Synthesis, Structure, DNA/BSA Binding, DNA Cleaving, Cytotoxic and SOD Mimetic Activities of Copper(II) Complexes Derived from Methoxybenzylamine Schiff Base Ligands
Abstract
1. Introduction
2. Results and Discussion
2.1. Synthesis
2.2. X-Ray Structure of Complex 4a
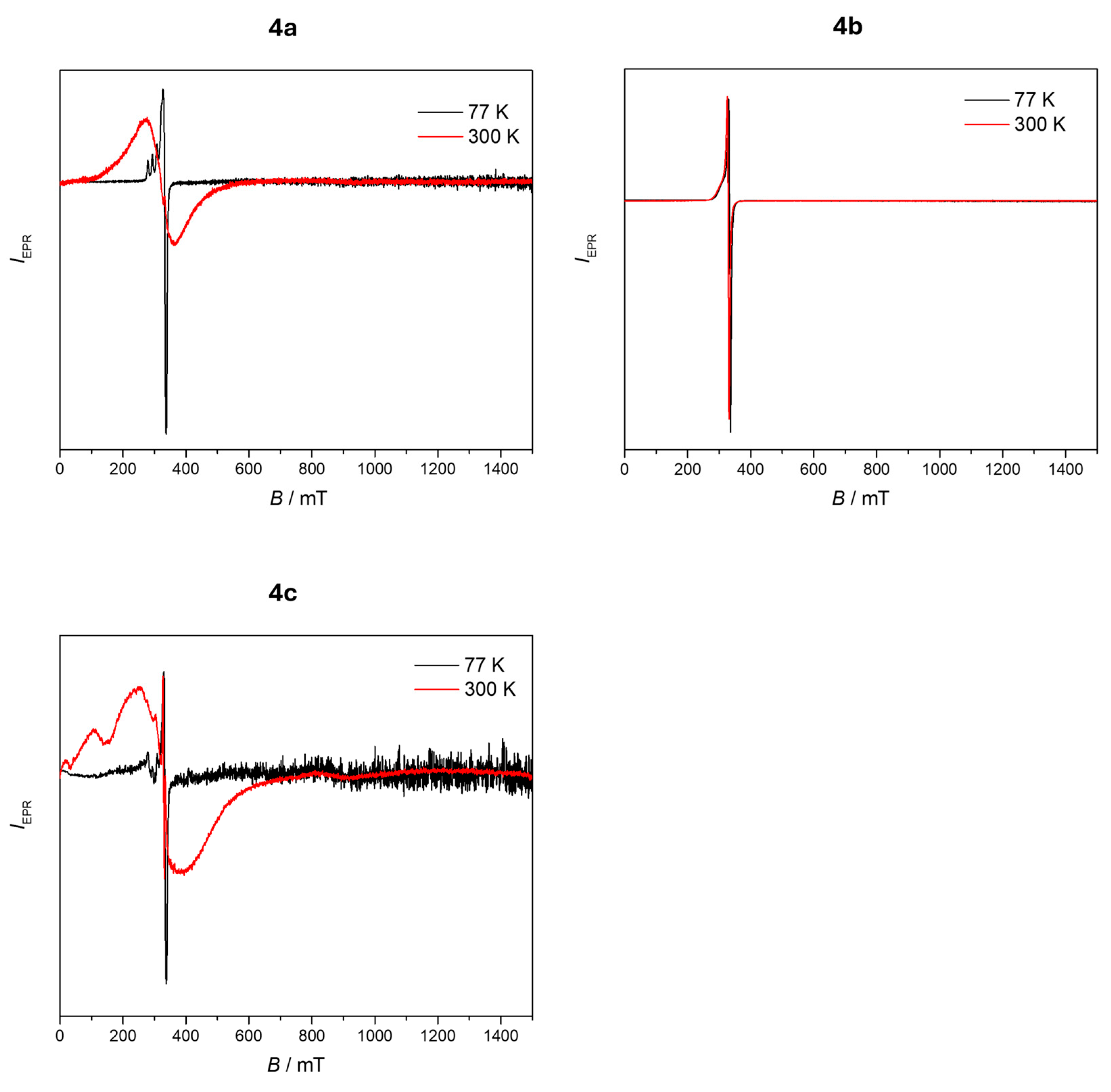
2.3. EPR Studies
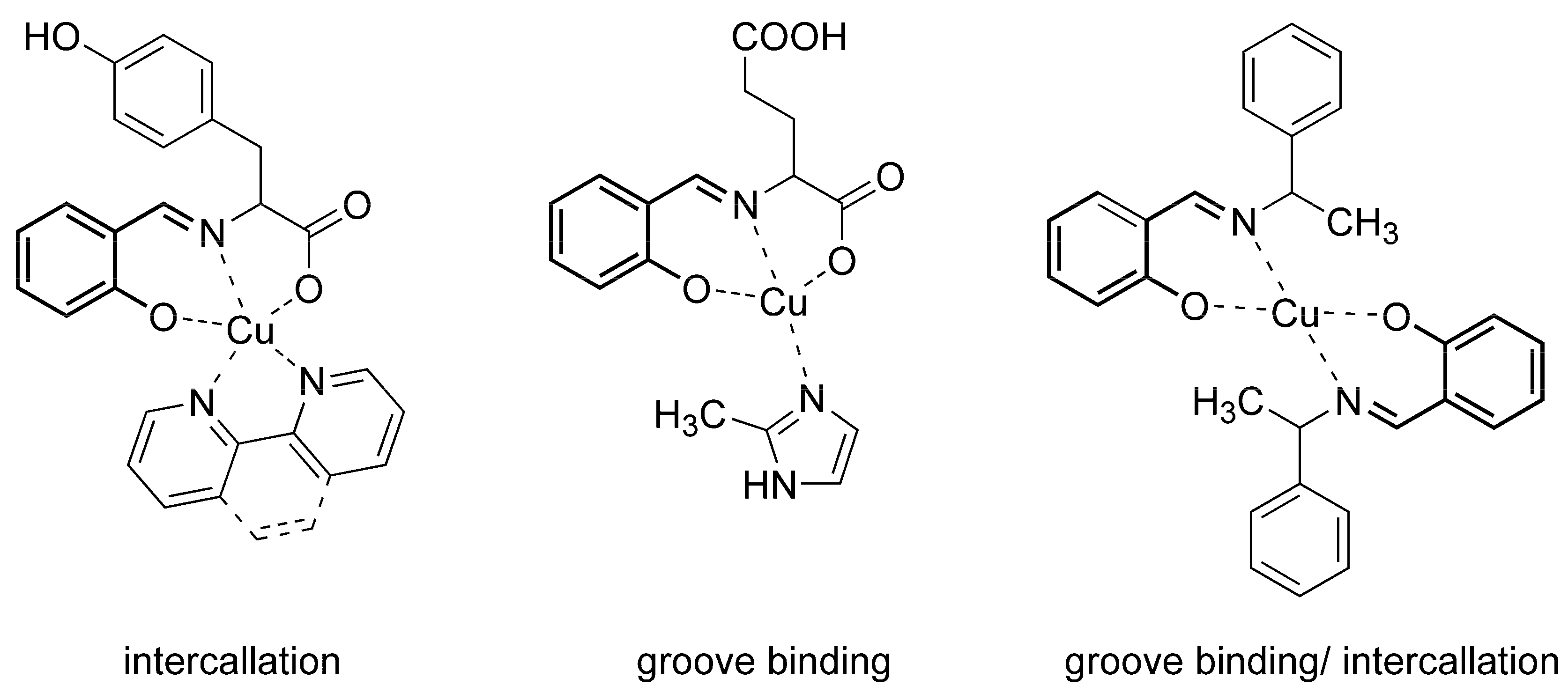
2.4. DNA Interaction Studies
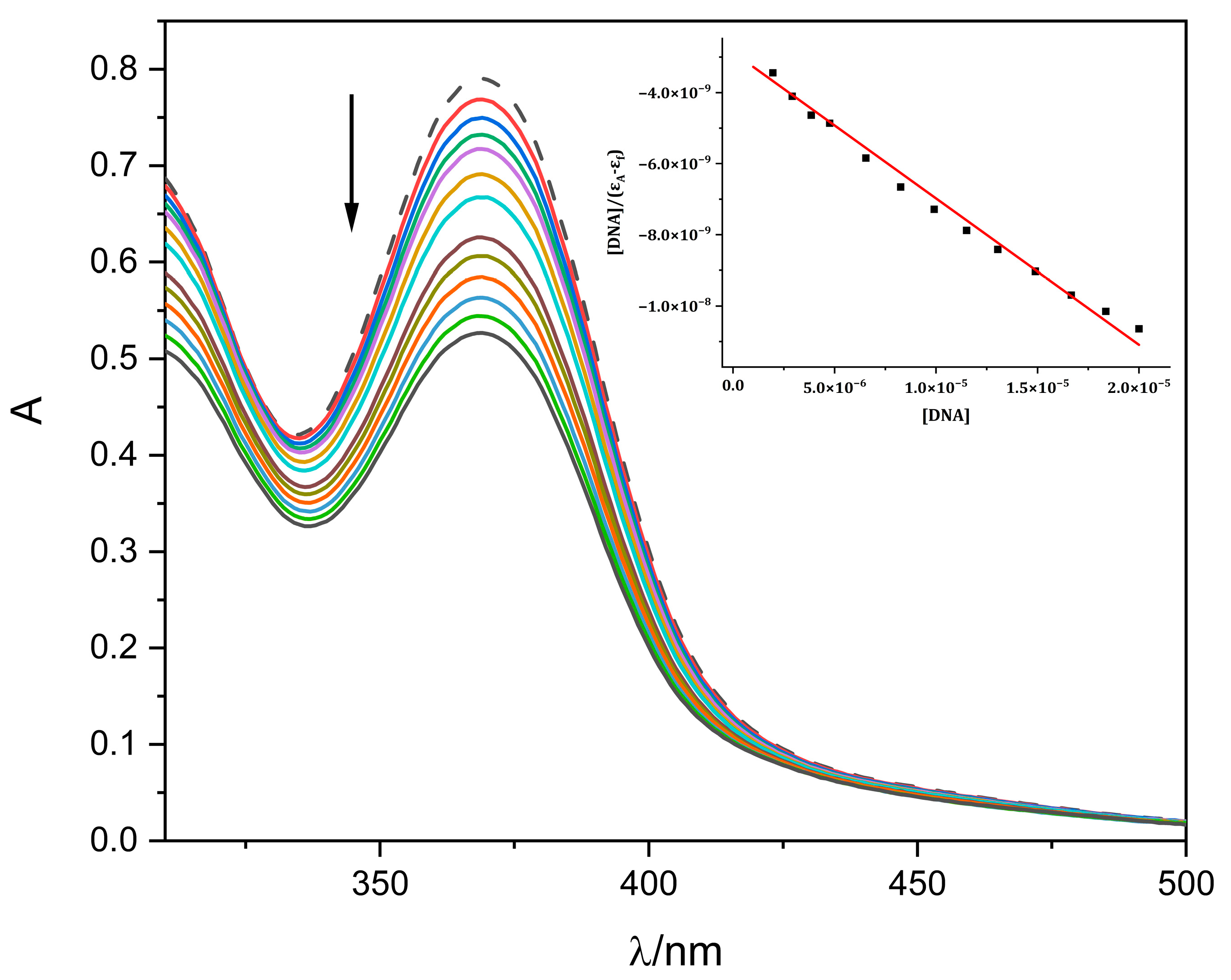
2.4.1. DNA Bnding Study by Absorption Titration
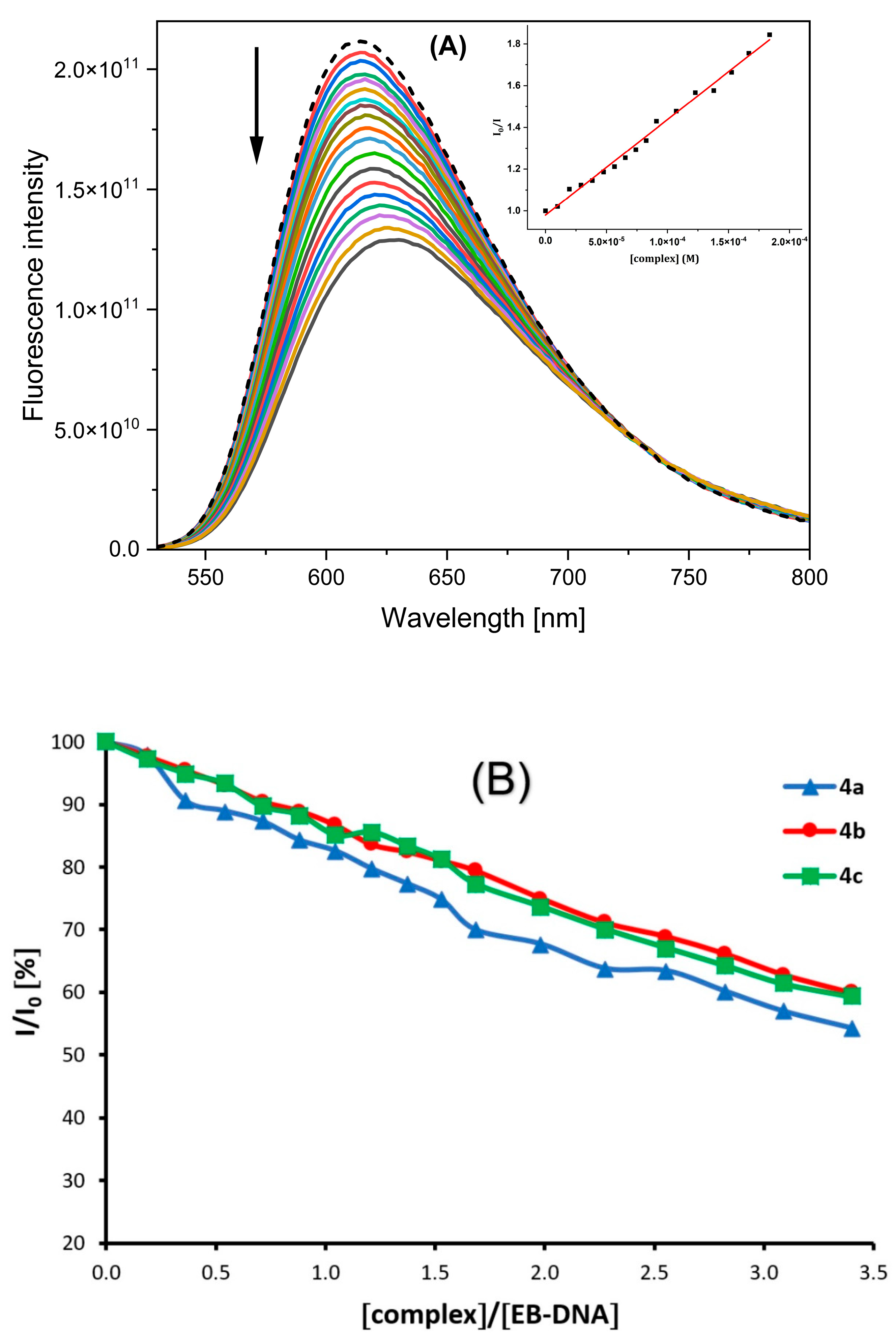
2.4.2. Competitive Studies with EB–DNA
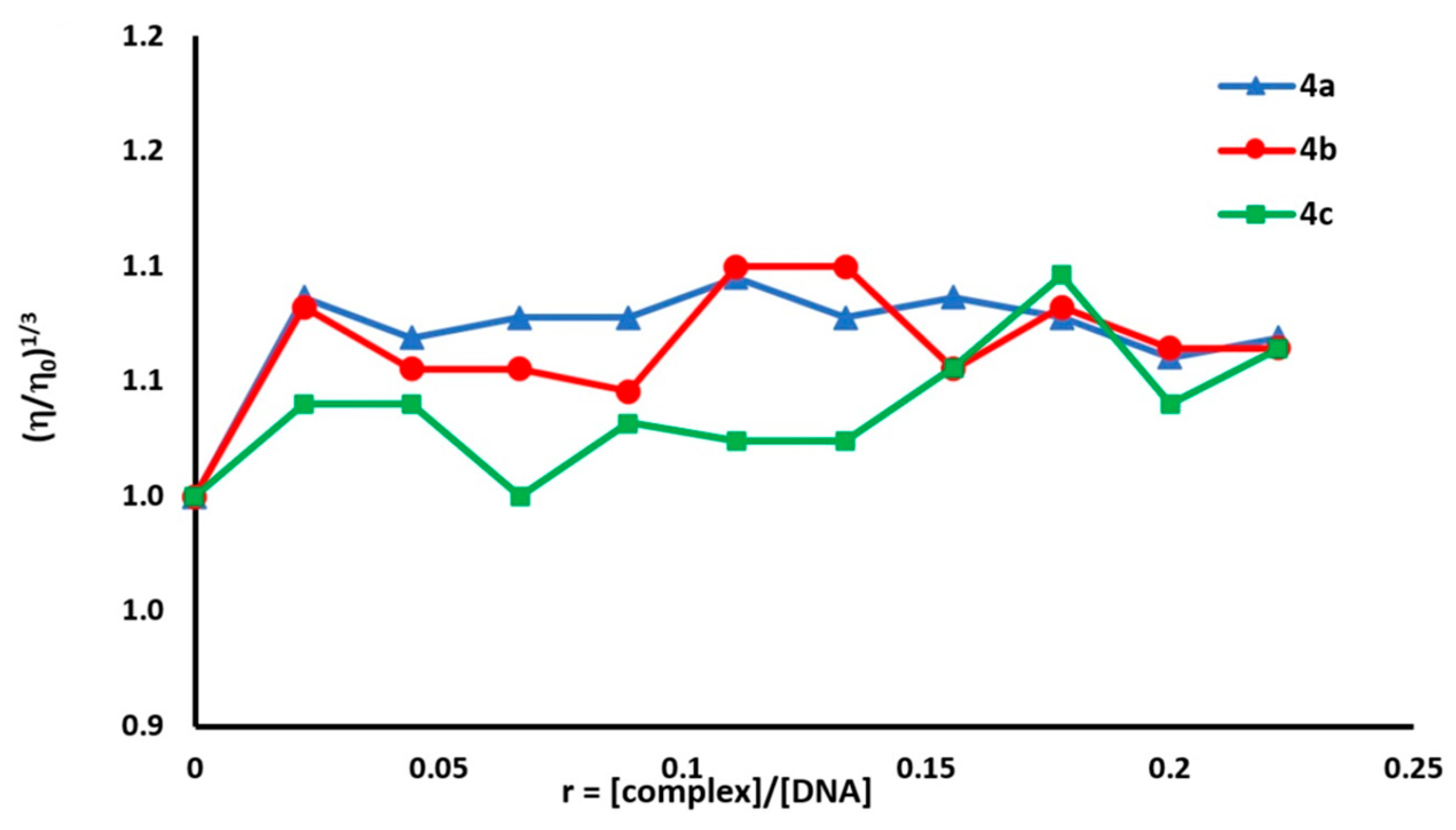
2.4.3. Viscosity Measurements
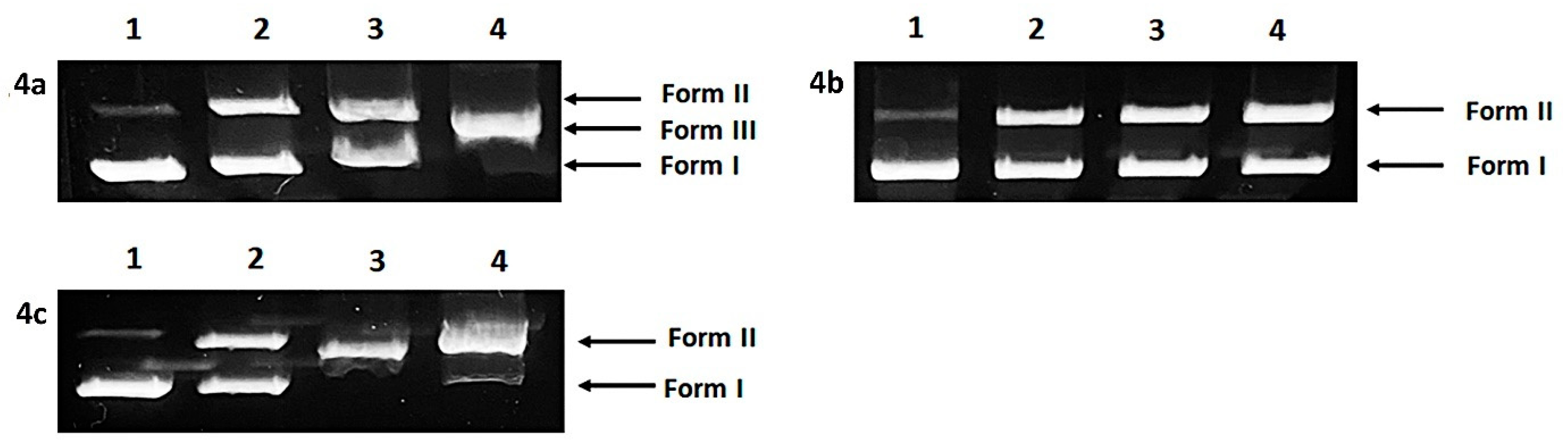
2.5. DNA Cleavage
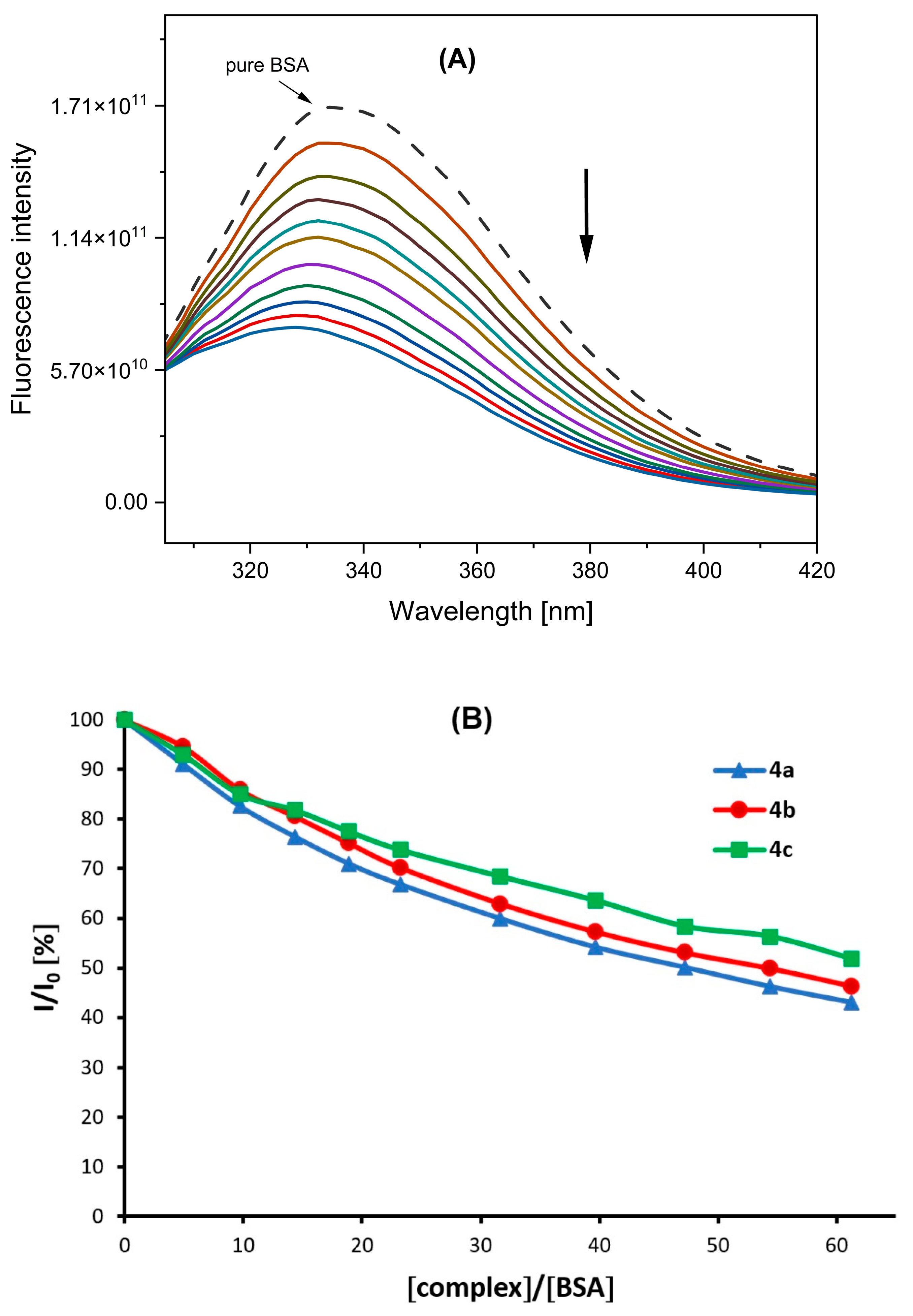
2.6. Interaction of Complexes Studied with BSA
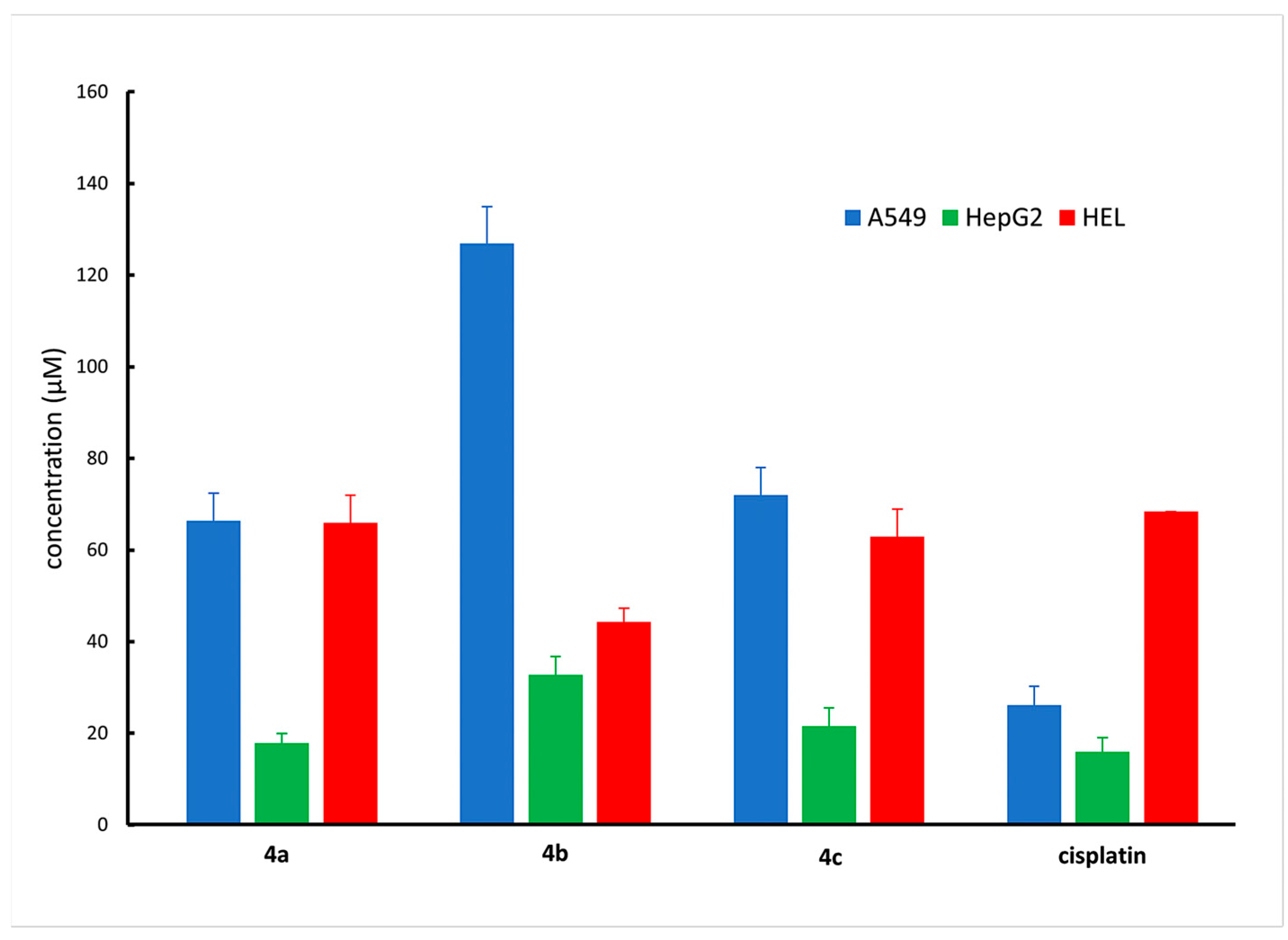
2.7. Cytotoxicity Based on Mitochondrial Activity
2.8. Cancer Cell Cytotoxicity by MTT Assay
2.9. SOD Mimetic Activity
3. Materials and Methods
3.1. Instrumentation and Materials Used in Synthesis
3.2. General Procedure for Synthesis of the Schiff Base Ligands (3)
3.3. General Procedure of Synthesis of Schiff Base Ligand Complexes (4)
3.4. Stability Measurements
3.5. X-Ray Structure Analysis
3.6. EPR Studies
3.7. DNA Cleavage Assay
3.8. DNA Binding Study by Absorption Titration
3.9. EB–DNA Displacement Method
3.10. Viscosimetric Studies
3.11. Bovine Serum Albumin Binding Studies
3.12. Resazurin Cytotoxicity Assay
| Wavelength | Reduced Resazurin (Resorufin) ƐRED | Oxidized Resazurin ƐOX |
| 570 nm | 155,677 | 80,586 |
| 600 nm | 14,652 | 117,216 |
3.13. MTT Cytotoxicity Assay
3.14. Radical Scavenging Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| A549 | adenocarcinoma human alveolar basal epithelial cell line |
| BSA | bovine serum albumin |
| ct-DNA | calf thymus DNA |
| DMSO | dimethyl sulfoxide |
| EB | ethidium bromide |
| HepG2 | human liver epithelial cell line, carcinoma |
| HEL | lung fibroblast cell line, noncancerous |
| INT | 2-(4-Iodophenyl)-3-(4-nitrophenyl)-5-phenyl-2H-tetrazolium chloride |
| NADH | nicotinamide adenine dinucleotide |
| MTT | 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide |
| pDNA | plasmid DNA |
| ROS | reactive oxygen species |
| SOD | superoxide dismutase |
References
- Arjmand, F.; Muddassir, M.; Yousuf, I. Design and synthesis of enantiomeric (R)- and (S)-copper(II) and diorganotin(IV)-based antitumor agents: Their in vitro DNA binding profile, cleavage efficiency and cytotoxicity studies. J. Photochem. Photobiol. B Biol. 2014, 136, 62–71. [Google Scholar] [CrossRef]
- Chen, Z.; Wu, Y.; Zhu, Z.; Zhang, Y. DNA cleavage, DNA/HSA binding study, and antiproliferative activity of a phenolate-bridged binuclear copper(II) complex. Biometals 2019, 32, 227–240. [Google Scholar] [CrossRef]
- Da Silva, D.A.; De Luca, A.; Squitti, R.; Rongioletti, M.; Rossi, L.; Machado, C.M.L.; Cerchiaro, G. Copper in tumors and the use of copper-based compounds in cancer treatment. J. Inorg. Biochem. 2022, 226, 111634. [Google Scholar] [CrossRef]
- Zehra, S.; Tabassum, S.; Arjmand, F. Biochemical Pathways of Copper Complexes: Progress over the past 5 years. Drug Discov. Today 2021, 26, 1086–1096. [Google Scholar] [CrossRef]
- Andrezálová, L.; Országhová, Z. Covalent and noncovalent interactions of coordination compounds with DNA: An overview. J. Inorg. Biochem. 2021, 225, 111624. [Google Scholar] [CrossRef]
