Molecular Insights into the Interaction of Orexin 1 Receptor Antagonists: A Comprehensive Study Using Classical and Quantum Computational Methods
Abstract
1. Introduction
2. Results
2.1. Ligand Optimization
2.2. Quantum Energies and Quantum Chemical Descriptors of Ligands
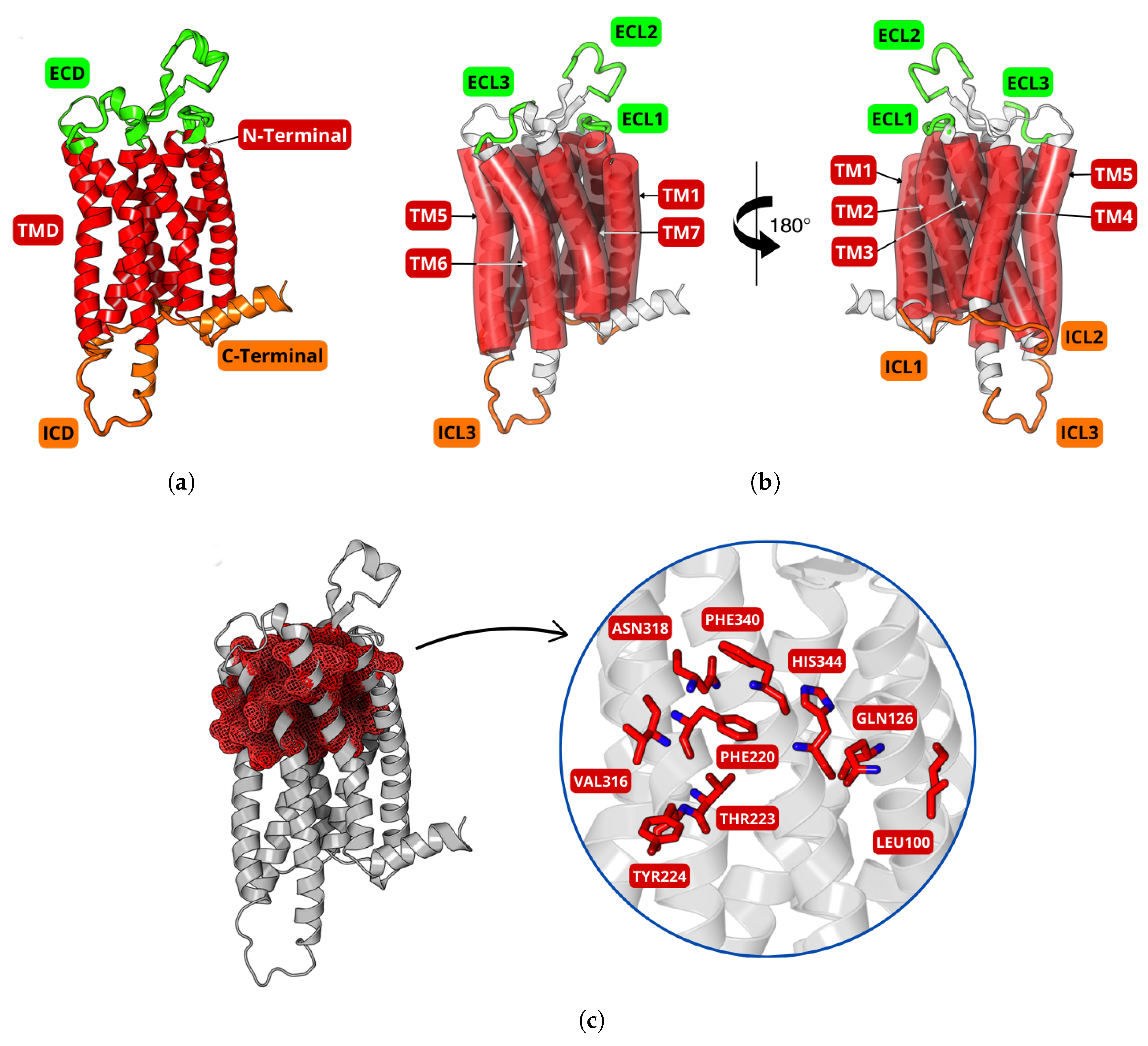
2.3. Molecular Docking
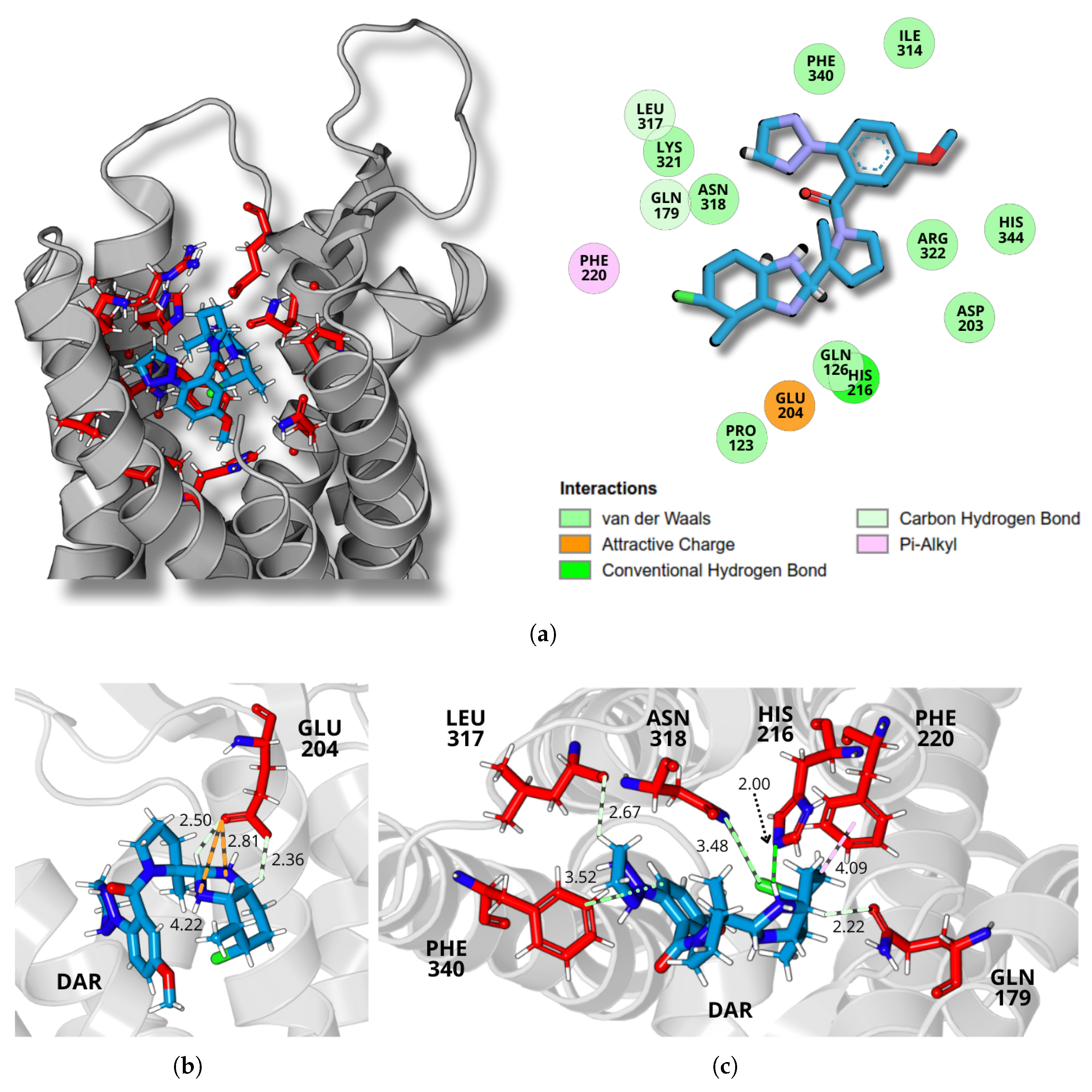
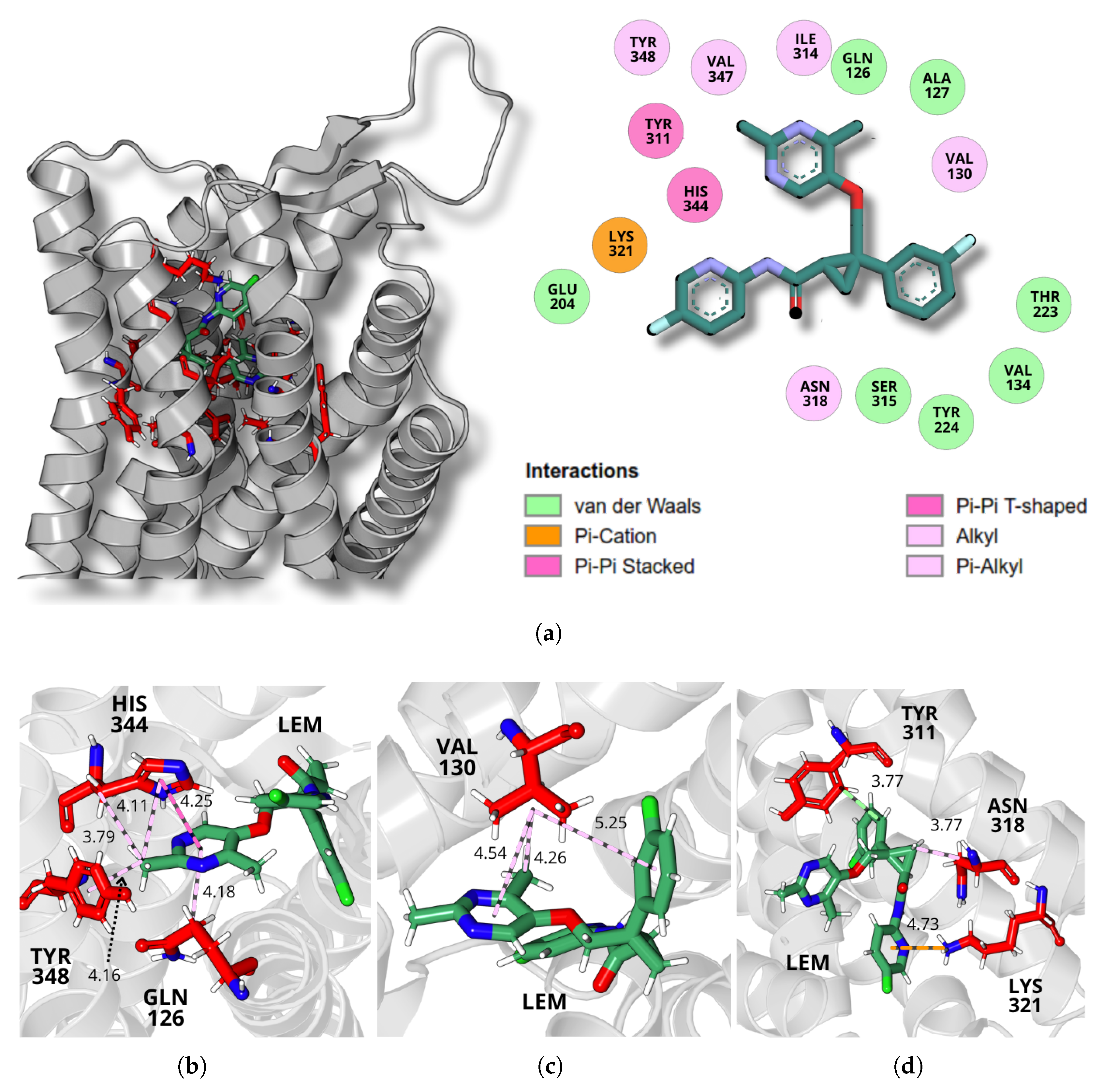
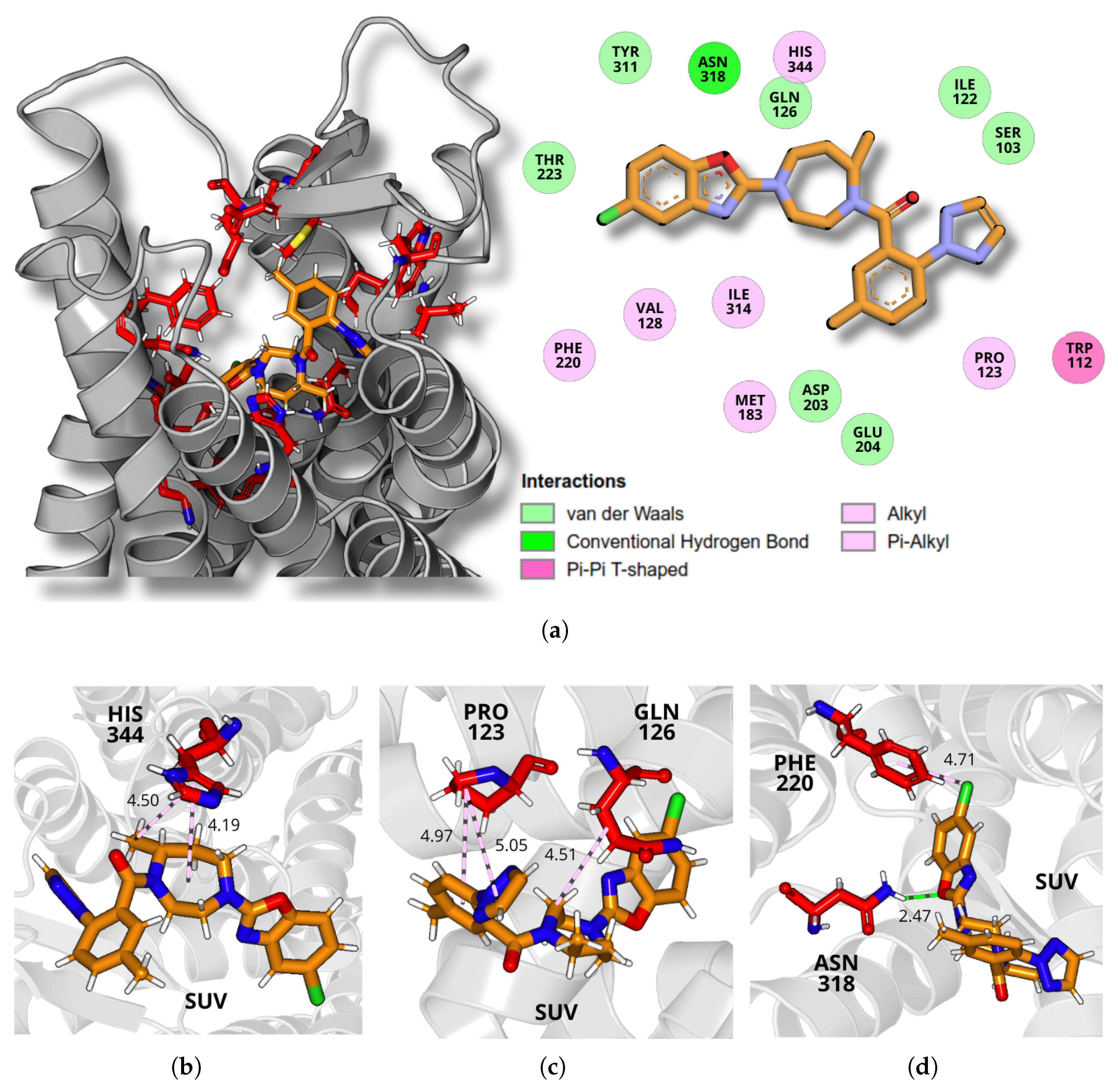
2.4. Dynamics and Intermolecular Interactions of DORA–OXR1 Complexes
3. Discussion
3.1. Conformational Analysis of Ligands and Electrostatic Potential Isosurface
3.2. Molecular Docking
3.3. Dynamics and Intermolecular Interactions of DORA–OXR1 Complexes
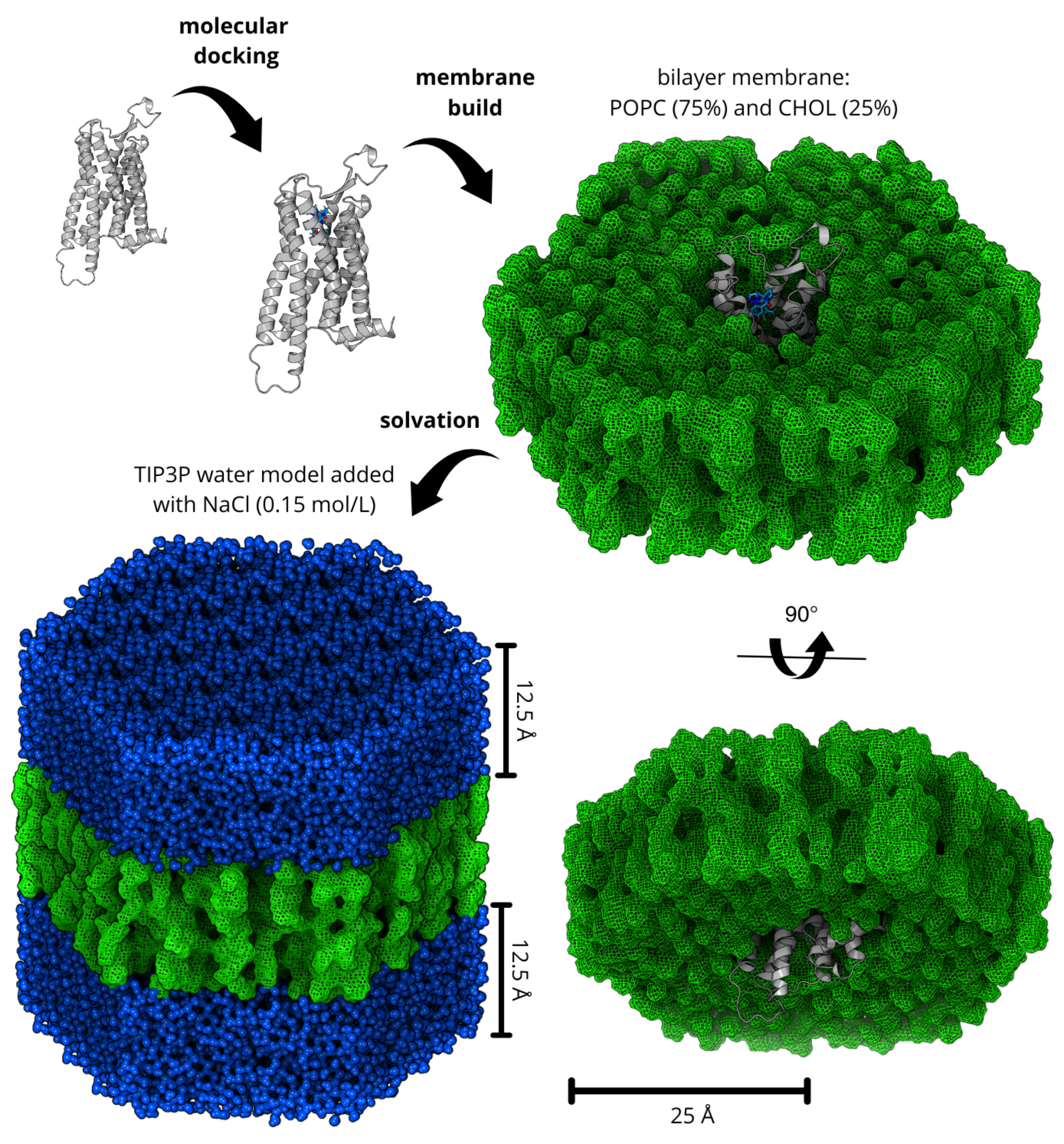
4. Materials and Methods
4.1. Ligand Molecular Optimization and DFT Calculation
4.2. Protein Preparation and Molecular Docking
4.3. Molecular Dynamics Simulation
4.4. Molecular Fractionation with Conjugate Cap
4.5. Quantum Calculations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| OXR1 | Orexin A receptor |
| OXR2 | Orexin B receptor |
| DORA | Dual orexin receptor antagonist |
| MD | Molecular dynamics |
| MFCC | Molecular fractionation with conjugate caps |
| DFT | Density functional theory |
| SASA | Solvent-accessible surface area |
| RMSD | Root-mean-square deviation |
| MO | Molecular orbitals |
| HOMO | Highest occupied molecular orbital |
| LUMO | Lowest unoccupied molecular orbital |
| RMSF | Root-mean-square fluctuation |
| GPCR | G-protein-coupled receptor |
References
- El Sayed, D.S.; Khalil, T.E.; Elbadawy, H.A. Rational and experimental investigation of antihypotensive midodrine-Fe(III) complex: Synthesis, spectroscopy, DFT, biological activity and molecular docking. J. Mol. Struct. 2024, 1311, 138421. [Google Scholar] [CrossRef]
- Naithani, U.; Guleria, V. Integrative computational approaches for discovery and evaluation of lead compound for drug design. Front. Drug Discov. 2024, 4, 1–11. [Google Scholar] [CrossRef]
- da Rocha, J.M.; Campos, D.M.O.; Esmaile, S.C.; Menezes, G.L.; Bezerra, K.S.; da Silva, R.A.; Junior, E.D.S.; Tayyeb, J.Z.; Akash, S.; Fulco, U.L.; et al. Quantum biochemical analysis of the binding interactions between a potential inhibitory drug and the Ebola viral glycoprotein. J. Biomol. Struct. Dyn. 2024, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Maldonado, A.M.; Durrant, J.D. From byte to bench to bedside: Molecular dynamics simulations and drug discovery. BMC Biol. 2023, 21, 299. [Google Scholar] [CrossRef]
- Singh, R.; Bhardwaj, V.K.; Das, P.; Purohit, R. A computational approach for rational discovery of inhibitors for non-structural protein 1 of SARS-CoV-2. Comput. Biol. Med. 2021, 135, 104555. [Google Scholar] [CrossRef]
