Cucurbitane Glycosides and Their Potential Anti-Inflammatory Activities from Hemsleya chinensis Tubers
Abstract
1. Introduction
2. Results and Discussion
2.1. Structural Elucidation of Isolated Compounds
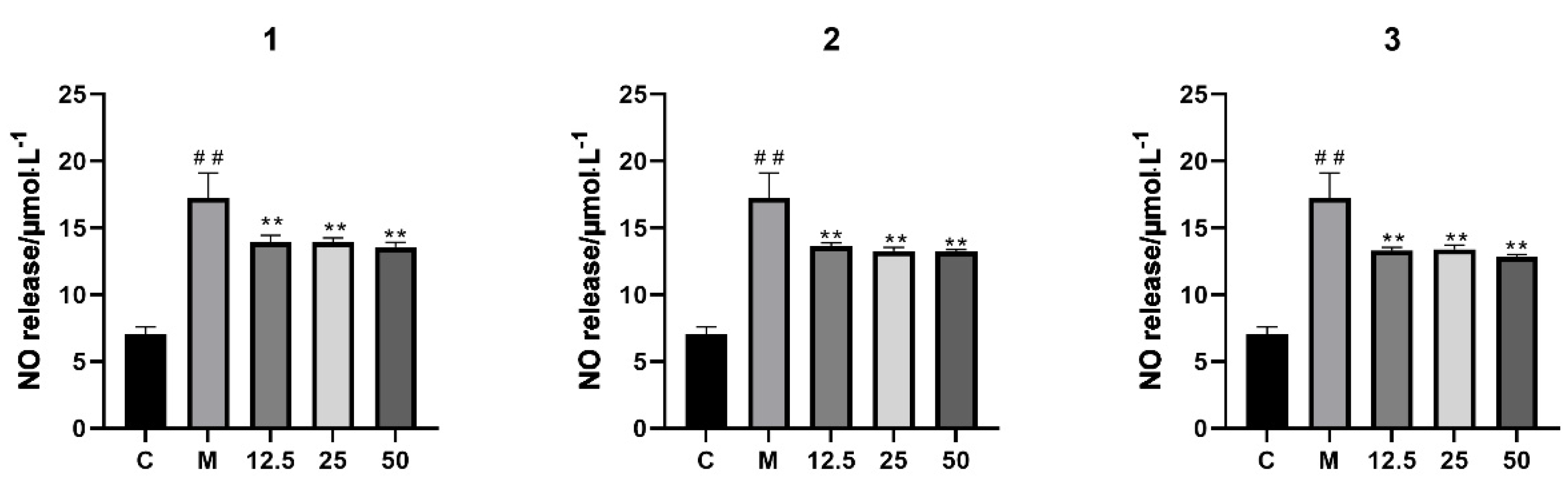
2.2. Anti-Inflammatory Effects of Compounds 1–3
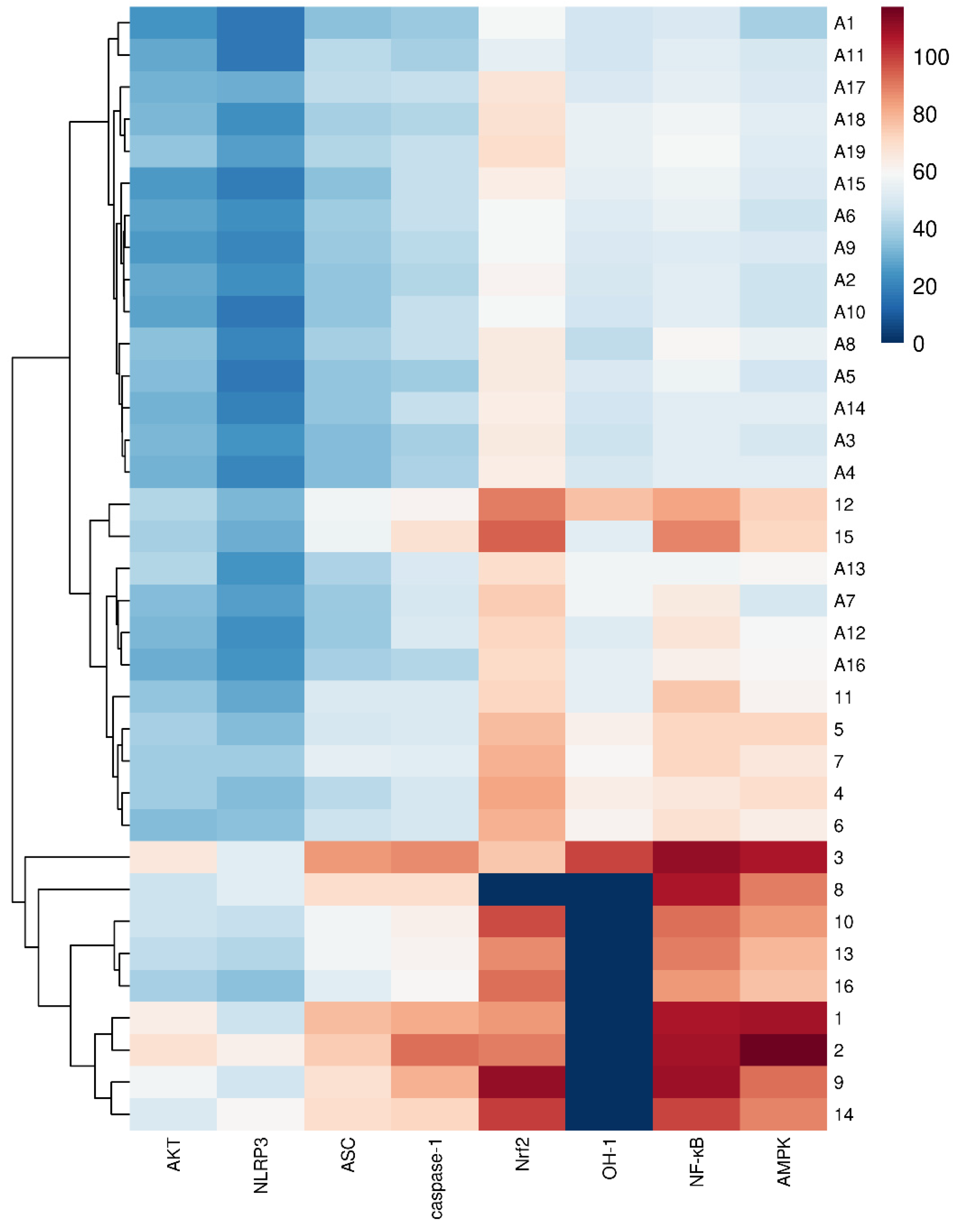
2.3. Molecular Docking
3. Materials and Methods
3.1. Materials and Reagents
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Spectral Data
3.5. Acid Hydrolysis for Compounds 1–3
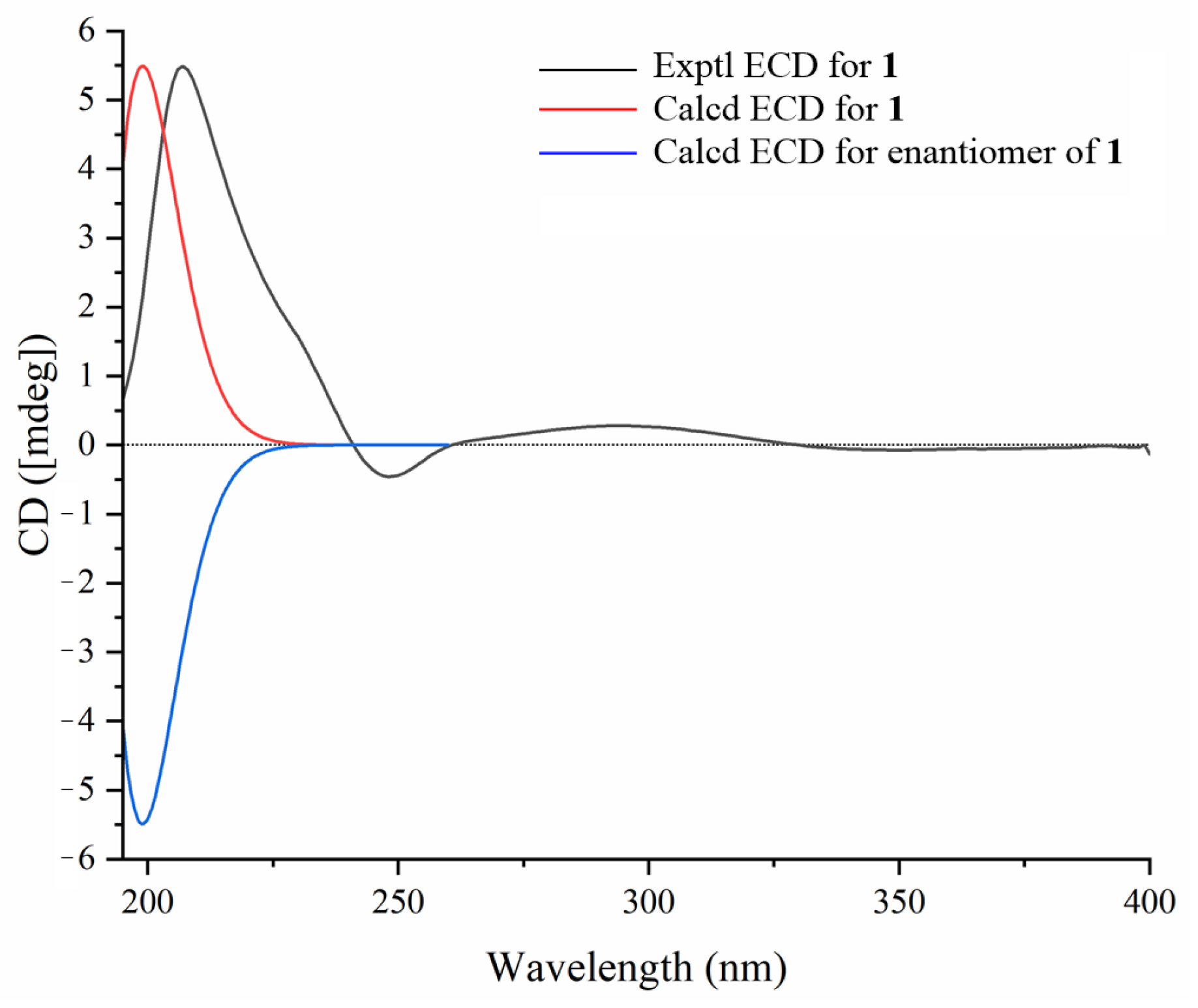
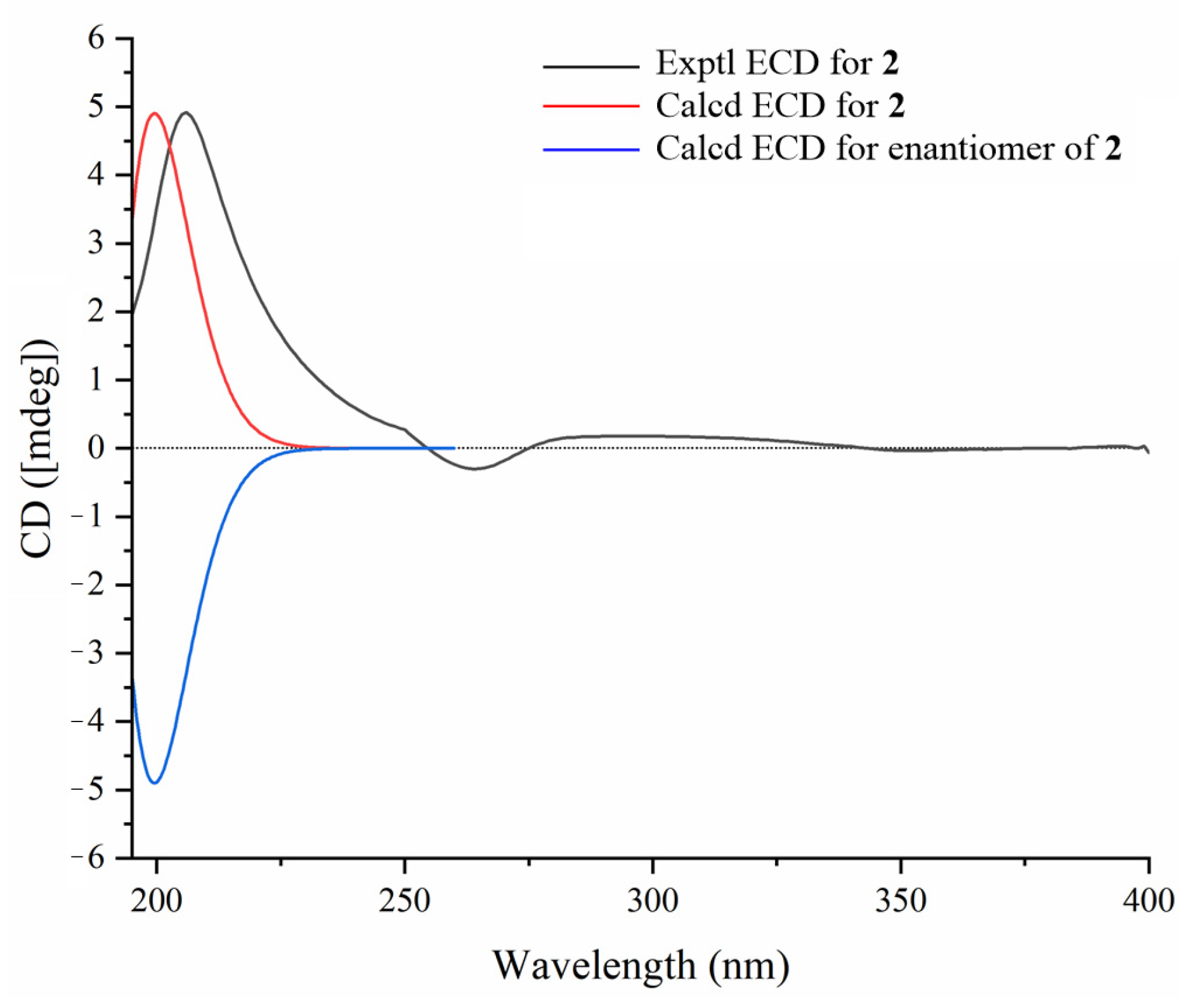
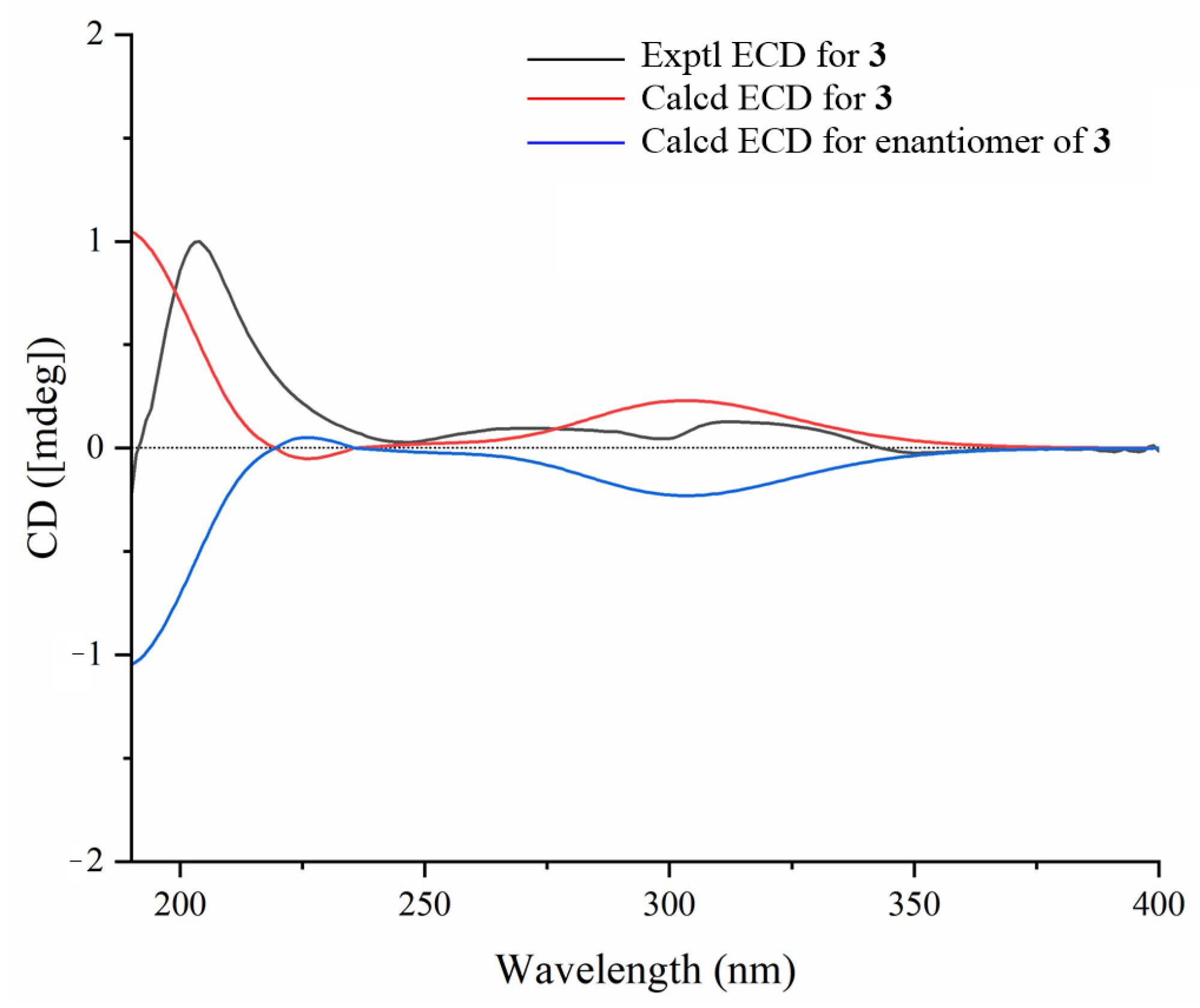
3.6. ECD Calculations of Compounds 1a–3a
3.7. Anti-Inflammatory Bioassays
3.7.1. Cell Culture and Cell Viability Assay
3.7.2. LPS-Induced NO Content in RAW 264.7 Cells
3.8. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- China Flora Editorial Board (Chinese Academy of Sciences). Flora of China; Science Press: Beijing, China, 1986; Volume 73, p. 122. [Google Scholar]