- Santini, C.; Pellei, M.; Gandin, V.; Porchia, M.; Tisato, F.; Marzano, C. Advances in Copper Complexes as Anticancer Agents. Chem. Rev. 2014, 114, 815–862. [Google Scholar] [CrossRef] [PubMed]
- Muniyandi, V.; Pravin, N.; Raman, N. Impact of metallonucleases on DNA interactions: Structural validation and in-vitro antibiogram assay. Inorg. Chem. Comm. 2014, 46, 60–64. [Google Scholar] [CrossRef]
- Erkkila, K.E.; Odom, D.T.; Barton, J.K. Recognition and Reaction of Metallointercalators with DNA. Chem. Rev. 1999, 99, 2777–2795. [Google Scholar] [CrossRef] [PubMed]
- Mehring, A.; Erdmann, N.; Walther, J.; Stiefelmaier, J.; Strieth, D.; Ulber, R. A simple and low-cost resazurin assay for vitality assessment across species. J. Biotechnol. 2021, 333, 63–66. [Google Scholar] [CrossRef]
- Erxleben, A. Interactions of copper complexes with nucleic acids. Coord. Chem. Rev. 2018, 360, 92–121. [Google Scholar] [CrossRef]
- Barone, G.; Terenzi, A.; Lauria, A.; Almerico, A.M.; Leal, J.M.; Busto, N.; Garcia, B. DNA-binding of nickel(II), copper(II) and zinc(II) complexes: Structure–affinity relationships. Coord. Chem. Rev. 2013, 257, 2848–2862. [Google Scholar] [CrossRef]
- Ghanghas, P.; Choudhary, A.; Kumar, D.; Poonia, K. Coordination metal complexes with Schiff bases: Useful pharmacophores with comprehensive biological applications. Inorg. Chem. Comm. 2021, 130, 108710. [Google Scholar] [CrossRef]
- Kargar, H.; Behjatmanesh-Ardakani, R.; Torabi, V.; Kashani, M.; Chavoshpour-Natanzi, Z.; Kazemi, Z.; Mirkhani, V.; Sahraei, A.; Tahir, M.N.; Ashfaq, M.; et al. Synthesis, characterization, crystal structures, DFT, TD-DFT, molecular docking and DNA binding studies of novel copper(II) and zinc(II) complexes bearing halogenated bidentate N,O-donor Schiff base ligands. Polyhedron 2021, 195, 114988. [Google Scholar] [CrossRef]
- Shahraki, S. Schiff base compounds as artificial metalloenzymes. Colloids Surf. B 2022, 218, 112727. [Google Scholar] [CrossRef]
- Shahraki, S.; Samareh Delarami, H.; Mansouri-Torshizi, H.; Nouri, H. Investigation of kinetics and thermodynamics in the interaction process between two pyridine derived Schiff base complexes and catalase. J. Mol. Liq. 2021, 334, 116527. [Google Scholar] [CrossRef]
- Tabassum, S.; Amir, S.; Arjmand, F.; Pettinari, C.; Marchetti, F.; Masciocchi, N.; Lupidi, G.; Pettinari, R. Mixed-ligand Cu(II)–vanillin Schiff base complexes; effect of coligands on their DNA binding, DNA cleavage, SOD mimetic and anticancer activity. Europ. J. Med. Chem. 2013, 60, 216–232. [Google Scholar] [CrossRef]
- Shahraki, S.; Heidari Majd, M.; Heydari, A. Novel tetradentate Schiff base zinc(II) complex as a potential antioxidant and cancer chemotherapeutic agent: Insights from the photophysical and computational approach. J. Mol. Struct. 2019, 1177, 536–544. [Google Scholar] [CrossRef]
- Chidambaram, B.; Anand, N.; Varma, S.R.; Ramamurthy, S.; Vichitra, C.; Sharma, A.; Mahalakshmi, A.M.; Essa, M.M. Superoxide dismutase and neurological disorders. IBRO Neurosci. Rep. 2024, 16, 373–394. [Google Scholar] [CrossRef]
- Policar, C.; Bouvet, J.; Bertrand, H.C.; Delsuc, N. SOD mimics: From the tool box of the chemists to cellular studies. Curr. Opi. Chem. Biol. 2022, 67, 102109. [Google Scholar] [CrossRef]
- Singh, S. Antioxidant nanozymes as next-generation therapeutics to free radical-mediated inflammatory diseases: A comprehensive review. Int. J. Biol. Macromol. 2024, 260, 129374. [Google Scholar] [CrossRef]
