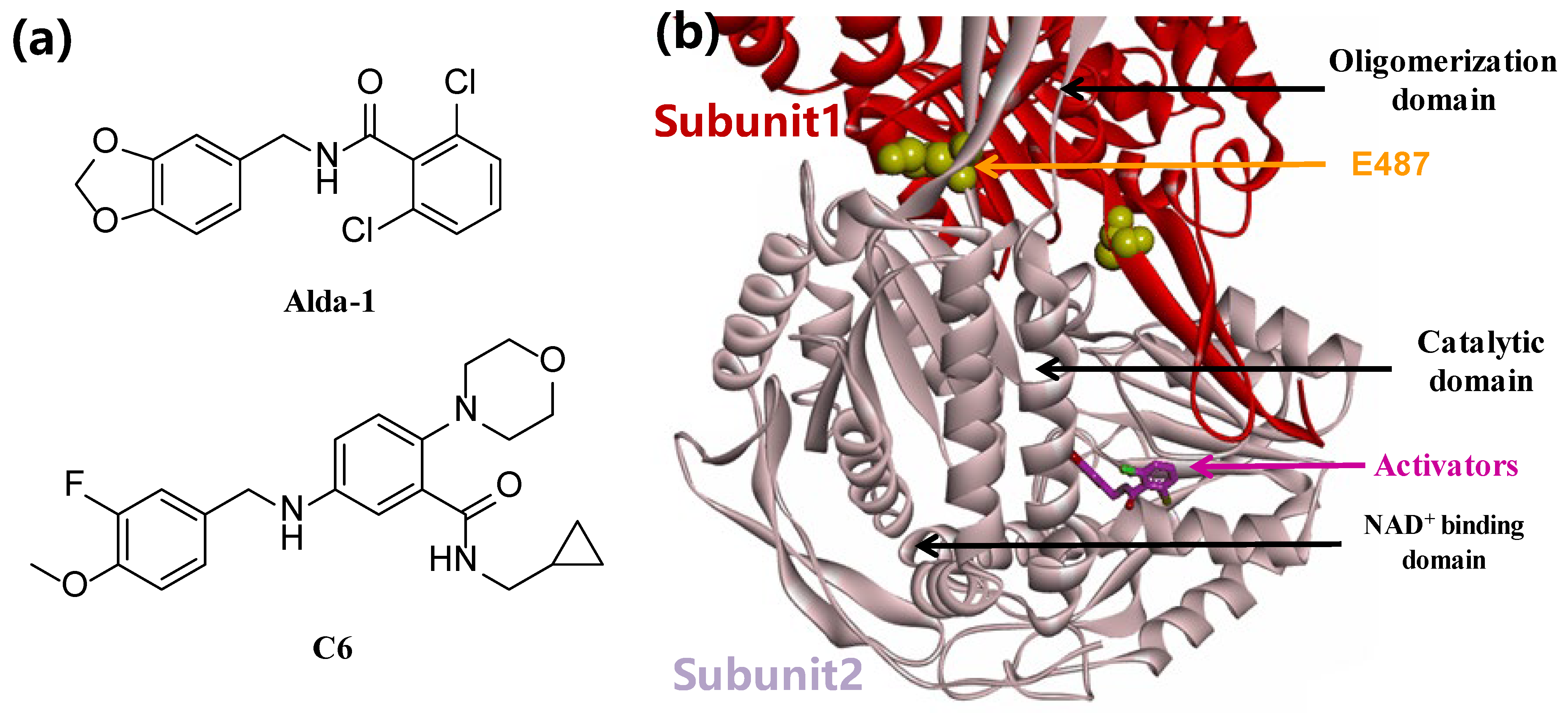
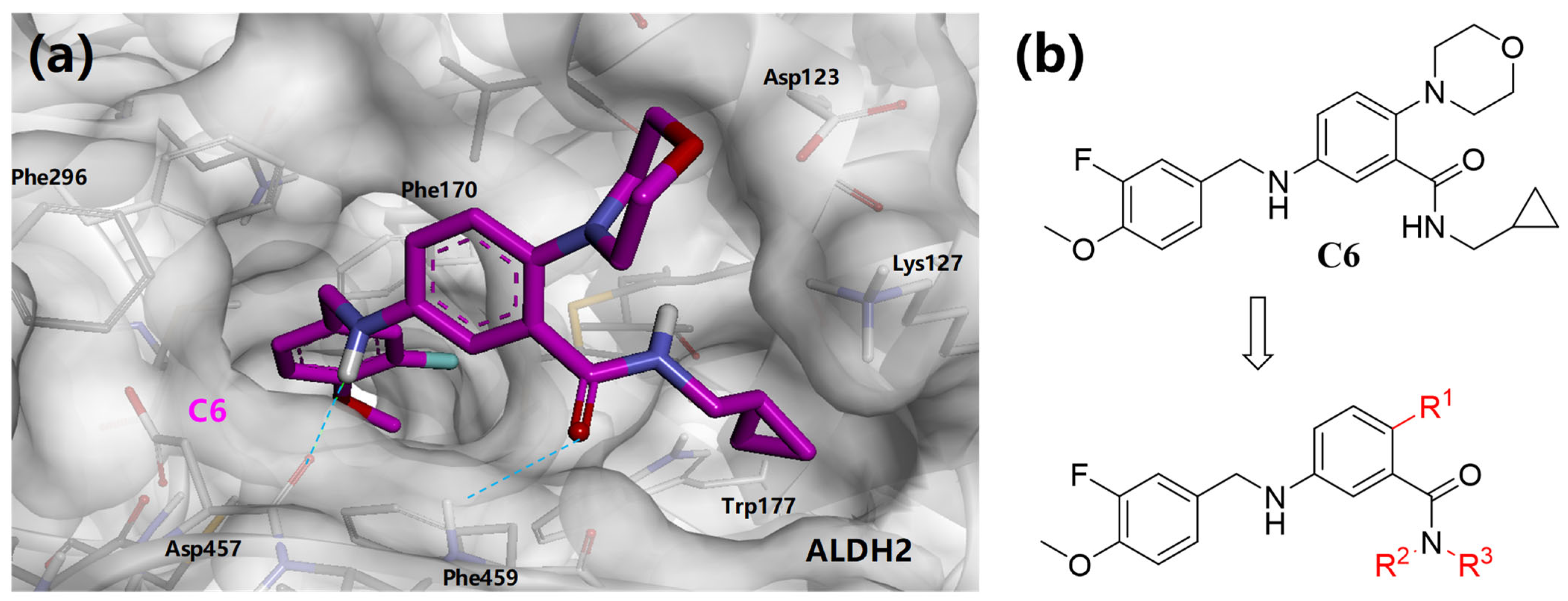
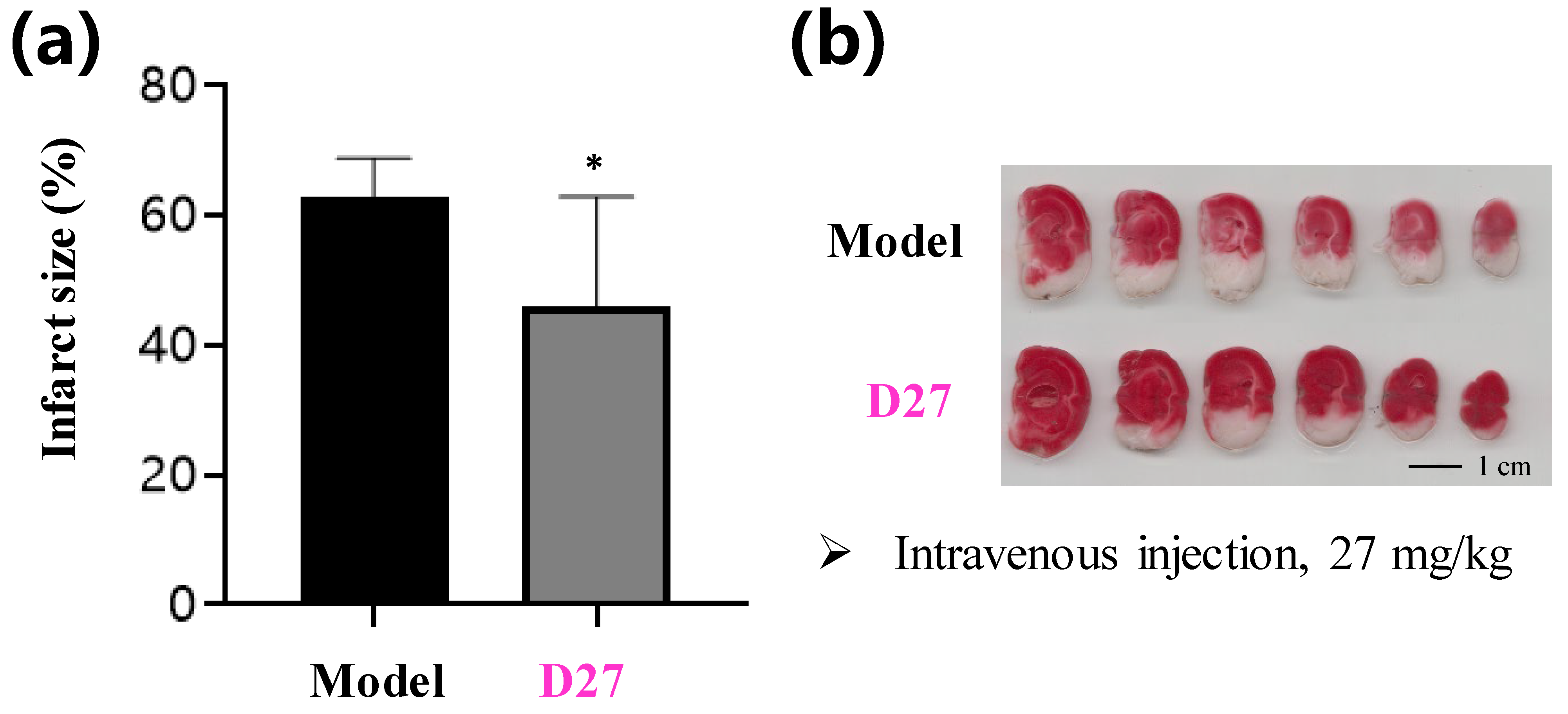
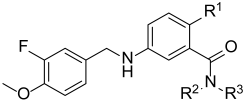
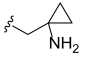
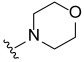
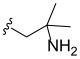
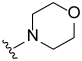
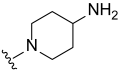
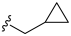
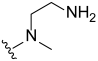
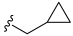
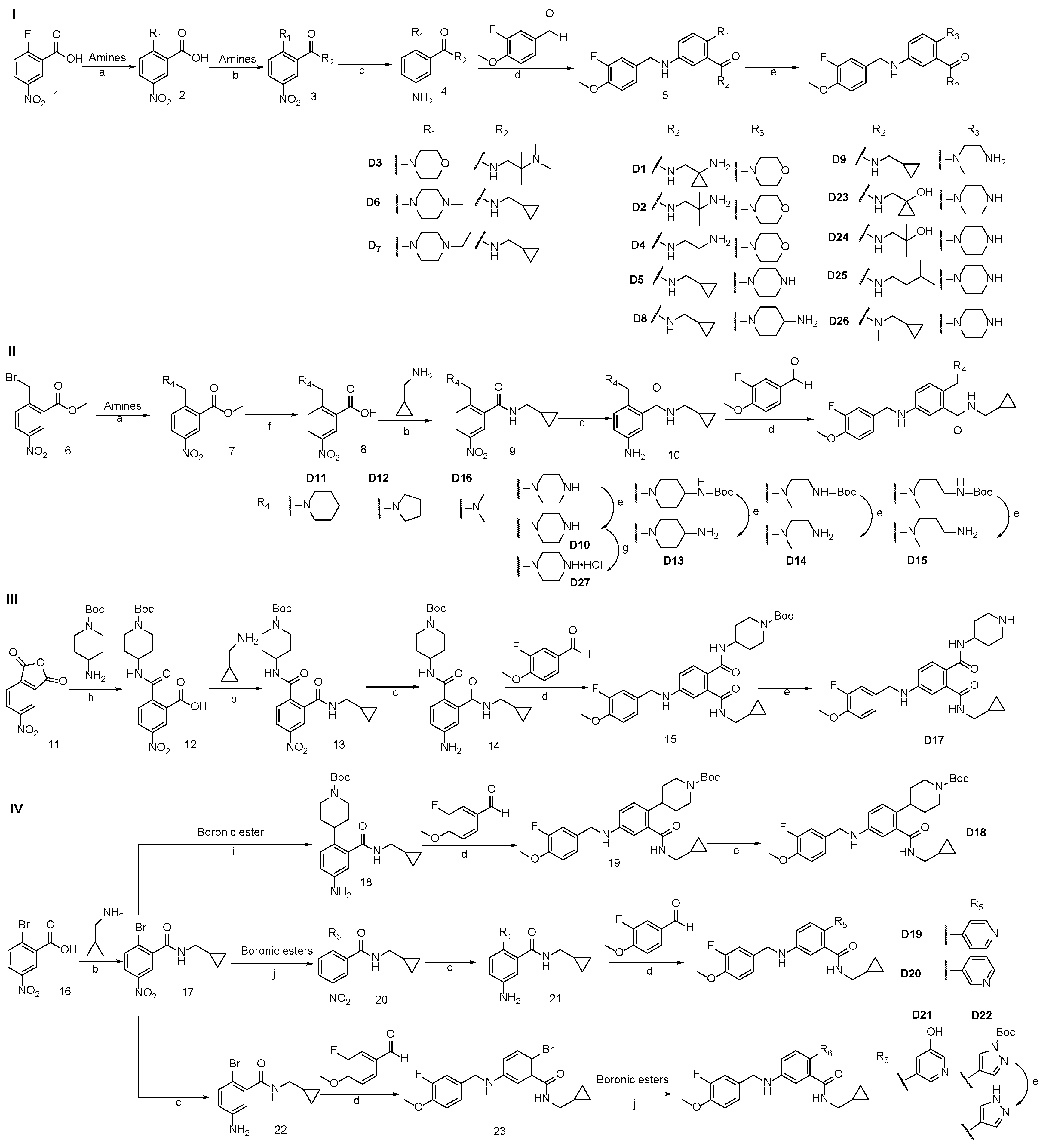3.2. Synthesis of Target Compounds
All reagents used in this study are of analytical purity. The mass spectrometry used for analyzing and verifying the structure of the compounds is Agilent 1290 Infinity-6538 UHD and Accurate-Mass QTOF/MS (Agilent, Santa Clara, CA, USA), and the proton nuclear magnetic resonance (1H NMR) and carbon nuclear magnetic resonance (13C NMR) spectra were obtained using a Bruker AV300/600 (Billerica, MA, USA). The purity was determined by an Agilent 1260 Infinity liquid chromatography system, with the chromatographic column being Agilent Eclipse Plus C18 (5 μm, 4.6 × 250mm). The purity of all the target compounds is >95%.
General method for step a: A solution of Halides (9.9 mmol), amines (9.9 mmol), and K2CO3 (18.8 mmol) in DMSO (15 mL) was stirred at 80 °C. Completion of the reaction was confirmed by TLC (Route Ι: DCM/MeOH system, and Route II: PE/EA system). The reaction mixture was cooled and added to water (50 mL), then