- Dong, J.Y.; Yue, L.; Shao, W.W.; Xu, J.Z. Chemical components in Hemsleya chensnsis (III). China J. Chin. Mater. Med. 2012, 37, 814–817. [Google Scholar]
- Li, X.S.; Wang, Q.L.; Xu, Z.P.; Liu, M.S.; Liang, X.Y.; Zheng Jia, C.; Deng, H.Y.; Liu, L.i.; Huang, Y.M.; Yang, M.X.; et al. Structurally diverse cucurbitane-type triterpenoids from the tubers of Hemsleya chinensis with cytotoxic activity. Phytochemistry 2024, 220, 114033. [Google Scholar] [CrossRef] [PubMed]
- Guizhou Medical Products Administration. Standards for Traditional Chinese Medicine Ethnic Medicinal Materials in Guizhou Province, 2019th ed.; Chinese Medical Science Press: Beijing, China, 2019; p. 256. [Google Scholar]
- Hubei Medical Products Administration. Quality Standards for Traditional Chinese Medicine in Hubei Province, 2018th ed.; Chinese Medical Science Press: Beijing, China, 2018; p. 203. [Google Scholar]
- Zhang, Y.; Guo, H.; Lian, F.H.; Xiang, X.; Li, Q.X.; Liu, X.Q.; Wang, Z.M.; Dai, L.P.; Xu, E.P. Protective effect of ethanolic extract of Hemsleya chinensis on HCl/ethanol-induced acute gastric ulcer in rats based on p38MAPK/NF-kB signaling pathway. Chin. J. Exp. Tradit. Med. Formulae 2023, 29, 37–44. [Google Scholar]
