Cea-Klapp, E.; Garrido, J.M.; Held, C.; Duarte, A.R.; Paiva, A. Improving the activity of horseradish peroxidase in betaine-based natural deep eutectic systems. RSC Sustain. 2023, 1, 886–897. [Google Scholar] [CrossRef]
- Gajardo-Parra, N.F.; Meneses, L.; Duarte, A.R.C.; Paiva, A.; Held, C. Assessing the Influence of Betaine-Based Natural Deep Eutectic Systems on Horseradish Peroxidase. ACS Sustain. Chem. Eng. 2022, 10, 12873–12881. [Google Scholar] [CrossRef]
- Miranda-Molina, A.; Xolalpa, W.; Strompen, S.; Arreola-Barroso, R.; Olvera, L.; López-Munguía, A.; Castillo, E.; Saab-Rincon, G. Deep eutectic solvents as new reaction media to produce alkyl-glycosides using alpha-amylase from Thermotoga maritima. Int. J. Mol. Sci. 2019, 20, 5439. [Google Scholar] [CrossRef]
- Cao, J.; Wu, R.; Zhu, F.; Dong, Q.; Su, E. Enzymes in nearly anhydrous deep eutectic solvents: Insight into the biocompatibility and thermal stability. Enzyme Microb. Technol. 2022, 157, 110022. [Google Scholar] [CrossRef]
- Toledo, M.L.; Pereira, M.M.; Freire, M.G.; Silva, J.P.A.; Coutinho, J.A.P.; Tavares, A.P.M. Laccase Activation in Deep Eutectic Solvents. ACS Sustain. Chem. Eng. 2019, 7, 11806–11814. [Google Scholar] [CrossRef]
- de Gonzalo, G.; Martin, C.; Fraaije, M.W. Positive impact of natural deep eutectic solvents on the biocatalytic performance of 5-hydroxymethyl-furfural oxidase. Catalysts 2020, 10, 447. [Google Scholar] [CrossRef]
- Juneidi, I.; Hayyan, M.; Hashim, M.A.; Hayyan, A. Pure and aqueous deep eutectic solvents for a lipase-catalysed hydrolysis reaction. Biochem. Eng. J. 2017, 117, 129–138. [Google Scholar] [CrossRef]
- Kim, S.H.; Park, S.; Yu, H.; Kim, J.H.; Kim, H.J.; Yang, Y.H.; Kim, Y.H.; Kim, K.J.; Kan, E.; Lee, S.H. Effect of deep eutectic solvent mixtures on lipase activity and stability. J. Mol. Catal. B Enzym. 2016, 128, 65–72. [Google Scholar] [CrossRef]
- Fredes, Y.; Chamorro, L.; Cabrera, Z. Increased selectivity of novozym 435 in the asymmetric hydrolysis of a substrate with high hydrophobicity through the use of deep eutectic solvents and high substrate concentrations. Molecules 2019, 24, 792. [Google Scholar] [CrossRef] [PubMed]
- Hümmer, M.; Kara, S.; Liese, A.; Huth, I.; Schrader, J.; Holtmann, D. Synthesis of (-)-menthol fatty acid esters in and from (-)-menthol and fatty acids–novel concept for lipase catalyzed esterification based on eutectic solvents. Mol. Catal. 2018, 458, 67–72. [Google Scholar] [CrossRef]
- Zeng, C.X.; Qi, S.J.; Xin, R.P.; Yang, B.; Wang, Y.H. Enzymatic selective synthesis of 1,3-DAG based on deep eutectic solvent acting as substrate and solvent. Bioprocess Biosyst. Eng. 2015, 38, 2053–2061. [Google Scholar] [CrossRef]
- Guajardo, N.; Müller, C.R.; Schrebler, R.; Carlesi, C.; de Domínguez María, P. Deep Eutectic Solvents for Organocatalysis, Biotransformations, and Multistep Organocatalyst/Enzyme Combinations. ChemCatChem 2016, 8, 1020–1027. [Google Scholar] [CrossRef]
- Mao, S.; Yu, L.; Ji, S.; Liu, X.; Lu, F. Evaluation of deep eutectic solvents as co-solvent for steroids 1-en-dehydrogenation biotransformation by Arthrobacter simplex. J. Chem. Technol. Biotechnol. 2016, 91, 1099–1104. [Google Scholar] [CrossRef]