- Shahraki, S.; Vaziri, E.; Saboury, A.A.; Fan, K. Biomedical potential of nanozymes: Harnessing redox enzyme mimicry for theranostic applications. Coord. Chem. Rev. 2024, 517, 215937. [Google Scholar] [CrossRef]
- Wang, C.; Li, S.; Shang, D.; Wang, X.; You, Z.; Li, H. Antihyperglycemic and neuroprotective effects of one novel Cu–Zn SOD mimetic. Bioorg. Med. Chem. Lett. 2011, 21, 4320–4324. [Google Scholar] [CrossRef] [PubMed]
- Klanicová, A.; Trávníček, Z.; Vančo, J.; Popa, I.; Šindelář, Z. Dinuclear copper(II) perchlorate complexes with 6-(benzylamino)purine derivatives: Synthesis, X-ray structure, magnetism and antiradical activity. Polyhedron 2010, 29, 2582–2589. [Google Scholar] [CrossRef]
- Vančo, J.; Švajlenová, O.; Račanská, E.; Muselík, J.; Valentová, J. Antiradical activity of different copper(II) Schiff base complexes and their effect on alloxan-induced diabetes. J. Trace Elem. Med. Biol. 2004, 18, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Lintnerová, L.; Valentová, L.; Herich, P.; Kožíšek, J.; Devínsky, F. Synthesis and antiradical activity of novel copper(II) complexes of long chain reduced Schiff base ligands. Monatsh. Chem. 2018, 149, 901–911. [Google Scholar] [CrossRef]
- Zhou, S.; Cai, H.; He, X.; Tang, Z.; Lu, S. Enzyme-mimetic antioxidant nanomaterials for ROS scavenging: Design, classification, and biological applications. Coord. Chem. Rev. 2024, 500, 215536. [Google Scholar] [CrossRef]
- Mapuskar, K.A.; Anderson, C.M.; Spitz, D.R.; Batinic-Haberle, I.; Allen, B.G.; Oberley-Deegan, R.E. Utilizing Superoxide Dismutase Mimetics to Enhance Radiation Therapy Response While Protecting Normal Tissues. Semin. Rad. Oncol. 2019, 29, 72–80. [Google Scholar] [CrossRef]
- Marzano, C.; Pellei, M.; Tisato, F.; Santini, C. Copper complexes as anticancer agents. Anticancer. Agents Med. Chem. 2009, 9, 185–211. [Google Scholar] [CrossRef]
- Matela, G. Schiff bases and complexes: A review on anti-cancer activity. Anticancer Agents Med. Chem. 2020, 20, 1908–1917. [Google Scholar] [CrossRef]
- Pages, B.J.; Ang, D.L.; Wright, E.P.; Aldrich-Wright, J.R. Metal complex interactions with DNA. Dalton Trans. 2015, 44, 3505–3526. [Google Scholar] [CrossRef]
- Aleksić, M.M.; Kapetanović, V. An overview of the optical and electrochemical methods for detection of DNA-drug interactions. Acta Chim. Slov. 2014, 61, 555–573. [Google Scholar]
- Andrezálová, L.; Plšíková, J.; Janočková, J.; Koňariková, K.; Žitňanová, I.; Kohútová, M.; Kožurková, M. DNA/BSA binding ability and genotoxic effect of mono- and binuclear copper (II) complexes containing a Schiff base derived from salicylaldehyde and D, L-glutamic acid. J. Organomet. Chem. 2017, 827, 67–77. [Google Scholar] [CrossRef]
- Reddy, P.R.; Shilpa, A. Oxidative and hydrolytic DNA cleavage by Cu(II) complexes of salicylidene tyrosine Schiff base and 1,10 phenanthroline/bipyridine. Polyhedron 2011, 30, 565–572. [Google Scholar] [CrossRef]
- Sundaravadivel, E.; Muthusamy, K.; Varghese, B. Synthesis, characterization and electrochemical behavior of new acyclic mono and binuclear copper (II) complexes: DNA binding and cleavage studies. Polyhedron 2013, 61, 33–44. [Google Scholar] [CrossRef]
- Dimitrakopoulou, A.; Dendrinou-Samara, C.; Pantazaki, A.A.; Alexiou, M.; Nordlander, E.; Kessissoglou, D.P. Synthesis, structure and interactions with DNA of novel tetranuclear, [Mn4(II/II/II/IV)] mixed valence complexes. J. Inorg. Biochem. 2008, 102, 618–628. [Google Scholar] [CrossRef]
- Sirajuddin, M.; Ali, S.; Badshah, A. Drug–DNA interactions and their study by UV–Visible, fluorescence spectroscopies and cyclic voltammetry. J. Photochem. Photobiol. B Biol. 2013, 124, 1–19. [Google Scholar] [CrossRef]