- Sakurai, T.; Amemiya, A.; Ishii, M.; Matsuzaki, I.; Chemelli, R.M.; Tanaka, H.; Williams, S.C.; Richardson, J.A.; Kozlowski, G.P.; Wilson, S.; et al. Orexins and Orexin Receptors: A Family of Hypothalamic Neuropeptides and G Protein-Coupled Receptors that Regulate Feeding Behavior. Cell 1998, 92, 573–585. [Google Scholar] [CrossRef]
- de Lecea, L.; Kilduff, T.S.; Peyron, C.; Gao, X.B.; Foye, P.E.; Danielson, P.E.; Fukuhara, C.; Battenberg, E.L.F.; Gautvik, V.T.; Bartlett, F.S.; et al. The hypocretins: Hypothalamus-specific peptides with neuroexcitatory activity. Proc. Natl. Acad. Sci. USA 1998, 95, 322–327. [Google Scholar] [CrossRef]
- Villano, I.; La Marra, M.; Di Maio, G.; Monda, V.; Chieffi, S.; Guatteo, E.; Messina, G.; Moscatelli, F.; Monda, M.; Messina, A. Physiological Role of Orexinergic System for Health. Int. J. Environ. Res. Public Health 2022, 19, 8353. [Google Scholar] [CrossRef]
- Mogavero, M.P.; Godos, J.; Grosso, G.; Caraci, F.; Ferri, R. Rethinking the Role of Orexin in the Regulation of REM Sleep and Appetite. Nutrients 2023, 15, 3679. [Google Scholar] [CrossRef]
- Pizza, F.; Barateau, L.; Dauvilliers, Y.; Plazzi, G. The orexin story, sleep and sleep disturbances. J. Sleep Res. 2022, 31, 13665. [Google Scholar] [CrossRef]
- Kakizaki, M.; Tsuneoka, Y.; Takase, K.; Kim, S.J.; Choi, J.; Ikkyu, A.; Abe, M.; Sakimura, K.; Yanagisawa, M.; Funato, H. Differential Roles of Each Orexin Receptor Signaling in Obesity. iScience 2019, 20, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kuang, H.; Zhu, Y.G.; Zhou, Z.F.; Yang, M.W.; Hong, F.F.; Yang, S.L. Sleep disorders in Alzheimer’s disease: The predictive roles and potential mechanisms. Neural Regen. Res. 2021, 16, 1965–1972. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Behl, T.; Sehgal, A.; Singh, S.; Sharma, N.; Bhatia, S.; Al-Harassi, A.; Abdel-Daim, M.M.; Bungau, S. Exploring the Role of Orexinergic Neurons in Parkinson’s Disease. Neurotox. Res. 2021, 39, 2141–2153. [Google Scholar] [CrossRef] [PubMed]
- Alster, P.; Madetko-Alster, N.; Otto-Ślusarczyk, D.; Migda, A.; Migda, B.; Struga, M.; Friedman, A. Role of orexin in pathogenesis of neurodegenerative parkinsonisms. Neurol. Neurochir. Pol. 2023, 57, 335–343. [Google Scholar] [CrossRef]
- Pavlova, M.K.; Latreille, V. Sleep Disorders. Am. J. Med. 2019, 132, 292–299. [Google Scholar] [CrossRef]
- Nicholson, W.C.; Pfeiffer, K. Sleep Disorders and Mood, Anxiety, and Post-Traumatic Stress Disorders: Overview of Clinical Treatments in the Context of Sleep Disturbances. Nurs. Clin. N. Am. 2021, 56, 229–247. [Google Scholar] [CrossRef]
- Dey, S.; Sun, E.; Frishman, W.H.; Aronow, W.S. Sleep Disorders and Coronary Artery Disease. Cardiol. Rev. 2023, 31, 219–224. [Google Scholar] [CrossRef]