extracted with EA, and the organic phase was dried over sodium sulfate anhydrous, filtered, and concentrated. The residue was partially purified by silica gel column (Route I: DCM/MeOH = 10:1, and Route II: PE/EA = 4:1) to afford the products with 79–99% yield.
General method for step b: Amines (1.1 mmol) were added in one portion at room temperature to a DCM solution (5 mL) of benzoic acids (1.0 mmol), EDCI (5.01 mmol), and DMAP (2.05 mmol). The resulting solution was allowed to stir at room temperature (1.5–6 h) under argon. The reaction was diluted with DCM, and washed with water (2×) and a saturated aq NaCl solution. The organic phase was dried over sodium sulfate anhydrous, filtered, and concentrated. The residue was purified by silica gel column (PE/EA = 3:1) to give the products with 67–95% yield.
General method for step c: Nitro compounds (0.06 mmol) were dissolved in EtOH (15 mL); NH4Cl (0.24 mmol) was dissolved in water (5 mL), and added to the reaction mixture; the system was refluxed at 80 °C for 0.5 h, and, then, iron powder (0.23 mmol) was added; and the reaction mixture and heated at 80 °C for 1–3 h. Completion of the reaction was confirmed by TLC. The reaction was cooled to room temperature, filtered over a plug of Celite, and the filtrate was combined and concentrated in vacuo. The residue was purified by silica gel column (PE/EA = 2:1) to afford the products with 71–97% yield.