- Bukonjić, A.M.; Tomović, D.L.; Stanković, A.S.; Jevtić, V.V.; Ratković, Z.R.; Bogojeski, J.V.; Milanović, J.Z.; Dordević, D.B.; Arsenijević, A.N.; Milanović, M.Z.; et al. Synthesis, characterization and biological activity of copper(II) complexes with ligands derived from β-amino acids. Transit. Met. Chem. 2019, 44, 65–76. [Google Scholar] [CrossRef]
- Wilson, W.D.; Ratmeyer, L.; Zhao, M.; Strekowski, L.; Boykin, D. The search for structure-specific nucleic acid-interactive drugs: Effects of compound structure on RNA versus DNA interaction strength. Biochemistry 1993, 32, 4098–4104. [Google Scholar] [CrossRef]
- Banerjee, D.; Pal, S.K. Simultaneous binding of minor groove binder and intercalator to dodecamer DNA: Importance of relative orientation of donor and acceptor in FRET. J. Phys. Chem. B 2007, 111, 5047–5052. [Google Scholar] [CrossRef]
- Dehkhodaei, M.; Sahihi, M.; Amiri Rudbari, H.; Momenbeik, F. DNA and HSA interaction of Vanadium (IV), Copper (II), and Zinc (II) complexes derived from an asymmetric bidentate Schiff-base ligand: Multi spectroscopic, viscosity measurements, molecular docking and ONIOM studies. J. Biol. Inorg. Chem. 2018, 23, 181–192. [Google Scholar] [CrossRef]
- Cole, K.D.; Tellez, C.M. Separation of large circular DNA by electrophoresis in agarose gels. Biotechnol. Prog. 2002, 18, 82–87. [Google Scholar] [CrossRef]
- Jaividhya, P.; Dhivya, R.; Akbarsha, M.A.; Palaniandavar, M. Efficient DNA cleavage mediated by mononuclear mixed ligand copper(II) phenolate complexes: The role of planarity on DNA binding and cleavage and anticancer activity. J. Inorg. Biochem. 2012, 114, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Mamindla, A.; Varadhan, M.; Kartikeyan, R.; Amuthamozhi, A.; Akbarsha, M.A.; Rajendiran, V. Comparative DNA-/BSA-binding, DNA cleavage, and cytotoxic properties of copper(II) amino/salicyl-phenolate Schiff bases (phen) complexes that induce generation of phenoxyl radicals. Polyhedron 2023, 243, 116534. [Google Scholar] [CrossRef]
- Sundaravadivel, I.; Reddy, G.R.; Manoj, D.; Rajendran, S.; Kandaswamy, M.; Janakiraman, M. DNA binding and cleavage studies of copper(II) complex containing N2O2 Schiff base ligand. Inorg. Chim. Acta 2018, 482, 170–178. [Google Scholar] [CrossRef]
- Kumar, M.P.; Ayodhya, D. Shivaraj Novel copper (II) binary complexes with N,O-donor isoxazole Schiff base ligands: Synthesis, characterization, DPPH scavenging, antimicrobial, and DNA binding and cleavage studies. Res. Chem. 2023, 5, 100845. [Google Scholar] [CrossRef]
- Kumar, A.; Ajay, A.K.; Bhat, M.K.; Rao, C.P. Cu(II) complexes of glyco-imino-aromatic conjugates in DNA binding, plasmid cleavage and cell cytotoxicity. J. Chem. Sci. 2012, 124, 1217–1228. [Google Scholar] [CrossRef]
- Ganeshpandian, M.; Ramakrishnan, S.; Palaniandavar, M.; Suresh, E.; Riyasdeen, A.; Akbarsha, M.A. Mixed ligand copper(II) complexes of 2,9-dimethyl-1,10-phenanthroline: Tridentate 3N primary ligands determine DNA binding and cleavage and cytotoxicity. J. Inorg. Biochem. 2014, 140, 202–212. [Google Scholar] [CrossRef]
- Siddiqui, S.; Ameen, F.; ur Rehman, S.; Sarwar, T.; Tabish, M. Studying the interaction of drug/ligand with serum albumin. J. Mol. Liq. 2021, 336, 116200. [Google Scholar] [CrossRef]
- Kratz, F.; Beyer, U. Serum proteins as drug carriers of anticancer agents: A review. Drug Deliv. 1998, 5, 281–299. [Google Scholar] [CrossRef]