- Haq, F.U.; Ali, A.; Khan, M.N.; Shah, S.M.Z.; Kandel, R.C.; Aziz, N.; Adhikari, A.; Choudhary, M.; Atta-Ur-Rahman; El-Seedi, H.R.; et al. Metabolite profiling and quantitation of cucurbitacins in cucurbitaceae plants by liquid chromatography coupled to tandem mass spectrometry. Sci. Rep. 2019, 9, 15992–16002. [Google Scholar] [CrossRef]
- Chen, J.C.; Chiu, H.M.; Nie, L.R.; Geoffrey, A.C.; Samuel, X.Q. Cucurbitacins and cucurbitane glycosides: Structures and biological activities. Nat. Prod. Rep. 2005, 22, 386–399. [Google Scholar] [CrossRef]
- Yu, K.; Liu, A.G.; Liu, B.; Yao, Q.Q. Progress on cucurbitacin IIa and its derivatives. Food Drug. 2019, 21, 161–165. [Google Scholar]
- Lian, F.H.; Chi, J.; Meng, Q.L.; Li, Q.X.; Chen, A.Y.; Wang, Z.M.; Dai, L.P. Cucurbitane triterpenes from Hemsleya chinensis tubers and their anti-inflammatory activities. Fitoterapia 2023, 166, 105441–105448. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Bai, H.; Zhou, L.; Deng, Z.; Zhong, H.; Wu, Z.; Yao, Q. Three new cucurbitane triterpenoids from Hemsleya penxianensis and their cytotoxic activities. Bioorg. Med. Chem. Lett. 2014, 24, 2159–2162. [Google Scholar] [CrossRef]