- Zhou, W.; Sun, L.; Zeng, L.; Wan, L. Mediation of the association between sleep disorders and cardiovascular disease by depressive symptoms: An analysis of the National health and Nutrition Examination Survey (NHANES) 2017–2020. Prev. Med. Rep. 2023, 33, 102183. [Google Scholar] [CrossRef]
- Sánchez-de-la-Torre, M.; Barbé, F. Sleep disorders and cardiovascular disease. Med. Clin. 2022, 158, 73–75. [Google Scholar] [CrossRef]
- Kadier, K.; Dilixiati, D.; Ainiwaer, A.; Liu, X.; Lu, J.; Liu, P.; Ainiwan, M.; Yesitayi, G.; Ma, X.; Ma, Y. Analysis of the relationship between sleep-related disorder and systemic immune-inflammation index in the US population. BMC Psychiatry 2023, 23, 773. [Google Scholar] [CrossRef]
- Benz, R.L.; Pressman, M.R.; Hovick, E.T.; Peterson, D.D. Potential novel predictors of mortality in end-stage renal disease patients with sleep disorders. Am. J. Kidney Dis. 2000, 35, 1052–1060. [Google Scholar] [CrossRef] [PubMed]
- Braley, T.J.; Shieu, M.M.; Zaheed, A.B.; Dunietz, G.L. Pathways between multiple sclerosis, sleep disorders, and cognitive function: Longitudinal findings from The Nurses’ Health Study. Mult. Scler. 2023, 29, 436–446. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Baek, J.H.; Shin, D.H.J.; Park, H.M.; Lee, Y.B.; Park, K.H.; Shin, D.H.J.; Noh, Y.; Sung, Y.H. Correlation of sleep disturbance and cognitive impairment in patients with Parkinson’s disease. J. Mov. Disord. 2014, 7, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Soyka, M.; Wild, I.; Caulet, B.; Leontiou, C.; Lugoboni, F.; Hajak, G. Long-term use of benzodiazepines in chronic insomnia: A European perspective. Front. Psychiatry 2023, 14, 1–12. [Google Scholar] [CrossRef]
- Zaami, S.; Graziano, S.; Tittarelli, R.; Beck, R.; Marinelli, E. BDZs, Designer BDZs and Z-drugs: Pharmacology and Misuse Insights. Curr. Pharm. Des. 2022, 28, 1221–1229. [Google Scholar] [CrossRef]
- Azevedo, K.; Johnson, M.; Wassermann, M.; Evans-Wall, J. Drugs of Abuse—Opioids, Sedatives, Hypnotics. Crit. Care Clin. 2021, 37, 501–516. [Google Scholar] [CrossRef]
- Scott, M.M.; Marcus, J.N.; Pettersen, A.; Birnbaum, S.G.; Mochizuki, T.; Scammell, T.E.; Nestler, E.J.; Elmquist, J.K.; Lutter, M. Hcrtr1 and 2 signaling differentially regulates depression-like behaviors. Behav. Brain Res. 2011, 222, 289–294. [Google Scholar] [CrossRef]
- Mohammadkhani, A.; Fragale, J.E.; Pantazis, C.B.; Bowrey, H.E.; James, M.H.; Aston-Jones, G. Orexin-1 Receptor Signaling in Ventral Pallidum Regulates Motivation for the Opioid Remifentanil. J. Neurosci. 2019, 39, 9831–9840. [Google Scholar] [CrossRef]
- Wiskerke, J.; James, M.H.; Aston-Jones, G. The orexin-1 receptor antagonist SB-334867 reduces motivation, but not inhibitory control, in a rat stop signal task. Brain Res. 2020, 1731, 146222. [Google Scholar] [CrossRef]