General method for step d: Amines (0.84 mmol) and 3-fluoro-4-methoxybenzaldehyde (0.84 mmol) in DCE were stirred at 80 °C for ~1 h and cooled to room temperature, and additional sodium triacetoxyhydroborate (1.67 mmol) was added and allowed to react at room temperature under argon atmosphere. The reaction was quenched by saturated sodium bicarbonate solution, and the reaction mixture was extracted with DCM (50 mL) three times. The combined organics were dried over anhydrous Na2SO4, and concentrated by rotary evaporator, and then purified by silica gel column (PE/EA = 1:1) to provide the products with 59–95% yield.
General method for step e: Tert butyl carboxylates were dissolved in DCM, and additional TFA was added; and the reaction was stirred at room temperature for 0.5–2.5 h. Completion of the reaction was confirmed by TLC. The reaction mixture was concentrated under reduced pressure and the residue was partially purified by reversed-phase chromatography (C18 column, gradient elution with MeOH/H2O), and the product was obtained after freeze-drying in 74–95% yield.
Method for step f: Esters (1.0 mmol) were dissolved in the mixed solvent with a MeOH:THF ratio of 1:1 (8 mL), and NaOH (10.0 mmol) in 5:3 MeOH:H2O was added dropwise to the reaction mixture at room temperature. Completion of the reaction was confirmed by TLC. The reaction mixture was concentrated by rotary evaporator; the residue was extracted with 1M diluted HCl and DCM; the combined organic phase was concentrated by rotary evaporator; and the solid was separated with 91% yield.