- Chen, D.L.; Xu, X.D.; Li, R.T.; Wang, B.W.; Yu, M.; Liu, Y.Y.; Ma, G.X. Five new cucurbitane-type triterpenoid glycosides from the rhizomes of Hemsleya penxianensis with cytotoxic activities. Molecules 2019, 24, 2937. [Google Scholar] [CrossRef]
- Kasai, R.; Matsumoto, K.; Nie, R.L.; Zhou, J.; Tanaka, O. Glycosides from Chinese medicinal plant, Hemsleya panacis-scandens, and structure-taste relationship to cucurbitane glycosides. Chem. Pharm. Bull. 1988, 36, 234–243. [Google Scholar] [CrossRef]
- Wei, H.; Liu, Y.H.; Tian, Y.G.; Xiang, F.F. Cucurbitane-type triterpenoids from the tubers of Hemsleya dolichocarpa. Acta Pharm. Sin. 2018, 53, 1526–1531. [Google Scholar]
- Kasai, R.; Matsumoto, K.; Nie, R.L.; Morita, T.; Awazu, A.; Zhou, J.; Tanaka, O. Sweet and bitter cucurbitane glycosides from hemsleya carnosiflora. Phytochemistry 1987, 26, 1371–1376. [Google Scholar] [CrossRef]