- Markham, A. Daridorexant: First Approval. Drugs 2022, 82, 601–607. [Google Scholar] [CrossRef]
- Ziemichód, W.; Grabowska, K.; Kurowska, A.; Biała, G. A Comprehensive Review of Daridorexant, a Dual-Orexin Receptor Antagonist as New Approach for the Treatment of Insomnia. Molecules 2022, 27, 6041. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.J. Lemborexant: First Approval. Drugs 2020, 80, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Waters, K. Review of the Efficacy and Safety of Lemborexant, a Dual Receptor Orexin Antagonist (DORA), in the Treatment of Adults With Insomnia Disorder. Ann. Pharmacother. 2022, 56, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.P.H. Suvorexant: First global approval. Drugs 2014, 74, 1817–1822. [Google Scholar] [CrossRef]
- Han, A.H.; Burroughs, C.R.; Falgoust, E.P.; Hasoon, J.; Hunt, G.; Kakazu, J.; Lee, T.; Kaye, A.M.; Kaye, A.D.; Ganti, L. Suvorexant, a Novel Dual Orexin Receptor Antagonist, for the Management of Insomnia. Health Psychol. Res. 2023, 10, 67898. [Google Scholar] [CrossRef]
- Ditchfield, R.; Hehre, W.J.; Pople, J.A. Self-Consistent Molecular-Orbital Methods. IX. An Extended Gaussian-Type Basis for Molecular-Orbital Studies of Organic Molecules. J. Chem. Phys. 1971, 54, 724–728. [Google Scholar] [CrossRef]
- Karhu, L.; Magarkar, A.; Bunker, A.; Xhaard, H. Determinants of Orexin Receptor Binding and Activation - A Molecular Dynamics Study. J. Phys. Chem. B 2019, 123, 2609–2622. [Google Scholar] [CrossRef]
- Waltenspühl, Y.; Ehrenmann, J.; Vacca, S.; Thom, C.; Medalia, O.; Plückthun, A. Structural basis for the activation and ligand recognition of the human oxytocin receptor. Nat. Commun. 2022, 13, 4153. [Google Scholar] [CrossRef]
- Koehl, A.; Hu, H.; Maeda, S.; Zhang, Y.; Qu, Q.; Paggi, J.M.; Latorraca, N.R.; Hilger, D.; Dawson, R.; Matile, H.; et al. Structure of the μ-opioid receptor–Gi protein complex. Nature 2018, 558, 547–552. [Google Scholar] [CrossRef]
- Asada, H.; Im, D.; Hotta, Y.; Yasuda, S.; Murata, T.; Suno, R.; Iwata, S. Molecular basis for anti-insomnia drug design from structure of lemborexant-bound orexin 2 receptor. Structure 2022, 30, 1582–1589. [Google Scholar] [CrossRef]
- Rappas, M.; Ali, A.A.E.; Bennett, K.A.; Brown, J.D.; Bucknell, S.J.; Congreve, M.; Cooke, R.M.; Cseke, G.; de Graaf, C.; Doré, A.S.; et al. Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis. J. Med. Chem. 2020, 63, 1528–1543. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Babaoglu, K.; Brautigam, C.A.; Clark, L.; Shao, Z.; Scheuermann, T.H.; Harrell, C.M.; Gotter, A.L.; Roecker, A.J.; Winrow, C.J.; et al. Structure and ligand-binding mechanism of the human OX1 and OX2 orexin receptors. Nat. Struct. Mol. Biol. 2016, 23, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Roch, C.; Bergamini, G.; Steiner, M.A.; Clozel, M. Nonclinical pharmacology of daridorexant: A new dual orexin receptor antagonist for the treatment of insomnia. Psychopharmacology 2021, 238, 2693–2708. [Google Scholar] [CrossRef] [PubMed]