- Cao, J.; Wu, R.; Dong, Q.; Zhao, L.; Cao, F.; Su, E. Effective Release of Intracellular Enzymes by Permeating the Cell Membrane with Hydrophobic Deep Eutectic Solvents. ChemBioChem 2020, 21, 672–680. [Google Scholar] [CrossRef]
- Torregrosa-Crespo, J.; Marset, X.; Guillena, G.; Ramón, D.J.; Martínez-Espinosa, R.M. New guidelines for testing “Deep eutectic solvents” toxicity and their effects on the environment and living beings. Sci. Total Environ. 2020, 704, 135382. [Google Scholar] [CrossRef]
- Bryant, S.J.; Awad, M.N.; Elbourne, A.; Christofferson, A.J.; Martin, A.V.; Meftahi, N.; Drummond, C.J.; Greaves, T.L.; Bryant, G. Deep eutectic solvents as cryoprotective agents for mammalian cells. J. Mater. Chem. B 2022, 10, 4546–4560. [Google Scholar] [CrossRef]
- Juneidi, I.; Hayyan, M.; Ali, O.M. Toxicity profile of choline chloride-based deep eutectic solvents for fungi and Cyprinus carpio fish. Environ. Sci. Pollut. Res. 2016, 23, 7648–7659. [Google Scholar] [CrossRef]
- Rodríguez-Juan, E.; López, S.; Abia, R.; Muriana, F.J.G.; Fernández-Bolaños, J.; García-Borrego, A. Antimicrobial activity on phytopathogenic bacteria and yeast, cytotoxicity and solubilizing capacity of deep eutectic solvents. J. Mol. Liq. 2021, 337, 116343. [Google Scholar] [CrossRef]
- Yang, T.X.; Zhao, L.Q.; Wang, J.; Song, G.L.; Liu, H.M.; Cheng, H.; Yang, Z. Improving Whole-Cell Biocatalysis by Addition of Deep Eutectic Solvents and Natural Deep Eutectic Solvents. ACS Sustain. Chem. Eng. 2017, 5, 5713–5722. [Google Scholar] [CrossRef]
- Daneshjou, S.; Khodaverdian, S.; Dabirmanesh, B.; Rahimi, F.; Daneshjoo, S.; Ghazi, F.; Khajeh, K. Improvement of chondroitinases ABCI stability in natural deep eutectic solvents. J. Mol. Liq. 2017, 227, 21–25. [Google Scholar] [CrossRef]
- Bubalo, M.C.; Andreou, T.; Panić, M.; Radović, M.; Radošević, K.; Redovniković, I.R. Natural multi-osmolyte cocktails form deep eutectic systems of unprecedented complexity: Discovery, affordances and perspectives. Green Chem. 2023, 25, 3398–3417. [Google Scholar] [CrossRef]
- Damjanović, A.; Logarušić, M.; Tumir, L.M.; Andreou, T.; Bubalo, M.C.; Redovniković, I.R. Enhancing protein stability under stress: Osmolyte-based deep eutectic solvents as a biocompatible and robust stabilizing medium for lysozyme under heat and cold shock. Phys. Chem. Chem. Phys. 2024, 26, 21040–21051. [Google Scholar] [CrossRef]
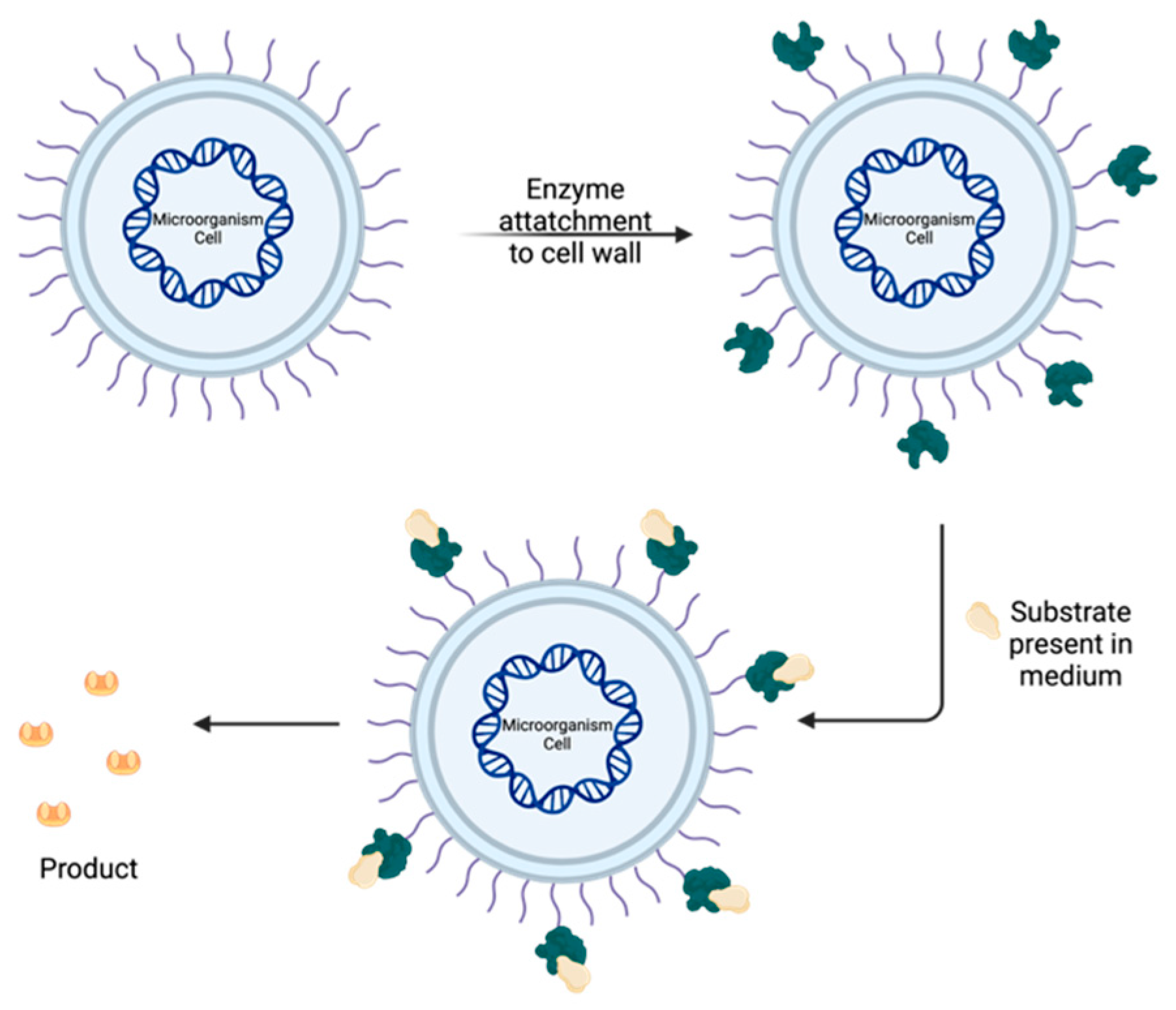

| Endoenzymes | Exoenzymes | |
|---|---|---|
| Production | Inside cells | Inside cells |
| Function location | Inside cells | Outside cells |
| Function | Facilitate biochemical reactions within the cell | Breakdown of the extremity of the polymer to form monomers one by one |
| Digestion | Inside the cell | Outside the cell |
| Enzymes | Function | Fermentation (a) | Application Field | Intracellular Enzyme | Extracellular Production |
|---|---|---|---|---|---|
| Proteases (Hydrolases, EC 3) | Hydrolysis of proteins | SMF [17] SSF ** [17] | Detergent; Pharmaceutical; Food | Protease Lon [18] e.g., Pseudomonas aeruginosa | Cysteine Proteases [19] e.g., Bacteria, archaea, and fungi |