Method for step g: Tert-butyl 4-(2-((cyclopropylmethyl)carbamoyl)-4-((3-fluoro-4-methoxybenzyl)amino)benzyl)piperazine-1-carboxylate (972 mg, 1.84 mmol) was dissolved in EA (10 mL); and concentrated HCl (0.5 mL) was added and stirred at room temperature for 1.5 h. The mixture was repeatedly dissolved with DCM, stirred, and concentrated to give a white solid (543 mg) in 69% yield.
Method for step h: Degassed DMF (6 mL) was added to 5-Nitroisobenzofuran-1,3-dione (300 mg, 1.6 mmol), tert-Butyl 4-aminopiperazine-1-carboxylate (311 mg, 1.6 mmol), HATU (2.36 g, 4.0 mmol), and DIPEA (1.08 mL, 4.0 mmol) under argon. The suspension was degassed by ultrasound for 30 min, refilled with argon, heated to 80 °C for 12 h, and then cooled to room temperature; the reaction mixture was extracted with diluted HCl and DCM; the combined organic phase was dried with anhydrous Na2SO4, concentrated by rotary evaporator; and the residue was purified by silica gel column (DCM:MeOH = 15:1) to afford a yellow solid (261.9 mg) with 42.8% yield.
Method for step i: 2-Bromo-N-(cyclopropylmethyl)-5-nitrobenzamide (50 mg, 0.17 mmol), tert-butyl 4-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-5,6-dihydropyridine-1(2H)-carboxylate (52 mg, 0.17 mmol), PdXPhosG2 (1.34 mg, 0.0017 mmol), Pd/C (0.02 mmol), and K3PO4 (107 mg, 0.50 mmol) were dissolved in 5 mL of a Dioxane: H2O mixed solvent with a ratio of 4:1 under argon atmosphere. The solution was heated to 80 °C. After 4 h, the reaction mixture was cooled to room temperature; and NH4COOH (106 mg, 1.68 mmol) was dissolved in MeOH (2 mL), poured into the reaction mixture, and reacted at room temperature for 16 h. Completion of the reaction was confirmed by TLC. The reaction was filtered through celite, concentrated by rotary evaporation, and purified by silica gel column (PE/EA = 1:1) to afford a white solid (47 mg) with 75% yield.
General method for step j: A degassed mixture of aryl bromides (0.17 mmol), boric acid pinacol esters (0.17 mmol), Pd(dppf)Cl2 (0.016 mmol), and K2CO3 (0.5 mmol) in a Dioxane:H2O mixed solvent with a ratio of 3:1 was heated at 100 °C for 12 h. The reaction mixture was diluted with EA and washed with water. The aqueous layer was extracted with EA. The combined organics were washed with brine, dried (Na2SO4), and concentrated. The residue was purified by silica gel column (PE/EA = 2:1) to provide a white solid with ~70% yield.
N-((1-aminocyclopropyl)methyl)-5-((3-fluoro-4-methoxybenzyl)amino)-2-morpholinobenzamide (D1): Brown solid, 1H NMR (600 MHz, DMSO-d6) δ 10.52 (t, J = 5.9 Hz, 1H), 8.40–8.28 (m, 3H), 7.24 (d, J = 2.9 Hz, 1H), 7.18–7.13 (m, 2H), 7.12–7.07 (m, 2H), 6.68 (dd, J = 8.7, 3.0 Hz, 1H), 4.20 (s, 2H), 3.78 (s, 3H), 3.75–3.70 (m, 4H), 3.57 (d, J = 6.1 Hz, 2H), 2.85–2.79 (m, 4H), 0.94–0.90 (m, 2H), 0.90–0.86 (m, 2H). 13C NMR (151 MHz, DMSO) δ 167.04, 151.44 (d, J = 243.8 Hz), 145.76 (d, J = 10.4 Hz), 145.70, 140.01, 133.12 (d, J = 4.3 Hz), 128.48, 123.05, 122.79, 115.36, 114.48 (d, J = 17.9 Hz), 113.82, 113.77, 66.65, 55.99, 53.30, 45.53, 42.84, 34.85, 8.96. HRMS (ESI): m/z calcd for C23H29FN4O3 (M + H)+: 429.2224, found 429.2326.
N-(2-amino-2-methylpropyl)-5-((3-fluoro-4-methoxybenzyl)amino)-2-morpholinobenzamide (D2): Yellow solid, 1H NMR (600 MHz, DMSO-d6) δ 10.61 (t, 1H), 7.86 (s, 3H), 7.25 (d, J = 3.0 Hz, 1H), 7.21 (d, J = 8.7 Hz, 1H), 7.15 (d, 1H), 7.12–7.07 (m, 2H), 6.70 (dd, J = 8.7, 3.0 Hz, 1H), 4.21 (s, 2H), 3.79 (s, 3H), 3.74–3.71 (m, 4H), 3.49 (d, J = 6.6 Hz, 2H), 2.87–2.80 (m, 4H), 1.26 (s, 6H). 13C NMR (151 MHz, DMSO) δ 167.16, 151.42 (d, J = 243.6 Hz), 145.77 (d, J = 3.9 Hz), 145.72, 139.86, 133.09 (d, J = 5.0 Hz), 128.51, 123.05, 122.98, 115.31, 114.47 (d, J = 17.9 Hz), 113.85, 113.76, 66.62, 55.98, 54.55, 53.31, 46.27, 45.52, 23.40. HRMS (ESI): m/z calcd for C23H31FN4O3 (M + H)+: 431.2477, found 431.2380.