- Treiber, A.; de Kanter, R.; Roch, C.; Gatfield, J.; Boss, C.; von Raumer, M.; Schindelholz, B.; Muehlan, C.; van Gerven, J.; Jenck, F. The Use of Physiology-Based Pharmacokinetic and Pharmacodynamic Modeling in the Discovery of the Dual Orexin Receptor Antagonist ACT-541468. J. Pharmacol. Exp. Ther. 2017, 362, 489–503. [Google Scholar] [CrossRef]
- Callander, G.E.; Olorunda, M.; Monna, D.; Schuepbach, E.; Langenegger, D.; Betschart, C.; Hintermann, S.; Behnke, D.; Cotesta, S.; Fendt, M.; et al. Kinetic properties of “dual” orexin receptor antagonists at OX1R and OX2R orexin receptors. Front. Neurosci. 2013, 7, 65900. [Google Scholar] [CrossRef]
- Winrow, C.J.; Gotter, A.L.; Cox, C.D.; Doran, S.M.; Tannenbaum, P.L.; Breslin, M.J.; Garson, S.L.; Fox, S.V.; Harrell, C.M.; Stevens, J.; et al. Promotion of sleep by suvorexant-a novel dual orexin receptor antagonist. J. Neurogenet. 2011, 25, 52–61. [Google Scholar] [CrossRef]
- Beuckmann, C.T.; Suzuki, M.; Ueno, T.; Nagaoka, K.; Arai, T.; Higashiyama, H. In Vitro and In Silico Characterization of Lemborexant (E2006), a Novel Dual Orexin Receptor Antagonist. J. Pharmacol. Exp. Ther. 2017, 362, 287–295. [Google Scholar] [CrossRef]
- Celik, S. DFT investigations and molecular docking as potent inhibitors of SARS-CoV-2 main protease of 4-phenylpyrimidine. J. Mol. Struct. 2023, 1277, 134895. [Google Scholar] [CrossRef]
- Salih, R.H.H.; Hasan, A.H.; Hussein, A.J.; Samad, M.K.; Shakya, S.; Jamalis, J.; Hawaiz, F.E.; Pratama, M.R.F. One-pot synthesis, molecular docking, ADMET, and DFT studies of novel pyrazolines as promising SARS-CoV-2 main protease inhibitors. Res. Chem. Intermed. 2022, 48, 4729–4751. [Google Scholar] [CrossRef]
- Mumit, M.A.; Pal, T.K.; Alam, M.A.; Islam, M.A.; Paul, S.; Sheikh, M.C. DFT studies on vibrational and electronic spectra, HOMO–LUMO, MEP, HOMA, NBO and molecular docking analysis of benzyl-3-N-(2,4,5-trimethoxyphenylmethylene)hydrazinecarbodithioate. J. Mol. Struct. 2020, 1220, 128715. [Google Scholar] [CrossRef]
- Tayyeb, J.Z.; Mondal, S.; Rahman, M.A.; Kumar, S.; Bayıl, I.; Akash, S.; Hossain, M.S.; Alqahtani, T.; Zaki, M.E.A.; Oliveira, J.I.N. Identification of Helicobacter pylori-carcinogenic TNF-alpha-inducing protein inhibitors via daidzein derivatives through computational approaches. J. Cell. Mol. Med. 2024, 28, e18358. [Google Scholar] [CrossRef] [PubMed]