- Chen, J.C.; Zhou, L.; Wang, Y.H.; Tian, R.R.; Yan, Y.X.; Nian, Y.; Sun, Y.; Zheng, Y.T.; Qiu, M.H. Cucurbitane triterpenoids from Hemsleya penxianensis. Nat. Prod. Bioprospect. 2012, 2, 138–144. [Google Scholar] [CrossRef]
- Li, Y.; Wang, W.X.; Zheng, Z.F.; Mu, Y.L.; Liu, Y.J.; Wang, H.Y.; Li, L.; Yao, Q.Q. Eight new cucurbitane triterpenoids from “Xue Dan”, the roots of Hemsleya pengxianensis. J. Asian Nat. Prod. Res. 2018, 20, 36–48. [Google Scholar] [CrossRef]
- Wang, W.X.; Mu, Y.L.; Li, Y.; Zheng, Z.F.; Chu, H.P.; Zhou, L.; Yao, Q.Q. Three new cucurbitane-type triterpenoid saponins from tubers of Hemsleya pengxianensis var. jinfushanensis. Chin. Trad. Herb. Drugs 2018, 49, 2959–2966. [Google Scholar]
- Li, Z.; Chen, M.; Chen, F.; Li, W.; Huang, G.; Xu, X.; Wang, S.; Ma, G.; Cui, P. Cucurbitane triterpenoid entities derived from Hemsleya penxianensis triggered glioma cell apoptosis via ER stress and MAPK signalling cross-talk. Bioorg. Chem. 2022, 127, 106013. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023–17031. [Google Scholar] [CrossRef]