N-(cyclopropylmethyl)-5-((3-fluoro-4-methoxybenzyl)amino)-2-(piperazin-1-yl)benzamide (D5): White solid, 1H NMR (300 MHz, Chloroform-d) δ 10.72 (t, J = 5.1 Hz, 1H), 7.57 (d, J = 3.0 Hz, 1H), 7.17–6.99 (m, 3H), 6.90 (t, J = 8.6 Hz, 1H), 6.62 (dd, J = 8.6, 3.0 Hz, 1H), 4.27 (s, 2H), 3.86 (s, 3H), 3.30 (dd, J = 7.1, 5.1 Hz, 2H), 3.07 (t, J = 4.7 Hz, 4H), 2.90 (t, J = 4.7 Hz, 4H), 2.48 (s, 1H), 1.06 (ddd, J = 12.4, 7.8, 4.7 Hz, 1H), 0.56 (dt, J = 8.4, 2.9 Hz, 2H), 0.29 (t, J = 5.0 Hz, 2H). 13C NMR (101 MHz, DMSO-d6) δ 165.62, 151.44 (d, J = 243.6 Hz), 145.73 (d, J = 11.1 Hz), 145.63, 139.55, 133.18 (d, J = 5.4 Hz), 129.39, 123.03 (d, J = 3.3 Hz), 122.01, 114.60, 114.47 (d, J = 19.2 Hz), 113.97, 113.75, 55.96, 51.37, 45.52, 44.41, 43.44, 10.86, 3.44. HRMS (ESI): m/z calcd for C23H29FN4O2 (M + H)+: 413.2275, found 413.44.
N-(cyclopropylmethyl)-5-((3-fluoro-4-methoxybenzyl)amino)-2-(piperazin-1-ylmethyl)benzamide (D10): White solid, 1H NMR (600 MHz, DMSO-d6) δ 9.97 (t, J = 5.2 Hz, 1H), 7.21–7.14 (m, 1H), 7.13–7.09 (m, 2H), 6.99–6.91 (m, 2H), 6.56 (dd, J = 8.3, 2.7 Hz, 1H), 6.42 (t, J = 6.1 Hz, 1H), 4.21 (d, J = 6.1 Hz, 2H), 3.81 (s, 3H), 3.43 (s, 2H), 3.13 (dd, J = 6.8, 5.3 Hz, 2H), 2.66 (s, 4H), 2.30 (s, 4H), 1.04–0.93 (m, 2H), 0.53–0.42 (m, 2H), 0.25–0.15 (m, 2H). 13C NMR (101 MHz, DMSO-d6) δ 168.26, 151.43 (d, J = 243.6 Hz), 147.83, 145.78 (d, J = 10.6 Hz), 138.09, 133.12 (d, J = 5.3 Hz), 132.32, 123.10 (d, J = 3.6 Hz), 120.37, 114.52 (d, J = 18.0 Hz), 113.75, 113.11, 112.51, 59.60, 55.97, 49.29, 45.34, 43.62, 43.32, 10.82, 3.36. HRMS (ESI): m/z calcd for C24H31FN4O2 (M + H)+: 427.2431, found 427.2554.
N-(cyclopropylmethyl)-5-((3-fluoro-4-methoxybenzyl)amino)-2-(piperidin-1-ylmethyl)benzamide (D11): White solid, 1H NMR (300 MHz, Chloroform-d) δ 9.41 (s, 1H), 7.17–7.02 (m, 4H), 6.89 (t, J = 8.6 Hz, 1H), 6.50 (dd, J = 8.2, 2.5 Hz, 1H), 4.25 (s, 2H), 3.85 (s, 3H), 3.66 (s, 2H), 3.23 (dd, J = 6.9, 5.1 Hz, 2H), 2.74 (s, 4H), 2.01 (s, 2H), 1.69–1.49 (m, 6H), 1.07 (qq, J = 7.4, 4.8, 3.6 Hz, 1H), 0.57–0.45 (m, 2H), 0.31–0.20 (m, 2H). 13C NMR (151 MHz, Chloroform-d) δ 170.09, 152.51 (d, J = 246.2 Hz), 149.04, 146.89 (d, J = 10.5 Hz), 137.48, 134.56, 131.53, 123.12, 115.61, 115.10 (d, J = 18.4 Hz), 114.33, 113.69, 113.26, 56.31 (d, J = 16.2 Hz), 52.01, 45.07 (d, J = 30.1 Hz), 23.27 (d, J = 38.7 Hz), 21.97 (d, J = 19.0 Hz), 10.67, 3.66 (d, J = 11.2 Hz). HRMS (ESI): m/z calcd for C25H32FN3O2 (M + H)+: 426.2479, found 426.2555.
N-(cyclopropylmethyl)-2-((dimethylamino)methyl)-5-((3-fluoro-4-methoxybenzyl)amino)benzamide (D16): Gray solid, 1H NMR (600 MHz, Chloroform-d) δ 8.45 (s, 1H), 7.29 (s, 2H), 7.07 (td, J = 7.5, 3.8 Hz, 3H), 6.91 (t, J = 8.6 Hz, 1H), 6.55 (dd, J = 8.3, 2.5 Hz, 1H), 4.28 (s, 2H), 3.97 (s, 2H), 3.87 (s, 3H), 3.23 (dd, J = 7.5, 4.1 Hz, 2H), 2.68 (s, 6H), 1.09 (s, 1H), 0.56–0.49 (m, 2H), 0.28 (dt, J = 6.1, 4.6 Hz, 2H). 13C NMR (151 MHz, Chloroform-d) δ 153.33, 151.69, 149.57, 146.89 (d, J = 10.9 Hz), 136.90, 134.14, 131.39, 122.95 (d, J = 3.6 Hz), 115.71, 115.06, 114.94, 113.95, 113.70, 113.48, 60.04, 56.34, 46.73, 45.15, 44.58, 41.78, 22.41 (d, J = 24.9 Hz), 10.40, 3.49. HRMS (ESI): m/z calcd for C22H28FN3O2 (M + H)+: 386.2166, found 386.2256.
N-(cyclopropylmethyl)-5-((3-fluoro-4-methoxybenzyl)amino)-2-(pyridin-3-yl)benzamide (D20): White solid, 1H NMR (600 MHz, Chloroform-d) δ 8.62 (d, J = 2.3 Hz, 1H), 8.52 (dd, J = 4.9, 1.6 Hz, 1H), 7.71 (dt, J = 7.9, 2.0 Hz, 1H), 7.30–7.28 (m, 1H), 7.17 (d, J = 8.3 Hz, 1H), 7.12–7.04 (m, 2H), 6.96–6.90 (m, 2H), 6.71 (dd, J = 8.4, 2.6 Hz, 1H), 5.45 (t, J = 5.4 Hz, 1H), 4.33 (s, 2H), 3.89 (s, 3H), 3.03 (dd, J = 7.2, 5.5 Hz, 2H), 0.73–0.58 (m, 1H), 0.38–0.27 (m, 2H), −0.00–−0.04 (m, 2H). HRMS (ESI): m/z calcd for C24H24FN3O2 (M + H)+: 406.1853, found 406.1921.