| Cellulases * (Hydrolases, EC 3) | Conversion of cellulose from plants into sugars | SMF [20] SSF [20] | Textile | Cellulases [21] e.g., Aspergillus oryzae | Cellulase [22] e.g., Trichoderma reesei |
| Xylanases (Hydrolases, EC 3) | Hydrolysis of hemicellulose | SMF [23] SSF [24] | Food; Pharmaceutical; Textiles; Paper | Xylanases I and II [25] e.g., Penicillium sclerotiorum | Xylanase IXT6 [26] e.g., Geobacillus stearothermophilus |
| Lipases (Hydrolases, EC 3) | Conversion of lipids and fats into fatty acids, glycerol, and other molecules | SMF [27] SSF [27] | Food; Detergent; Pharmaceutical; Leather; Textile; Cosmetic; Paper | Hormone-sensitive Lipase [28] e.g., humans and mouse | Triacylglycerol acyl hydrolase [29] e.g., Bacillus subtilis |
| Amylases * (Hydrolases, EC 3) | Hydrolysis of complex carbohydrates into sugars | SMF [30] SSF [30] | Food; Fermentation; Textile; Paper; Detergent; Pharmaceutical | α-amylase [31] e.g., Paenibacillus sp. | α-amylase [32] e.g., Pseudogymnoascus sp. |
| Phytases * (Hydrolases, EC 3) | Converts phytate into phosphorus | SMF [33] | Food | Phytases [34] e.g., Lactobacillus plantarum | Phytases [34] e.g., Lactobacillus plantarum |
| Fermentation Process | Solid State Fermentation (SSF) | Submerged Fermentation (SMF) |
|---|---|---|
| Microorganism preference | Fungi | Bacteria and yeast |
| Medium composition | Agro-waste nutrients | Liquid substrate (e.g., molasses and broths) rich in oxygen and carbon dioxide |
| Regulation | Low | High (medium, pH, temperature) |
| Costs | Low costs due to the usage of agro-waste nutrients | High cost due to the required media components |
| Effluent | Less effluent waste | Higher effluent waste |
| Enzyme production | High volumetric production | Low volumetric production |
| Method | Immobilization | Molecules Used for Immobilization | Advantages |
|---|---|---|---|
| Adsorption | Hydrophobic interaction and salt linkages [78] | Coconut fibers [79]; microcrystalline cellulose [80]; micro/mesoporous with thiol functionalized [81] | Enzyme shield from aggregation, proteolyss, and interaction with hydrophobic surfaces [78] |
| Covalent Binding | Association between side chain amino acids (arginine, aspartic acid, histidine) with functional groups [75] | Functional groups: imidazole, indolyl, and phenolic hydroxyl [75] | Higher specific activity and stability [82]; increase in half-life and thermal stability [83] |
| Cross-Linking | Two distinct methods: 1-Enzyme conjugated with a unit with high affinity to the matrix [84] 2-Matrix precoupled to an affinity ligand for target enzyme [84] | Alkali stable chitosan-coated porous silica beads [85]; Agarose-linked multilayered concanavalin [86] | Can be used for simultaneous enzyme purification through cross-linked enzyme aggregates (CLEAs) and cross-linking enzyme crystals (CLECs) [87]; Ability to harbor higher amounts of enzymes, increasing the stability and efficiency [85,86] |
| Entrapment | Cage by covalent or non-covalent bonds within gels or fibers [88] | Encapsulation with alginate-gelatin-calcium hybrids Nanostructured supports such as electro spun nanofibers and pristine materials [89]; Entrapment by mesoporous silica [83]; Sol-gel matrices [90] | High thermostability [91], high affinity, and enhancement of activity [92] |