- Gałgańska, H.; Jarmuszkiewicz, W.; Gałgański, Ł. Carbon dioxide and MAPK signalling: Towards therapy for inflammation. Cell Commun. Signal. 2023, 21, 280–308. [Google Scholar] [CrossRef]
- Manning, B.D.; Toker, A. AKT/PKB signaling: Navigating the network. Cell 2017, 169, 381–405. [Google Scholar] [CrossRef]
- Swanson, K.V.; Deng, M.; Ting, J.P.Y. The NLRP3 inflammasome: Molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 2019, 19, 477–489. [Google Scholar] [CrossRef]
- Li, B.Y.; Wang, Y.X.; Jiang, X.L.; Du, H.W.; Shi, Y.; Xiu, M.H.; Liu, Y.Q.; He, J.Z. Natural products targeting Nrf2/ARE signaling pathway in the treatment of inflammatory bowel disease. Biomed. Pharmacother. 2023, 164, 114950–114968. [Google Scholar] [CrossRef] [PubMed]
- Echizen, K.; Hirose, O.; Maeda, Y.; Oshima, M. Inflammation in gastric cancer: Interplay of the COX-2/prostaglandin E2 and Toll-like receptor/MyD88 pathways. Cancer Sci. 2016, 107, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.E. Non-Smad pathways in TGF-β signaling. Cell Res. 2017, 19, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.L.; Chi, J.; Li, Y.X.; Zhang, W.J.; Zhang, L.X.; Wang, Z.M.; Dai, L.P. Diarylheptanoid glycosides from Dioscorea nipponica rhizomes. Fitoterapia 2024, 177, 106078–106083. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, H.Y.; Chao, L.P.; Qu, L.; Ruan, J.Y.; Liu, Y.X.; Dong, Y.Z.; Han, L.F.; Wang, T. Anti-inflammatory steroids from the rhizomes of Dioscorea septemloba Thunb. Steroids 2016, 112, 95–102. [Google Scholar] [CrossRef]
 ), HMBC (
), HMBC ( ), and NOESY (
), and NOESY ( ) correlations of compound 1. (A) 1H-1H COSY and HMBC; (B) NOESY.
) correlations of compound 1. (A) 1H-1H COSY and HMBC; (B) NOESY.
 ), HMBC (
), HMBC ( ), and NOESY (
), and NOESY ( ) correlations of compound 1. (A) 1H-1H COSY and HMBC; (B) NOESY.
) correlations of compound 1. (A) 1H-1H COSY and HMBC; (B) NOESY.
 ), HMBC (
), HMBC ( ), and NOESY (
), and NOESY ( ) correlations of compound 2. (A) 1H-1H COSY and HMBC; (B) NOESY.
) correlations of compound 2. (A) 1H-1H COSY and HMBC; (B) NOESY.
 ), HMBC (
), HMBC ( ), and NOESY (
), and NOESY ( ) correlations of compound 2. (A) 1H-1H COSY and HMBC; (B) NOESY.