5-((3-fluoro-4-methoxybenzyl)amino)-N-isopentyl-2-(piperazin-1-yl)benzamide (D25): White solid, 1H NMR (600 MHz, DMSO-d6) δ 10.44 (s, 1H), 7.23 (d, J = 3.0 Hz, 1H), 7.15 (d, J = 12.4 Hz, 1H), 7.09 (d, J = 16.8 Hz, 3H), 6.63 (d, J = 8.7 Hz, 1H), 6.32 (s, 1H), 4.18 (d, J = 6.3 Hz, 2H), 3.79 (s, 3H), 2.81 (s, 4H), 2.69 (s, 4H), 2.61 (s, 1H), 2.38 (s, 1H), 1.66–1.60 (m, 1H), 1.43 (d, J = 7.3 Hz, 2H), 1.23 (s, 2H), 0.91(d, J = 6.6 Hz, 5H). 13C NMR (151 MHz, Chloroform-d) δ 166.16 (d, J = 11.9 Hz), 153.34, 151.71, 145.81, 140.69, 140.10, 132.18, 129.02, 122.97 (d, J = 3.4 Hz), 115.58, 115.39 (d, J = 11.6 Hz), 115.19, 115.07, 113.59, 56.35, 53.28, 51.83, 47.45 (d, J = 43.3 Hz), 45.15, 38.81 (d, J = 4.2 Hz), 37.86 (d, J = 11.2 Hz), 26.17, 22.53. HRMS (ESI): m/z calcd for C24H33FN4O2 (M + H)+: 429.2588, found 429.2675.
N-(2-(dimethylamino)-2-methylpropyl)-5-((3-fluoro-4-methoxybenzyl)amino)-2-morpholinobenzamide (D3): White solid, 1H NMR (600 MHz, DMSO-d6) δ 10.75 (t, J = 6.8 Hz, 1H), 7.27 (d, J = 3.0 Hz, 1H), 7.22 (d, J = 8.7 Hz, 1H), 7.17–7.13 (m, 1H), 7.12–7.07 (m, 2H), 6.70 (dd, J = 8.7, 3.0 Hz, 1H), 6.45 (t, J = 6.2 Hz, 1H), 4.22 (d, J = 6.1 Hz, 2H), 3.79 (s, 3H), 3.75–3.70 (m, 4H), 3.67 (d, J = 7.4 Hz, 2H), 2.85–2.80 (m, 4H), 2.77 (s, 6H), 1.33 (s, 6H). HRMS (ESI): m/z calcd for C25H35FN4O3 (M + H)+: 459.2693, found 459.2273.
N-(2-aminoethyl)-5-((3-fluoro-4-methoxybenzyl)amino)-2-morpholinobenzamide (D4): White solid, 1H NMR (600 MHz, DMSO-d6) δ 10.17–10.01 (m, 1H), 7.21–7.13 (m, 2H), 7.11–7.03 (m, 3H), 6.62 (td, J = 8.6, 3.0 Hz, 1H), 6.33 (dt, J = 13.0, 6.2 Hz, 1H), 4.18 (dd, J = 6.2, 2.5 Hz, 2H), 3.79 (s, 3H), 3.73 (dt, J = 21.5, 4.7 Hz, 4H), 3.28 (dd, J = 10.5, 5.6 Hz, 3H), 3.07 (q, J = 6.0 Hz, 1H), 2.81–2.73 (m, 4H), 2.70 (t, J = 6.2 Hz, 1H). HRMS (ESI): m/z calcd for C21H27FN4O3 (M + H)+: 403.2067, found 403.2174.
N-(cyclopropylmethyl)-5-((3-fluoro-4-methoxybenzyl)amino)-2-(4-methylpiperazin-1-yl)benzamide (D6): White solid, 1H NMR (300 MHz, Chloroform-d) δ 10.53 (s, 1H), 7.56 (d, J = 2.8 Hz, 1H), 7.17–7.01 (m, 3H), 6.97–6.84 (m, 1H), 6.69–06.56 (m, 1H), 4.28 (s, 2H), 3.87 (s, 3H), 3.31 (t, J = 6.2 Hz, 2H), 3.11–2.92 (m, 4H), 2.70 (s, 4H), 2.42 (s, 3H), 1.14–0.98 (m, 1H), 0.67–0.50 (m, 2H), 0.36–0.21 (m, 2H). HRMS (ESI): m/z calcd for C24H31FN4O2 (M + H)+: 427.2631, found 427.2506.
N-(cyclopropylmethyl)-2-(4-ethylpiperazin-1-yl)-5-((3-fluoro-4-methoxybenzyl)amino)benzamide (D7): White solid, 1H NMR (600 MHz, Chloroform-d) δ 10.71 (s, 1H), 7.58 (d, J = 3.0 Hz, 1H), 7.14 (d, J = 8.6 Hz, 1H), 7.10–7.05 (m, 2H), 6.92 (t, J = 8.4 Hz, 1H), 6.64 (dd, J = 8.6, 3.0 Hz, 1H), 4.29 (s, 2H), 3.88 (s, 3H), 3.32 (dd, J = 7.1, 5.1 Hz, 2H), 3.00 (t, J = 4.9 Hz, 4H), 2.68 (s, 4H), 2.52 (q, J = 7.2 Hz, 2H), 1.16 (t, J = 7.2 Hz, 3H), 1.11–1.02 (m, 1H), 0.63–0.54 (m, 2H), 0.34–0.25 (m, 2H). HRMS (ESI): m/z calcd for C25H33FN4O2 (M + H)+: 441.2588, found 441.2677.