- Shahab, M.; Morais, G.C.F.; Akash, S.; Fulco, U.L.; Oliveira, J.I.N.; Zheng, G.; Akter, S. A robust computational quest: Discovering potential hits to improve the treatment of pyrazinamide-resistant Mycobacterium tuberculosis. J. Cell. Mol. Med. 2024, 28, e18279. [Google Scholar] [CrossRef] [PubMed]
- Thanikaivelan, P.; Subramanian, V.; Rao, J.R.; Nair, B.U. Application of quantum chemical descriptor in quantitative structure activity and structure property relationship. Chem. Phys. Lett. 2000, 323, 59–70. [Google Scholar] [CrossRef]
- Eswar, N.; John, B.; Mirkovic, N.; Fiser, A.; Ilyin, V.A.; Pieper, U.; Stuart, A.C.; Marti-Renom, M.A.; Madhusudhan, M.S.; Yerkovich, B.; et al. Tools for comparative protein structure modeling and analysis. Nucleic Acids Res. 2003, 31, 3375–3380. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Honorato, R.V.; Trellet, M.E.; Jiménez-García, B.; Schaarschmidt, J.J.; Giulini, M.; Reys, V.; Koukos, P.I.; Rodrigues, J.P.G.L.M.; Karaca, E.; van Zundert, G.C.P.; et al. The HADDOCK2.4 web server for integrative modeling of biomolecular complexes. Nat. Protoc. 2024, 19, 3219–3241. [Google Scholar] [CrossRef]
- Honorato, R.V.; Koukos, P.I.; Jiménez-García, B.; Tsaregorodtsev, A.; Verlato, M.; Giachetti, A.; Rosato, A.; Bonvin, A.M.J.J. Structural Biology in the Clouds: The WeNMR-EOSC Ecosystem. Front. Mol. Biosci. 2021, 8, 729513. [Google Scholar] [CrossRef]
- van Zundert, G.C.P.; Rodrigues, J.P.G.L.M.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A.M.J.J. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Gutiérrez, I.S.; Lin, F.Y.; Vanommeslaeghe, K.; Lemkul, J.A.; Armacost, K.A.; Brooks, C.L.; MacKerell, A.D. Parametrization of halogen bonds in the CHARMM general force field: Improved treatment of ligand–protein interactions. Bioorg. Med. Chem. 2016, 24, 4812–4825. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; MacKerell, A.D. Automation of the CHARMM General Force Field (CGenFF) I: Bond Perception and Atom Typing. J. Chem. Inf. Model. 2012, 52, 3144–3154. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Choi, Y.K.; Kim, S.; Lee, J.; Im, W. CHARMM-GUI Membrane Builder for Lipid Nanoparticles with Ionizable Cationic Lipids and PEGylated Lipids. J. Chem. Inf. Model. 2021, 61, 5192–5202. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Patel, D.S.; Ståhle, J.; Park, S.J.; Kern, N.R.; Kim, S.; Lee, J.; Cheng, X.; Valvano, M.A.; Holst, O.; et al. CHARMM-GUI Membrane Builder for Complex Biological Membrane Simulations with Glycolipids and Lipoglycans. J. Chem. Theory Comput. 2019, 15, 775–786. [Google Scholar] [CrossRef] [PubMed]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Dávila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- van Gunsteren, W.F.; Berendsen, H.J.C. A Leap-frog Algorithm for Stochastic Dynamics. Mol. Simul. 1988, 1, 173–185. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An Nlog(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1998, 18, 1463–1472. [Google Scholar] [CrossRef]