- Shiri, F.; Rahimi-Nasrabadi, M.; Ahmadi, F.; Ehrlich, H. Multispectroscopic and molecular modeling studies on the interaction of copper-ibuprofenate complex with bovine serum albumin (BSA). Spectrochim. Acta A Mol. Biomol. Spectrosc. 2018, 203, 510–521. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, X.; Ji, R.; Bian, H.; Guo, X.; He, Y.; Chen, H.; Abdalla, A.N. Studies of Interactions between Beta-Cyfluthrin and BSA Based on Fluorescence Spectrometry and Ultraviolet Degradation. Photonics 2023, 10, 1079. [Google Scholar] [CrossRef]
- Tan, C.; Liu, J.; Li, H.; Zheng, W.; Shi, S.; Chen, L.; Ji, L. Differences in structure, physiological stability, electrochemistry, cytotoxicity, DNA and protein binding properties between two Ru(III) complexes. J. Inorg. Biochem. 2008, 102, 347–358. [Google Scholar] [CrossRef]
- Varlan, A.; Hillebrand, M. Bovine and human serum albumin interactions with 3-carboxyphenoxathiin studied by fluorescence and circular dichroism spectroscopy. Molecules 2010, 15, 3905–3919. [Google Scholar] [CrossRef] [PubMed]
- Keikha, A.O.; Mansouri-Torshizi, H.; Shahraki, S.; Dehghanian, E. Zn (II) and Pd (II) complexes derived from novel benzohydrazide-based Schiff base ligand as multi-target agents. J. Mol. Liq. 2023, 391, 123272. [Google Scholar] [CrossRef]
- Oboňová, B.; Valentová, J.; Litecká, M.; Pašková, Ľ.; Hricovíniová, J.; Bilková, A.; Horváth, B.; Habala, L. Novel Copper (II) Complexes with Fluorine-Containing Reduced Schiff Base Ligands Showing Marked Cytotoxicity in the HepG2 Cancer Cell Line. Int. J. Mol. Sci. 2024, 25, 9166. [Google Scholar] [CrossRef]
- Niu, M.; Hong, M.; Chang, G.; Li, X.; Li, Z. A comparative study of cytotoxicity and interaction with DNA/protein of five transition metal complexes with Schiff base ligands. J. Photochem. Photobiol. B. 2015, 148, 232–241. [Google Scholar] [CrossRef]
- Li, Z.; Yan, H.; Chang, G.; Hong, M.; Dou, J.; Niu, M. Cu(II), Ni(II) complexes derived from chiral Schiff-base ligands: Synthesis, characterization, cytotoxicity, protein and DNA–binding properties. J. Photochem. Photobiol. B 2016, 163, 403–412. [Google Scholar] [CrossRef]
- Alhazmi, H.A.; Al Bratty, M.; Meraya, A.M.; Najmi, A.; Alam, M.S.; Javed, S.A.; Ahsan, W. Spectroscopic characterization of the interactions of bovine serum albumin with medicinally important metal ions: Platinum(IV), iridium(III) and iron(II). Acta Biochim. Pol. 2021, 68, 99–107. [Google Scholar] [CrossRef]
- Shahabadi, N.; Hakimi, M.; Morovati, T.; Hadidi, S.; Moeini, K. Spectroscopic investigation into the interaction of a diazacyclam-based macrocyclic copper (ii) complex with bovine serum albumin. Luminescence 2017, 32, 43–50. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Zhang, X.P.; Hou, H.N.; Dai, J.; Liu, Y. Study on the interaction between Cu phen2+3 and bovine serum albumin by spectroscopic methods. Biol. Trace Elem. Res. 2008, 121, 276–287. [Google Scholar] [CrossRef]
- Smolková, R.; Smolko, L.; Samoľová, E.; Dušek, M. Co(II) fenamato, tolfenamato and niflumato complexes with neocuproine: Synthesis, crystal structure, spectral characterization and biological activity. J. Mol. Struct. 2023, 1272, 134172. [Google Scholar] [CrossRef]
- Bitacura, J.G. The Use of Baker’s Yeast in the Resazurin Reduction Test: A Simple, Low-Cost Method for Determining Cell Viability in Proliferation and Cytotoxicity Assays. J. Microbiol. Biol. Educ. 2018, 19, 19–87. [Google Scholar] [CrossRef]