2-(4-aminopiperidin-1-yl)-N-(cyclopropylmethyl)-5-((3-fluoro-4-methoxybenzyl)amino)benzamide (D8): White solid, 1H NMR (300 MHz, Methanol-d4) δ 7.48 (dd, J = 9.2, 4.1 Hz, 1H), 7.25 (d, J = 3.4 Hz, 1H), 7.11 (d, J = 12.9 Hz, 3H), 6.91 (dt, J = 8.4, 3.4 Hz, 1H), 4.45–4.28 (m, 2H), 3.93–3.79 (m, 3H), 3.53 (s, 5H), 2.37 (d, J = 13.5 Hz, 2H), 2.03 (d, J = 11.9 Hz, 2H), 1.15 (s, 1H), 0.59 (t, J = 5.7 Hz, 2H), 0.33 (d, J = 4.6 Hz, 2H). HRMS (ESI): m/z calcd for C24H31FN4O2 (M + H)+: 427.2431, found 427.2508.
2-((2-aminoethyl)(methyl)amino)-N-(cyclopropylmethyl)-5-((3-fluoro-4-methoxybenzyl)amino)benzamide (D9): Gray solid, 1H NMR (600 MHz, DMSO-d6) δ 9.46 (s, 1H), 7.19–7.09 (m, 1H), 6.82 (d, J = 2.9 Hz, 4H), 6.64 (dd, J = 8.7, 2.9 Hz, 1H), 6.38 (t, J = 6.1 Hz, 1H), 4.19 (d, J = 6.1 Hz, 2H), 3.80 (s, 3H), 3.16–3.10 (m, 2H), 2.99 (t, J = 6.1 Hz, 2H), 2.68 (t, J = 6.1 Hz, 2H), 2.53 (s, 3H), 1.04–0.96 (m, 1H), 0.47–0.38 (m, 2H), 0.24–0.18 (m, 2H). HRMS (ESI): m/z calcd for C22H29FN4O2 (M + H)+: 401.2275, found 401.2386.
5-((3-fluoro-4-methoxybenzyl)amino)-N-((1-hydroxycyclopropyl)methyl)-2-(piperazin-1-yl)benzamide (D23): White solid, 1H NMR (600 MHz, DMSO-d6) δ 10.10 (t, J = 5.1 Hz, 1H), 8.98 (s, 2H), 7.23 (d, J = 2.9 Hz, 1H), 7.14 (d, 1H), 7.11–7.05 (m, 3H), 6.66 (dd, J = 8.7, 3.0 Hz, 1H), 4.19 (s, 2H), 3.78 (s, 3H), 3.36 (d, J = 5.0 Hz, 2H), 3.31–3.27 (m, 4H), 3.02–2.97 (m, 4H), 1.22 (s, 1H), 0.66–0.62 (m, 2H), 0.56–0.52 (m, 2H). HRMS (ESI): m/z calcd for C23H29FN4O3 (M + H)+: 429.2224, found 429.2313.
5-((3-fluoro-4-methoxybenzyl)amino)-N-(2-hydroxy-2-methylpropyl)-2-(piperazin-1-yl)benzamide (D24): White solid, 1H NMR (600 MHz, DMSO-d6) δ 10.13 (t, J = 5.5 Hz, 1H), 8.90 (s, 2H), 7.28 (d, J = 3.0 Hz, 1H), 7.16 (d, J = 12.8 Hz, 1H), 7.12–7.07 (m, 3H), 6.67 (dd, J = 8.7, 3.0 Hz, 1H), 4.20 (s, 2H), 3.79 (s, 3H), 3.34–3.28 (m, 4H), 3.27 (d, J = 5.5 Hz, 2H), 3.02–2.96 (m, 4H), 1.13 (s, 6H). HRMS (ESI): m/z calcd for C23H31FN4O3 (M + H)+: 431.2380, found 431.2478.
N-(cyclopropylmethyl)-5-((3-fluoro-4-methoxybenzyl)amino)-N-methyl-2-(piperazin-1-yl)benzamide (D26): White solid, 1H NMR (600 MHz, DMSO-d6) δ 7.19–7.06 (m, 3H), 6.94–6.85 (dd, J = 8.7, 22.6 Hz, 1H), 6.59–6.53 (ddd, J = 2.8, 8.7, 14.9 Hz, 1H), 6.36–6.20 (m, 2H), 4.21–4.13 (t, J = 5.9 Hz, 2H), 3.83–3.76 (d, J = 8.3 Hz, 3H), 3.14–2.95 (m, 8H), 2.85–2.69 (m, 4H), 1.09–1.01 (s, 1H), 0.69–0.21 (m, 4H), 0.06–0.22 (m, 2H). HRMS (ESI): m/z calcd for C24H31FN4O2 (M + H)+: 427.2431, found 427.2514.
N-(cyclopropylmethyl)-5-((3-fluoro-4-methoxybenzyl)amino)-2-(pyrrolidin-1-ylmethyl)benzamide (D12): White solid, 1H NMR (600 MHz, Chloroform-d) δ 8.91 (s, 1H), 7.28 (d, J = 8.3 Hz, 1H), 7.09–7.00 (m, 3H), 6.91 (t, J = 8.6 Hz, 1H), 6.57 (dd, J = 8.3, 2.6 Hz, 1H), 4.26 (s, 2H), 3.86 (s, 3H), 3.24 (dd, J = 7.1, 4.5 Hz, 2H), 2.89 (d, J = 6.7 Hz, 4H), 2.03 (s, 2H), 1.92 (p, J = 3.3 Hz, 4H), 1.06–0.99 (m, 1H), 0.55–0.49 (m, 2H), 0.25 (dd, J = 4.7, 1.3 Hz, 2H). HRMS (ESI): m/z calcd for C24H30FN3O2 (M + H)+: 412.2322, found 412.2410.
2-(((2-aminoethyl)(methyl)amino)methyl)-N-(cyclopropylmethyl)-5-((3-fluoro-4-methoxybenzyl)amino)benzamide (D14): Light yellow solid powder, 1H NMR (600 MHz, DMSO-d6) δ 9.37–9.18 (m, 1H), 9.01–8.90 (d, J = 7.0 Hz, 1H), 8.28–8.12 (s, 2H), 7.28–7.07 (m, 4H), 6.95–6.85 (s, 1H), 6.73–6.64 (d, J = 8.3 Hz, 1H), 4.34–4.24 (s, 2H), 4.11–3.97 (s, 1H), 3.83 –3.77 (s, 3H), 3.30–3.06 (m, 7H), 2.73–2.58 (s, 3H), 1.11–0.95 (s, 1H), 0.52–0.38 (d, J = 7.7 Hz, 2H), 0.29–0.16 (d, J = 5.0 Hz, 2H). HRMS (ESI): m/z calcd for C23H31FN4O2 (M + H)+: 415.2431, found 415.2519.