- Meza, J.C. Steepest descent. WIREs Comp. Stat. 2010, 2, 719–722. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Nosé, S.; Klein, M.L. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1983, 50, 1055–1076. [Google Scholar] [CrossRef]
- Gómez-Jeria, J.S.; Robles-Navarro, A.; Kpotin, G.; Gómez-Jeria, J.S.; Kpotin, G.A.; Garrido-Sáez, N.; Gatica-Díaz, N. Some remarks about the relationships between the common skeleton concept within the Klopman-Peradejordi-Gómez QSAR method and the weak molecule-site interactions. Chem. Res. J. 2020, 2, 32–52. [Google Scholar] [CrossRef]
- Zhang, D.W.; Zhang, J.Z.H. Molecular fractionation with conjugate caps for full quantum mechanical calculation of protein–molecule interaction energy. J. Chem. Phys. 2003, 119, 3599–3605. [Google Scholar] [CrossRef]
- Albuquerque, A.C.C.; Bezerra, K.S.; Vianna, J.F.; Batista, S.O.; Lima Neto, J.X.; Campos, D.M.O.; Oliveira, J.I.N.; Galvão, D.S.; Fulco, U.L. In Silico Evaluation of the Binding Energies of Androgen Receptor Agonists in Wild-Type and Mutational Models. J. Phys. Chem. B 2023, 127, 5005–5017. [Google Scholar] [CrossRef]
- Santos, J.L.S.; Bezerra, K.S.; Barbosa, E.D.; Pereira, A.C.L.; Meurer, Y.S.R.; Oliveira, J.I.N.; Gavioli, E.C.; Fulco, U.L. In silico analysis of energy interactions between nociceptin/orfanin FQ receptor and two antagonists with potential antidepressive action. New J. Chem. 2022, 7950–7959, 2022. [Google Scholar] [CrossRef]
- Barbosa, E.D.; Lima Neto, J.X.; Bezerra, K.S.; Oliveira, J.I.N.; Machado, L.D.; Fulco, U.L. Quantum Biochemical Investigation of Lys49-PLA 2 from Bothrops moojeni. J. Phys. Chem. B 2022, 125, 12972–12980. [Google Scholar] [CrossRef]
- Campos, D.M.O.; Bezerra, K.S.; Esmaile, S.C.; Fulco, U.L.; Albuquerque, E.L.; Oliveira, J.I.N. Intermolecular interactions of cn-716 and acyl-KR-aldehyde dipeptide inhibitors against Zika virus. Phys. Chem. Chem. Phys. 2020, 22, 15683–15695. [Google Scholar] [CrossRef]
- Bezerra, K.S.; Vianna, J.F.; Lima Neto, J.X.; Oliveira, J.I.N.; Albuquerque, E.L.; Fulco, U.L. Interaction energies between two antiandrogenic and one androgenic agonist receptor in the presence of a T877A mutation in prostate cancer: A quantum chemistry analysis. New J. Chem. 2020, 44, 5903–5912. [Google Scholar] [CrossRef]
- de Sousa, B.G.; Oliveira, J.I.N.; Albuquerque, E.L.; Fulco, U.L.; Amaro, V.E.; Blaha, C.A.G. Molecular modelling and quantum biochemistry computations of a naturally occurring bioremediation enzyme: Alkane hydroxylase from Pseudomonas putida P1. J. Mol. Graph. Model. 2017, 77, 232–239. [Google Scholar] [CrossRef]