- Botstein, D.; Chervitz, S.A.; Cherry, J.M. Yeast as a model organism. Science 1997, 277, 1259–1260. [Google Scholar] [CrossRef]
- Botstein, D.; Fink, G.R. Yeast: An experimental organism for 21st century biology. Genetics 2011, 189, 695–704. [Google Scholar] [CrossRef]
- Matuo, R.; Sousa, F.G.; Soares, D.G.; Bonatto, D.; Saffi, J.; Escargueil, A.E.; Larsen, A.K.; Henriques, J.A. Saccharomyces cerevisiae as a model system to study the response to anticancer agents. Cancer Chemother. Pharmacol. 2012, 70, 491–502. [Google Scholar] [CrossRef]
- Trávníček, Z.; Vančo, J.; Belza, J.; Zoppellaro, G.; Dvořák, Z. Dinuclear copper(II) complexes with a bridging bis(chalcone) ligand reveal considerable in vitro cytotoxicity on human cancer cells and enhanced selectivity. J. Inorg. Biochem. 2024, 252, 112481. [Google Scholar] [CrossRef]
- Yenigun, V.B.; Kocyigit, A.; Kanimdan, E.; Balkan, E.; Gul, A.Z. Copper (II) increases anti-Proliferative activity of thymoquinone in colon cancer cells by increasing genotoxic, apoptotic, and reactive oxygen species generating effects. Toxicon 2024, 250, 108103. [Google Scholar] [CrossRef]
- Chen, Z.; Li, Y.; Liu, X. Copper homeostasis and copper-induced cell death: Novel targeting for intervention in the pathogenesis of vascular aging. Biomed. Pharmacother. 2023, 169, 115839. [Google Scholar] [CrossRef]
- Zhang, X.; Tao, T.; Qiu, Y.; Guo, X.; Zhu, X.; Zhou, X. Copper-mediated novel cell death pathway in tumor cells and implications for innovative cancer therapies. Biomed. Pharmacother. 2023, 168, 115730. [Google Scholar] [CrossRef]
- Ghorbanpour, M.; Shayanfar, A.; Soltani, B. Copper pyrazole complexes as potential anticancer agents: Evaluation of cytotoxic response against cancer cells and their mechanistic action at the molecular level. Coord. Chem. Rev. 2024, 498, 215459. [Google Scholar] [CrossRef]
- Dolomanov, H.; Bouhris, O.V.; Gildea, L.J.; Howard, R.J.; Puschmann, J.A.K. OLEX 2: A complete structure solution, refinement and analysis program. J. Appl. Cryst. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT-Integrated space-group and crystal-structure determination. Acta Crystallogr. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Bergerhoff, G.; Berndt, M.; Brandenburg, K. Crystallographic Data with the Program DIAMOND. J. Res. Natl. Inst. Stand. Technol. 1996, 101, 221–225. [Google Scholar] [CrossRef]
- Macrae, C.F.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Shields, G.P.; Taylor, R.; Towler, M.; van de Streek, J. Mercury: Visualization and analysis of crystal structures. J. Appl. Cryst. 2006, 39, 453–457. [Google Scholar] [CrossRef]
- Stoll, S.; Schweiger, A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson. 2006, 178, 42–55. [Google Scholar] [CrossRef]
- Stoll, S. CW-EPR Spectral Simulations: Solid State. Methods Enzymol. 2015, 563, 121–142. [Google Scholar] [CrossRef] [PubMed]
- Molton, F. Simultispin: A versatile graphical user interface for the simulation of solid-state continuous wave EPR spectra. Magn. Reson. Chem. 2020, 58, 718–726. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Agarose gel electrophoresis. Cold Spring Harbor Protoc. 2019. [Google Scholar] [CrossRef]
- Schmidt, T.; Friehs, K.; Flaschel, E. Chapter 2: Structures of plasmid DNA. In Plasmids for Therapy and Vaccination; Schleef, M., Ed.; WILEY-VCH Verlag GmbH: Weinheim, Germany, 2001; pp. 29–43. Available online: https://onlinelibrary.wiley.com/doi/book/10.1002/9783527612833 (accessed on 26 April 2023).