2-(((3-aminopropyl)(methyl)amino)methyl)-N-(cyclopropylmethyl)-5-((3-fluoro-4-methoxybenzyl)amino)benzamide (D15): White solid, 1H NMR (600 MHz, DMSO-d6) δ 9.21 (s, 1H), 8.04–7.65 (m, 3H), 7.20–7.00 (m, 4H), 6.85 (d, J = 2.5 Hz, 1H), 6.60 (s, 1H), 4.23 (d, J = 5.9 Hz, 2H), 3.80 (s, 3H), 3.36 (s, 4H), 3.12 (t, J = 6.2 Hz, 2H), 2.76 (d, J = 20.6 Hz, 2H), 2.41–2.08 (m, 3H), 1.78 (s, 2H), 1.00 (s, 1H), 0.49–0.42 (m, 2H), 0.21 (dt, J = 6.1, 2.9 Hz, 2H). HRMS (ESI): m/z calcd for C24H33FN4O2 (M + H)+: 429.2588, found 429.2674.
N2-(cyclopropylmethyl)-4-((3-fluoro-4-methoxybenzyl)amino)-N1-(piperidin-4-yl)phthalamide (D17): 1H NMR (600 MHz, DMSO-d6) δ 8.26–8.22 (m, 1H), 8.04–7.94 (m, 1H), 7.30 (dd, J = 32.7, 8.4 Hz, 1H), 7.18–7.07 (m, 3H), 6.79 (dt, J = 12.5, 6.2 Hz, 1H), 6.62–6.51 (m, 2H), 4.26 (d, J = 6.2 Hz, 2H), 3.95–3.87 (m, 1H), 3.80 (d, J = 1.2 Hz, 3H), 3.28–3.20 (m, 2H), 3.11–2.94 (m, 4H), 2.01–1.85 (m, 2H), 1.68–1.51 (m, 2H), 1.00–0.89 (m, 1H), 0.39 (dddd, J = 13.8, 8.2, 5.9, 4.1 Hz, 2H), 0.25–0.14 (m, 2H). HRMS (ESI): m/z calcd for C25H31FN4O3 (M + H)+: 455.2380, found 455.2470.
N-(cyclopropylmethyl)-5-((3-fluoro-4-methoxybenzyl)amino)-2-(pyridin-4-yl)benzamide (D19): White solid, 1H NMR (400 MHz, Chloroform-d) δ 8.77–8.24 (m, 2H), 7.36–7.29 (m, 2H), 7.20 (dd, J = 8.7, 2.1 Hz, 1H), 7.08 (m, 2H), 6.94 (t, J = 8.0 Hz, 1H), 6.88 (d, J = 2.9 Hz, 1H), 6.75–6.66 (m, 1H), 5.42 (d, J = 5.8 Hz, 1H), 4.33 (s, 2H), 3.89 (s, 3H), 3.04 (t, J = 6.5 Hz, 2H), 0.75–0.56 (m, 1H), 0.40–0.28 (m, 2H), 0.04–−0.10 (m, 2H). HRMS (ESI): m/z calcd for C24H24FN3O2 (M + H)+: 406.1853, found 406.1934.
N-(cyclopropylmethyl)-5-((3-fluoro-4-methoxybenzyl)amino)-2-(piperidin-4-yl)benzamide (D18): White solid, 1H NMR (600 MHz, Methanol-d4) δ 7.14–7.06 (m, 3H), 7.01 (t, J = 8.6 Hz, 1H), 6.74 (dd, J = 8.6, 2.6 Hz, 1H), 6.67 (d, J = 2.5 Hz, 1H), 4.29 (s, 2H), 3.83 (s, 3H), 3.49–3.39 (m, 2H), 3.19 (d, J = 7.0 Hz, 2H), 3.06–2.97 (m, 3H), 2.02 (d, J = 14.1 Hz, 2H), 1.91–1.77 (m, 2H), 1.13–1.00 (m, 1H), 0.58–0.44 (m, 2H), 0.28–0.22 (m, 2H). HRMS (ESI): m/z calcd for C24H30FN3O2 (M + H)+: 412.2322, found 412.2396.
N-(cyclopropylmethyl)-5-((3-fluoro-4-methoxybenzyl)amino)-2-(5-hydroxypyridin-3-yl)benzamide (D21): White solid, 1H NMR (300 MHz, Chloroform-d) δ 8.16 (dd, J = 11.6, 2.2 Hz, 1H), 7.41 (t, J = 2.1 Hz, 1H), 7.28 (s, 1H), 7.20 (t, J = 7.8 Hz, 1H), 7.12–6.97 (m, 4H), 6.91 (t, J = 8.6 Hz, 1H), 6.71 (dd, J = 8.1, 2.4 Hz, 1H), 6.37 (s, 1H), 4.28 (s, 1H), 3.87 (s, 3H), 3.29 (dd, J = 7.2, 5.3 Hz, 2H), 1.17–0.95 (m, 1H), 0.62–0.49 (m, 2H), 0.37–0.18 (m, 2H). HRMS (ESI): m/z calcd for C24H24FN3O3 (M + H)+: 422.1802, found 422.16.
N-(cyclopropylmethyl)-5-((3-fluoro-4-methoxybenzyl)amino)-2-(1H-pyrazol-4-yl)benzamide (D22): White solid, 1H NMR (600 MHz, Chloroform-d) δ 7.62 (s, 2H), 7.51 (d, J = 7.5 Hz, 1H), 7.12–7.04 (m, 2H), 7.01–6.91 (m, 3H), 6.46 (s, 1H), 4.66 (s, 1H), 4.32 (s, 2H), 3.84 (s, 3H), 3.32 (d, J = 7.0 Hz, 2H), 1.01 (hept, J = 7.0 Hz, 1H), 0.45 (ddd, J = 7.1, 6.0, 4.2 Hz, 2H), 0.28–0.20 (m, 2H). HRMS (ESI): m/z calcd for C22H23FN4O2 (M − H)−: 393.1805, found 393.44.