- Li, A.; Muddana, H.S.; Gilson, M.K. Quantum mechanical calculation of noncovalent interactions: A large-scale evaluation of PMx, DFT, and SAPT approaches. J. Chem. Theory Comput. 2014, 10, 1563–1575. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16 Revision C.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Grimme, S. Semiempirical GGA-type density functional constructed with a long-range dispersion correction. J. Comput. Chem. 2006, 27, 1787–1799. [Google Scholar] [CrossRef]
- Cossi, M.; Rega, N.; Scalmani, G.; Barone, V. Energies, structures, and electronic properties of molecules in solution with the C–PCM solvation model. J. Comput. Chem. 2003, 24, 669–681. [Google Scholar] [CrossRef]
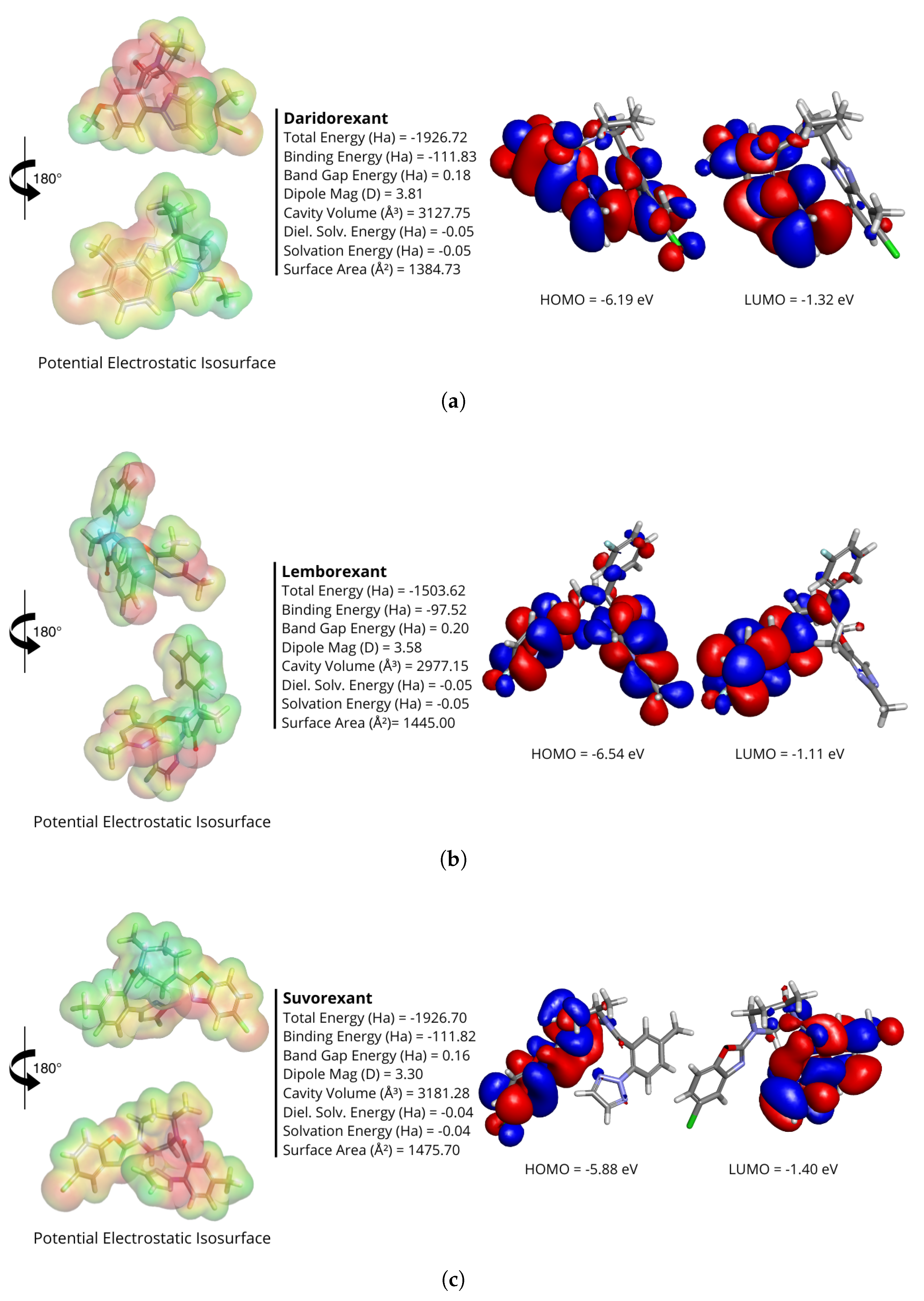
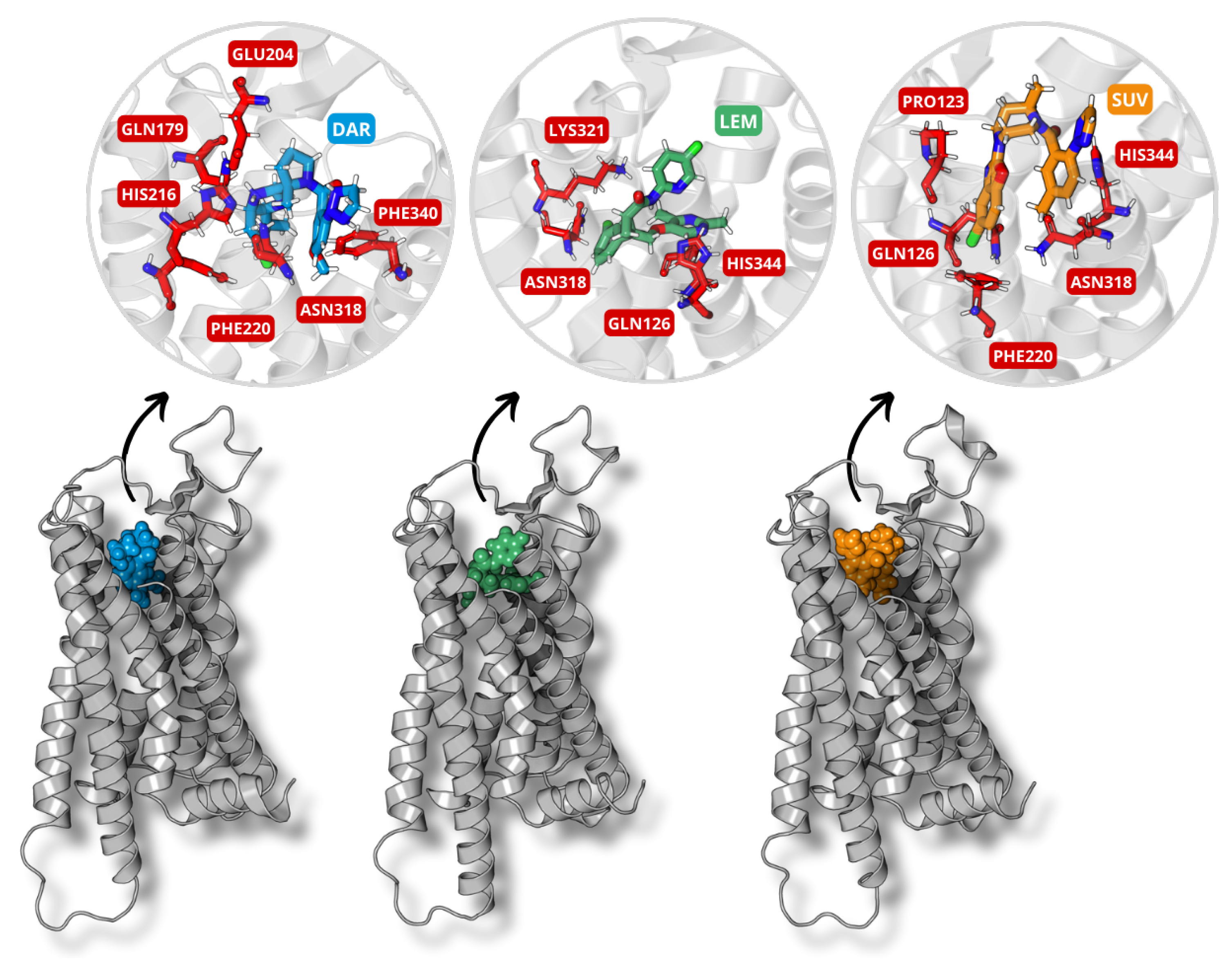
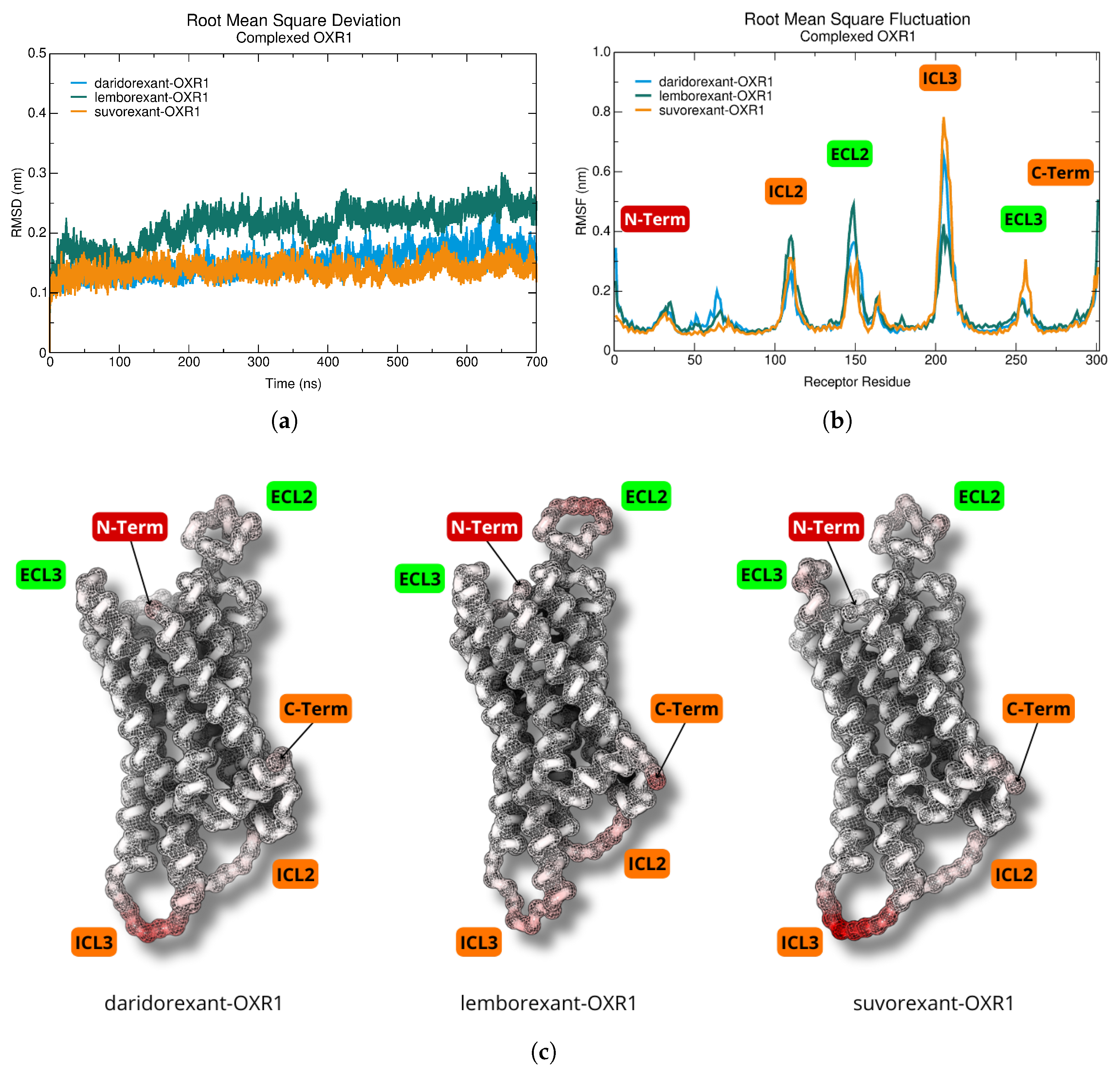
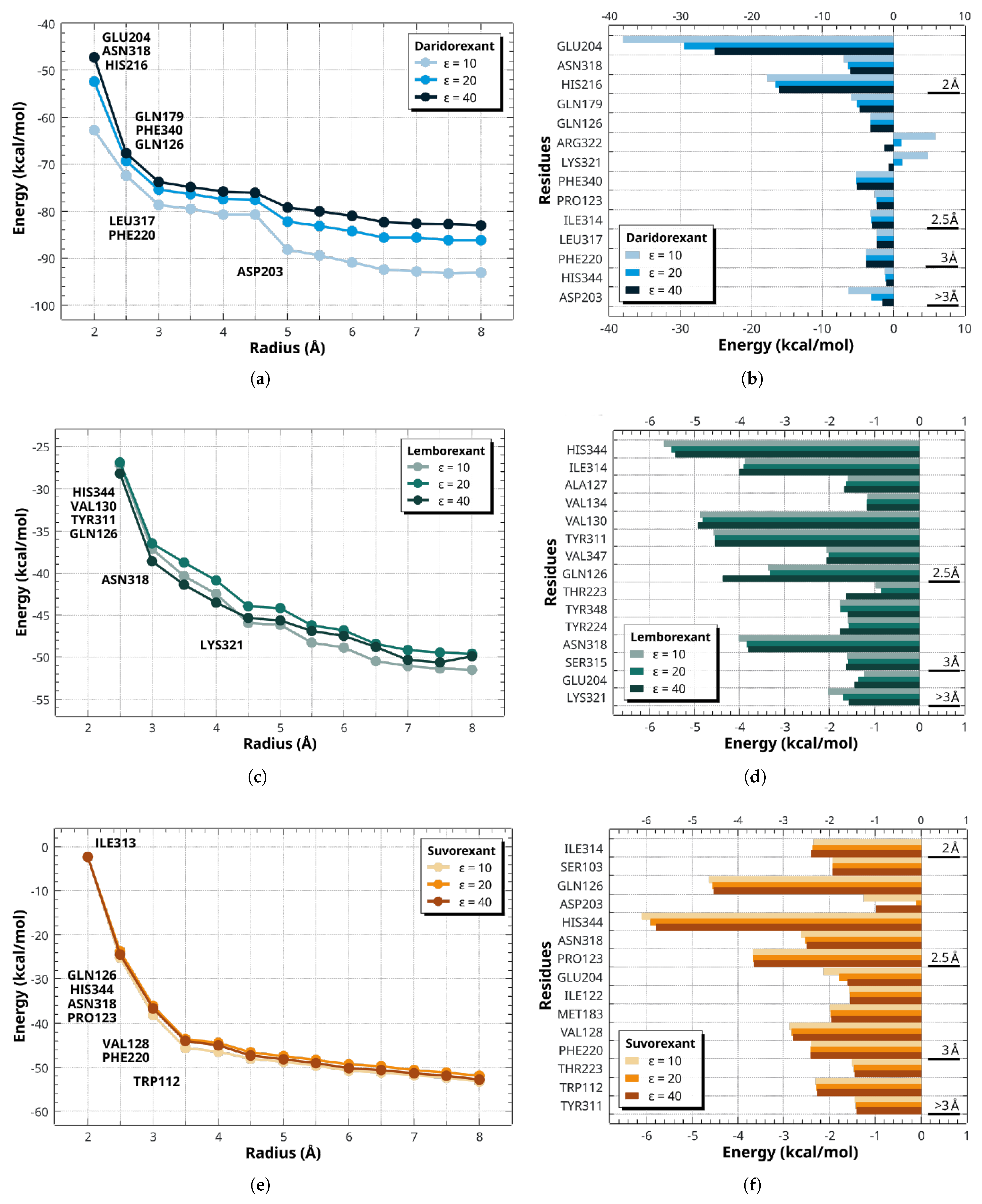
| DORA | GAP | I | A | |||||
|---|---|---|---|---|---|---|---|---|
| Daridorexant | 4.86721 | 6.19316 | 1.32595 | 2.43360 | 0.41091 | 3.75956 | −3.75956 | 17.19860 |
| Lemborexant | 5.42455 | 6.53966 | 1.11510 | 2.71228 | 0.36869 | 3.82738 | −3.82738 | 19.84586 |
| Suvorexant | 4.47390 | 5.87751 | 1.40361 | 2.23695 | 0.44704 | 3.64056 | −3.64056 | 14.82392 |
| DORA | Cluster Size | HS | RMSD | EVDW | EELEC | Z-Score |
|---|---|---|---|---|---|---|
| Daridorexant | 199 | −44.8 ± 0.9 | 0.2 ± 0.1 | −25.2 ± 4.0 | −59.6 ± 16.7 | 0.0 |
| Lemborexant | 200 | −39.9 ± 1.0 | 0.3 ± 0.2 | −23.2 ± 2.3 | −33.1 ± 15.6 | 0.0 |
| Suvorexant | 200 | −43.7 ± 1.7 | 0.3 ± 0.2 | −25.2 ± 0.7 | −53.0 ± 10.3 | 0.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sena, C.; Albuquerque, P.; Oliveira, J.; Vieira, D. Molecular Insights into the Interaction of Orexin 1 Receptor Antagonists: A Comprehensive Study Using Classical and Quantum Computational Methods. Molecules 2025, 30, 2790. https://doi.org/10.3390/molecules30132790
Sena C, Albuquerque P, Oliveira J, Vieira D. Molecular Insights into the Interaction of Orexin 1 Receptor Antagonists: A Comprehensive Study Using Classical and Quantum Computational Methods. Molecules. 2025; 30(13):2790. https://doi.org/10.3390/molecules30132790
Chicago/Turabian StyleSena, Caio, Pedro Albuquerque, Jonas Oliveira, and Davi Vieira. 2025. "Molecular Insights into the Interaction of Orexin 1 Receptor Antagonists: A Comprehensive Study Using Classical and Quantum Computational Methods" Molecules 30, no. 13: 2790. https://doi.org/10.3390/molecules30132790
APA StyleSena, C., Albuquerque, P., Oliveira, J., & Vieira, D. (2025). Molecular Insights into the Interaction of Orexin 1 Receptor Antagonists: A Comprehensive Study Using Classical and Quantum Computational Methods. Molecules, 30(13), 2790. https://doi.org/10.3390/molecules30132790