- Jozefíková, F.; Perontsis, S.; Koňáriková, K.; Švorc, Ľ.; Mazúr, M.; Psomas, G.; Moncol, J. In vitro biological activity of copper (II) complexes with NSAIDs and nicotinamide: Characterization, DNA-and BSA-interaction study and anticancer activity. J. Inor. Biochem. 2022, 228, 111696. [Google Scholar] [CrossRef] [PubMed]
- Liu, D. A rapid biochemical test for measuring chemical toxicity. Bull. Environ. Contam. Toxicol. 1981, 26, 145–149. [Google Scholar] [CrossRef]
- Piroš, M.; Schoeller, M.; Koňáriková, K.; Sumbalová, Z.; Valentová, J.; Moncoľ, J.; May, N.V.; Pap, J.S.; Švorec, J. Synthesis, characterization and biological evaluation of three novel copper (II) clonixinate complexes with methylpyridines: Insights into structure, DNA and BSA binding properties and anticancer activity. Polyhedron. 2023, 245, 116619. [Google Scholar] [CrossRef]
- Strotmann, U.J.; Butz, B.; Bias, W.R. The dehydrogenase assay with resazurin: Practical performance as a monitoring system and pH-dependent toxicity of phenolic compounds. Ecotoxicol. Environ. Saf. 1993, 25, 79–89. [Google Scholar] [CrossRef]
- Candeias, L.P.; MacFarlane, D.P.S.; McWhinnie, S.L.W.; Maidwell, N.L.; Roesechlaub, C.A.; Sammes, P.G.; Whittlesey, R. The catalyzed NADH reduction of resazurin to resorufin. J. Chem. Soc. Perkin Trans. 1998, 2, 2333–2334. [Google Scholar] [CrossRef]
- Borra, R.C.; Lotufo, M.A.; Gagioti, S.M.; Barros, F.D.M.; Andrade, P.M. A simple method to measure cell viability in proliferation and cytotoxicity assays. Braz. Oral Res. 2009, 23, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Lavogina, D.; Lust, H.; Tahk, M.J.; Laasfeld, T.; Vellama, H.; Nasirova, N.; Vardja, M.; Eskla, K.L.; Salumets, A.; Rinken, A.; et al. Revisiting the Resazurin-Based Sensing of Cellular Viability: Widening the Application Horizon. Biosensors 2022, 12, 196. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef] [PubMed]

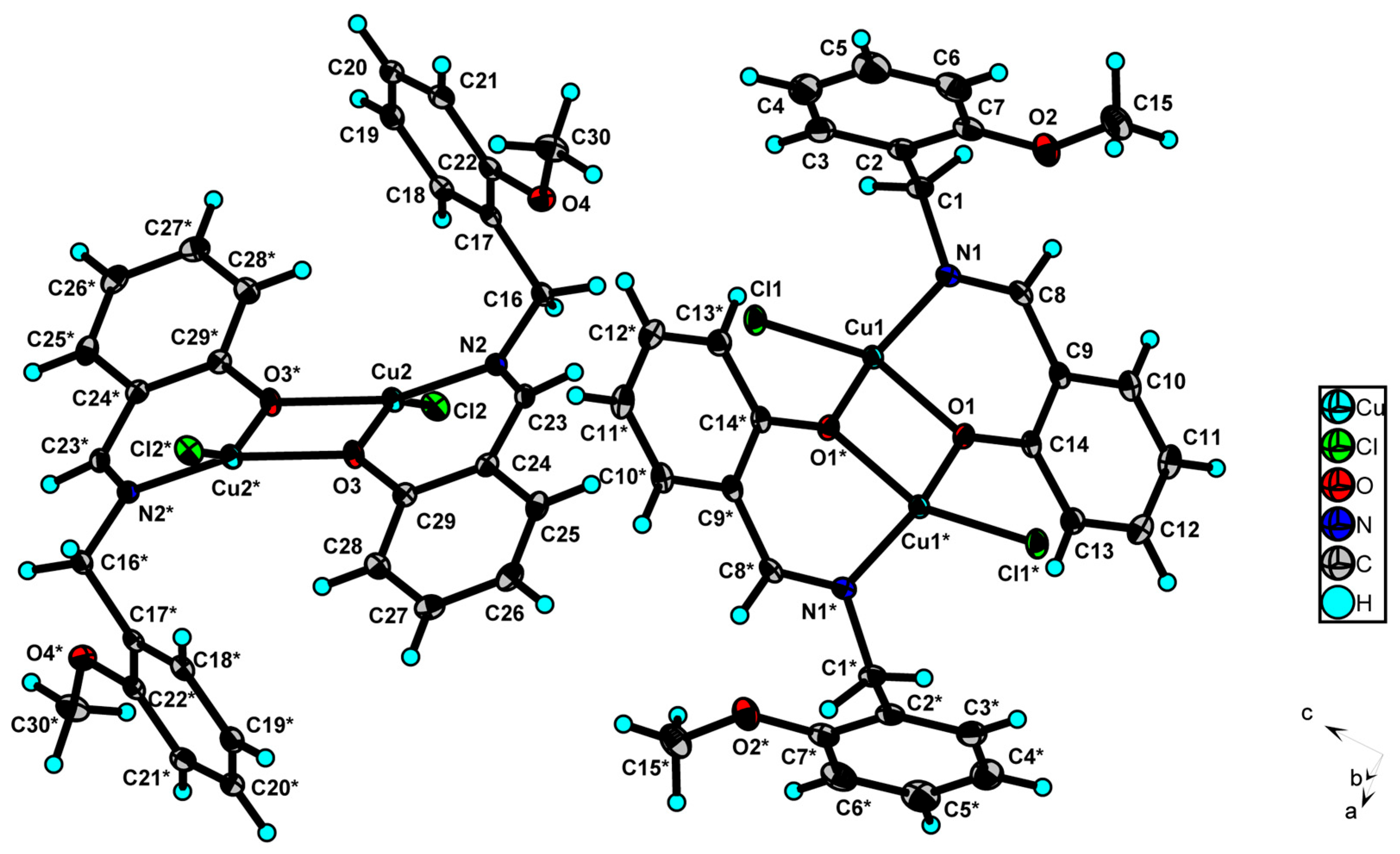

| Empirical Formula | C30H28Cl2Cu2N2O4 |
|---|---|
| Temperature [K] | 100(1) |
| Wavelength [Å] | 1.54186 |
| Crystal system | Monoclinic |
| Space group | P21/c |
| Unit cell dimensions [Å], [°] | a = 9.2198(1) b = 17.4213(3) c = 17.4576(2) β = 92.412(1) |
| Formula weight | 678.52 |
| Volume [Å3] | 2801.57(7) |
| Z/Calculated density [Mg/m3] | 4/1.609 |
| Absorption coeff. [mm−1] | 3.955 |
| F(000) | 1384.0 |
| Crystal size [mm] | 0.2 × 0.04 × 0.02 |
| Theta range for data collection | 7.172 to 143.116° |
| Index ranges | −11 ≤ h ≤ 5 −20 ≤ k ≤ 21 −21 ≤ l ≤ 21 |
| Reflections collected/ Independent reflections | 83,351/5393 [R(int) = 0.0325, Rσ = 0.0166] |
| Refinement method | Full-matrix least-squares on F2 |
| Data/restraints/parameters | 5393/0/363 |
| Goodness-of-fit on F2 | 1.088 |
| Final R indices [I > 2σ(I)] | R1 = 0.0259, wR2 = 0.0775 |
| R indices (all data) | R1 = 0.0303, wR2 = 0.0791 |
| Largest diff. peak and hole [e.Å−3] | 0.36/−0.55 |
| Atom | Atom | Length/Å | Atom | Atom | Length/Å | ||
|---|---|---|---|---|---|---|---|
| Cu1 | Cu1 * | 3.0485(5) | Cu2 | Cu2 ** | 3.0501(5) | ||
| Cu1 | Cl1 | 2.1975(5) | Cu2 | Cl2 | 2.1946(5) | ||
| Cu1 | O1 | 1.9435(12) | Cu2 | N2 | 1.9433(15) | ||
| Cu1 | O1 * | 1.9650(12) | Cu2 | O3 | 1.9304(13) | ||
| Cu1 | N1 | 1.9374(15) | Cu2 | O3 ** | 1.9671(12) | ||
| Atom | Atom | Atom | Angle/˚ | Atom | Atom | Atom | Angle/˚ |
| Cl1 | Cu1 | Cu1 * | 132.188(19) | Cl2 | Cu2 | Cu2 ** | 131.859(19) |
| O1 | Cu1 | Cu1 * | 38.99(4) | N2 | Cu2 | Cu2 ** | 127.18(4) |
| O1 * | Cu1 | Cu1 * | 38.49(4) | N2 | Cu2 | Cl2 | 100.23(5) |
| O1 * | Cu1 | Cl1 | 99.93(4) | N2 | Cu2 | O3 ** | 154.41(6) |
| O1 | Cu1 | Cl1 | 151.53(5) | O3 ** | Cu2 | Cu2 ** | 38.07(4) |
| O1 | Cu1 | O1 * | 77.48(5) | O3 | Cu2 | Cu2 ** | 38.93(3) |
| N1 | Cu1 | Cu1 * | 127.56(5) | O3 ** | Cu2 | Cl2 | 100.51(4) |
| N1 | Cu1 | Cl1 | 99.65(5) | O3 | Cu2 | Cl2 | 150.29(4) |
| N1 | Cu1 | O1 * | 155.18(6) | O3 | Cu2 | N2 | 92.05(6) |
| N1 | Cu1 | O1 | 92.20(6) | O3 | Cu2 | O3 ** | 77.00(5) |