| Enzyme | Study | Advantages |
|---|---|---|
| α-amylase | DES used as a reaction medium and co-solvent for converting starch or maltotriose into alkyl glucosides | DESs showed that can be used for selective reactions |
| β-glucosidase and Candida antarctica lipase B | DES used to explore biocompatibility and thermostability of the enzymes | Increase enzyme biocompatibility and thermal stability |
| Laccase | The influence of DES in the stability and activity to increase storage time | Enzyme activity stable at 60 °C after two days of incubation; At −80 °C and over 20 days in storage, activity levels increased |
| 5-hydroxymethylfurfural oxidase (HMFO) | DES used for the production of furan-2,5-dicarboxylic acid (FDCA) | Strong stabilization for HMFO and increased the thermostability |
| Burkholderia cepacia lipase (BCL) | Evaluation of the stability, activity, and thermostability | Enzymatic activity increased (by up to 2.6 times over the buffer) and improved kinetics |
| Candida rugosa lipase | Effect on the activity and stability | Improve enzyme activity and stability; half-life increased by 9.2 times compared to buffer |
| Novozym 435 | Synthesis of chiral drugs from hydrophobic substrates | Enhance enzyme selectivity by 16%; 99% purity of enantiomeric products |
| Candida rugosa lipase | Esterification reactions | Increased the esterification of fatty acids |
| Novozym 435 | Enzymatic selective esterification | Increased the esterification of 1,3-DAG |
| Immobilized whole cells from Arthrobacter simplex | Bioconversion efficiency of cortisone acetate (CA) to prednisone acetate (PA) | High potential of DESs for bio-dehydrogenation reactions |
| Lipases | Selective enzymatic synthesis of α-MBG (α-monobenzoate glycerol) | High conversion rate (99%) |
| Phospholipase D (PLD) | Release of intracellular enzymes using hydrophobic DESs | Intracellular components could be extracted without cell disruption |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monteiro, H.; Meneses, L.; Paiva, A.; Galamba, N.; Duarte, A.R.C. A Perspective Towards More Sustainable Production of Biotechnologically Relevant Enzymes Using DESs. Molecules 2025, 30, 3915. https://doi.org/10.3390/molecules30193915
Monteiro H, Meneses L, Paiva A, Galamba N, Duarte ARC. A Perspective Towards More Sustainable Production of Biotechnologically Relevant Enzymes Using DESs. Molecules. 2025; 30(19):3915. https://doi.org/10.3390/molecules30193915
Chicago/Turabian StyleMonteiro, Hugo, Liane Meneses, Alexandre Paiva, Nuno Galamba, and Ana Rita C. Duarte. 2025. "A Perspective Towards More Sustainable Production of Biotechnologically Relevant Enzymes Using DESs" Molecules 30, no. 19: 3915. https://doi.org/10.3390/molecules30193915
APA StyleMonteiro, H., Meneses, L., Paiva, A., Galamba, N., & Duarte, A. R. C. (2025). A Perspective Towards More Sustainable Production of Biotechnologically Relevant Enzymes Using DESs. Molecules, 30(19), 3915. https://doi.org/10.3390/molecules30193915