| Cu1 | O1 | Cu1 * | 102.51(5) | C16 | N2 | Cu2 | 116.84(11) |
| C14 | O1 | Cu1 * | 132.25(11) | C23 | N2 | Cu2 | 124.35(13) |
| C14 | O1 | Cu1 | 124.25(11) | Cu2 | O3 | Cu2 ** | 103.00(5) |
| C1 | N1 | Cu1 | 118.54(12) | C29 | O3 | Cu2 ** | 132.17(11) |
| C8 | N1 | Cu1 | 123.93(13) | C29 | O3 | Cu2 | 123.37(11) |
| Complex | Kb [M−1] | λ (nm) | (ΔA/A0, %) |
|---|---|---|---|
| 4a | 1.43 (±0.84) × 105 | 369 | 32 |
| 4b | 1.02 (±0.82) × 105 | 369 | 26 |
| 4c | 3.25 (±0.83) × 105 | 370 | 28 |
| Complex | Δ I/I0 [%] | Ksv [M−1] × 103 | kq [M−1 s−1] × 1011 |
|---|---|---|---|
| 4a | 45.7 | 4.58 (±0.10) | 1.98 (±0.05) |
| 4b | 40.1 | 3.61 (±0.09) | 1.56 (±0.04) |
| 4c | 40.7 | 3.82 (±0.12) | 1.66 (±0.05) |
| Complex | c (M) | Form I (%) | Form II (%) | Form III (%) |
|---|---|---|---|---|
| 4a | 0 | 90.7 | 9.7 | 0 |
| 1 × 10−3 | 58.8 | 41.2 | 0 | |
| 3 × 10−3 | 53.8 | 46.2 | 0 | |
| 5 × 10−3 | 0 | 0 | 100 | |
| 4b | 0 | 89.7 | 10.3 | 0 |
| 1 × 10−3 | 58.5 | 41.5 | 0 | |
| 3 × 10−3 | 56.2 | 43.8 | 0 | |
| 5 × 10−3 | 48.2 | 51.8 | 0 | |
| 4c | 0 | 93.6 | 6.4 | 0 |
| 1 × 10−3 | 49.8 | 50.2 | 0 | |
| 3 × 10−3 | 0 | 100 | 0 | |
| 5 × 10−3 | 4.7 | 95.3 | 0 |
| Complex | Ksv (M−1) × 104 | kq (M−1s−1) × 1012 | KBSA (M−1) × 104 | n |
|---|---|---|---|---|
| 4a | 7.16 (±0.03) | 7.16 (±0.03) | 6.66 (±0.30) | 1.03 |
| 4b | 6.39 (±0.06) | 6.39 (±0.06) | 5.37 (±0.25) | 0.94 |
| 4c | 4.86 (±0.15) | 4.86 (±0.15) | 4.77 (±0.20) | 1.16 |
| Sample | Absorbance * 570 nm | Absorbance * 600 nm | Reduction Percentage (%) | Cell Death (%) |
|---|---|---|---|---|
| Positive control C+ | 1.10 | 0.12 | 88.6 | 12.4 |
| Negative control C− | 0.69 | 0.93 | 4.4 | 95.6 |
| 4a (1 mM) | 0.55 | 0.25 | 32.9 | 67.1 |
| 4a (3 mM) | 0.44 | 0.5 | 8.4 | 91.6 |
| 4a (5 mM) | 0.32 | 0.26 | 3.5 | 96.5 |
| 4b (1 mM) | 0.75 | 0.69 | 24.0 | 76.0 |
| 4b (3 mM) | 0.69 | 0.88 | 7.4 | 92.6 |
| 4b (5 mM) | 0.63 | 0.90 | 1.0 | 99.0 |
| 4c (1 mM) | 0.60 | 0.77 | 5.2 | 94.8 |
| 4c (3 mM) | 0.40 | 0.44 | 8.5 | 91.5 |
| 4c (5 mM) | 0.33 | 0.27 | 2.6 | 97.4 |
| Compound | IC50—A549 | IC50—HepG2 | IC50—HEL |
|---|---|---|---|
| 4a | 66.4 ± 6.2 | 17.9 ± 2.6 | 65.9 ± 6.9 |
| 4b | 126.9 ± 8.1 | 32.8 ± 4.9 | 44.3 ± 3.6 |
| 4c | 72.0 ± 6.5 | 21.6 ± 4.9 | 62.9 ± 6.2 |
| cisplatin | 26.2 ± 4.1 | 16.0 ± 3.4 | 68.4 ± 0.3 |
| Compound | Activity at c1/c1* (%) | Activity at c2 (%) | SOD-IC50 (μM) |
|---|---|---|---|
| 4a | 95.8 ± 2.2 | 14.3 ± 3.3 | 242 ± 1 |
| 4b | 40.1 ± 1.9 * | 35.8 ± 1.3 | 1090 ± 56 |
| 4c | 98.9 ± 0.5 | 23.9 ± 1.3 | 189 ± 11 |
| cystamine | 22.1 ± 1.8 | 2.0 ± 0.0 | 1700 ± 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lintnerová, L.; Herich, P.; Korcová, J.; Svitková, B.; Jozefíková, F.; Valentová, J. Synthesis, Structure, DNA/BSA Binding, DNA Cleaving, Cytotoxic and SOD Mimetic Activities of Copper(II) Complexes Derived from Methoxybenzylamine Schiff Base Ligands. Molecules 2025, 30, 3461. https://doi.org/10.3390/molecules30173461
Lintnerová L, Herich P, Korcová J, Svitková B, Jozefíková F, Valentová J. Synthesis, Structure, DNA/BSA Binding, DNA Cleaving, Cytotoxic and SOD Mimetic Activities of Copper(II) Complexes Derived from Methoxybenzylamine Schiff Base Ligands. Molecules. 2025; 30(17):3461. https://doi.org/10.3390/molecules30173461
Chicago/Turabian StyleLintnerová, Lucia, Peter Herich, Jana Korcová, Barbora Svitková, Flóra Jozefíková, and Jindra Valentová. 2025. "Synthesis, Structure, DNA/BSA Binding, DNA Cleaving, Cytotoxic and SOD Mimetic Activities of Copper(II) Complexes Derived from Methoxybenzylamine Schiff Base Ligands" Molecules 30, no. 17: 3461. https://doi.org/10.3390/molecules30173461
APA StyleLintnerová, L., Herich, P., Korcová, J., Svitková, B., Jozefíková, F., & Valentová, J. (2025). Synthesis, Structure, DNA/BSA Binding, DNA Cleaving, Cytotoxic and SOD Mimetic Activities of Copper(II) Complexes Derived from Methoxybenzylamine Schiff Base Ligands. Molecules, 30(17), 3461. https://doi.org/10.3390/molecules30173461