) correlations of compound 2. (A) 1H-1H COSY and HMBC; (B) NOESY.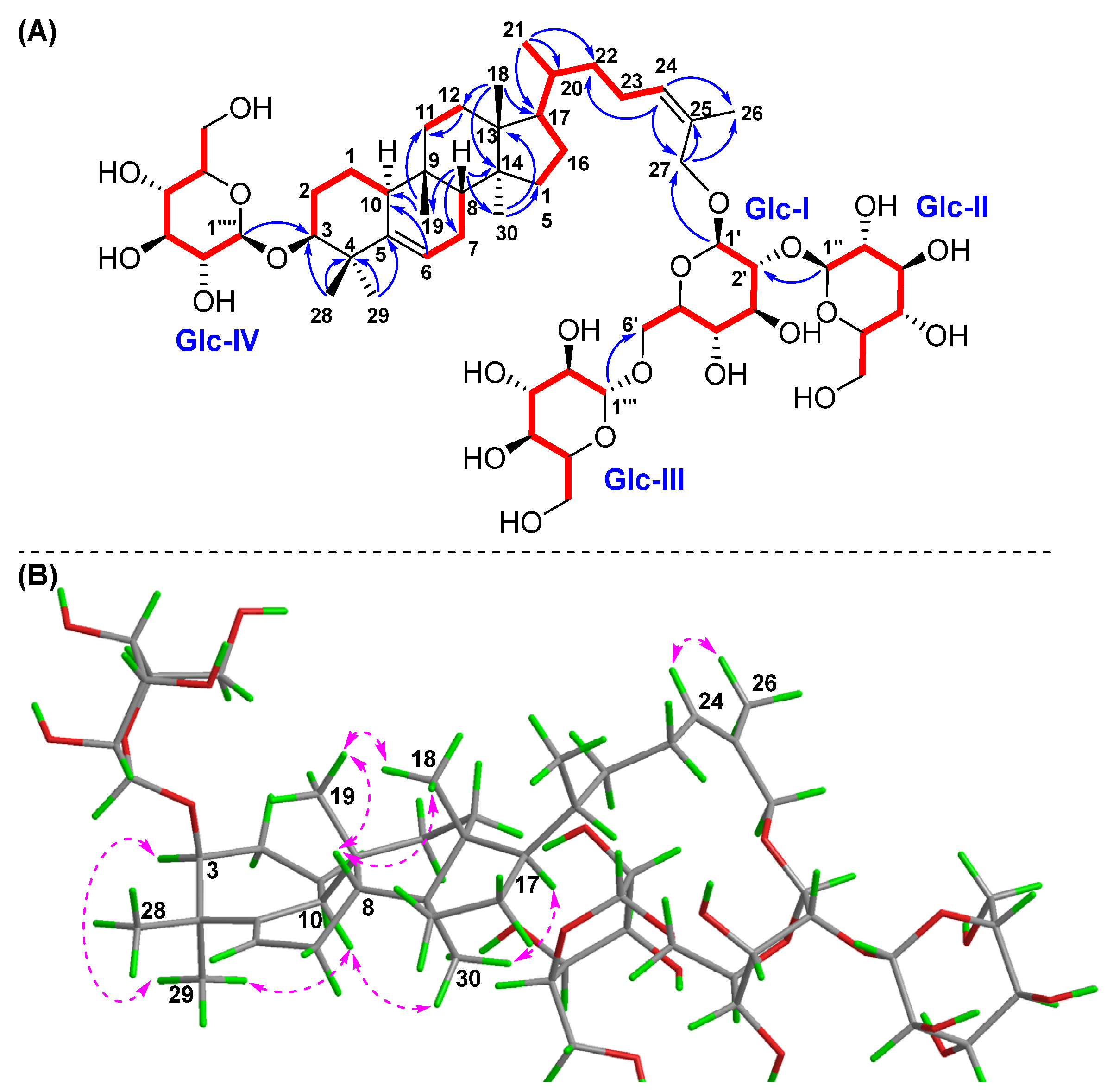
 ), HMBC (
), HMBC ( ), and NOESY (
), and NOESY ( ) correlations of compound 3. (A) 1H-1H COSY and HMBC; (B) NOESY.
) correlations of compound 3. (A) 1H-1H COSY and HMBC; (B) NOESY.
 ), HMBC (
), HMBC ( ), and NOESY (
), and NOESY ( ) correlations of compound 3. (A) 1H-1H COSY and HMBC; (B) NOESY.
) correlations of compound 3. (A) 1H-1H COSY and HMBC; (B) NOESY.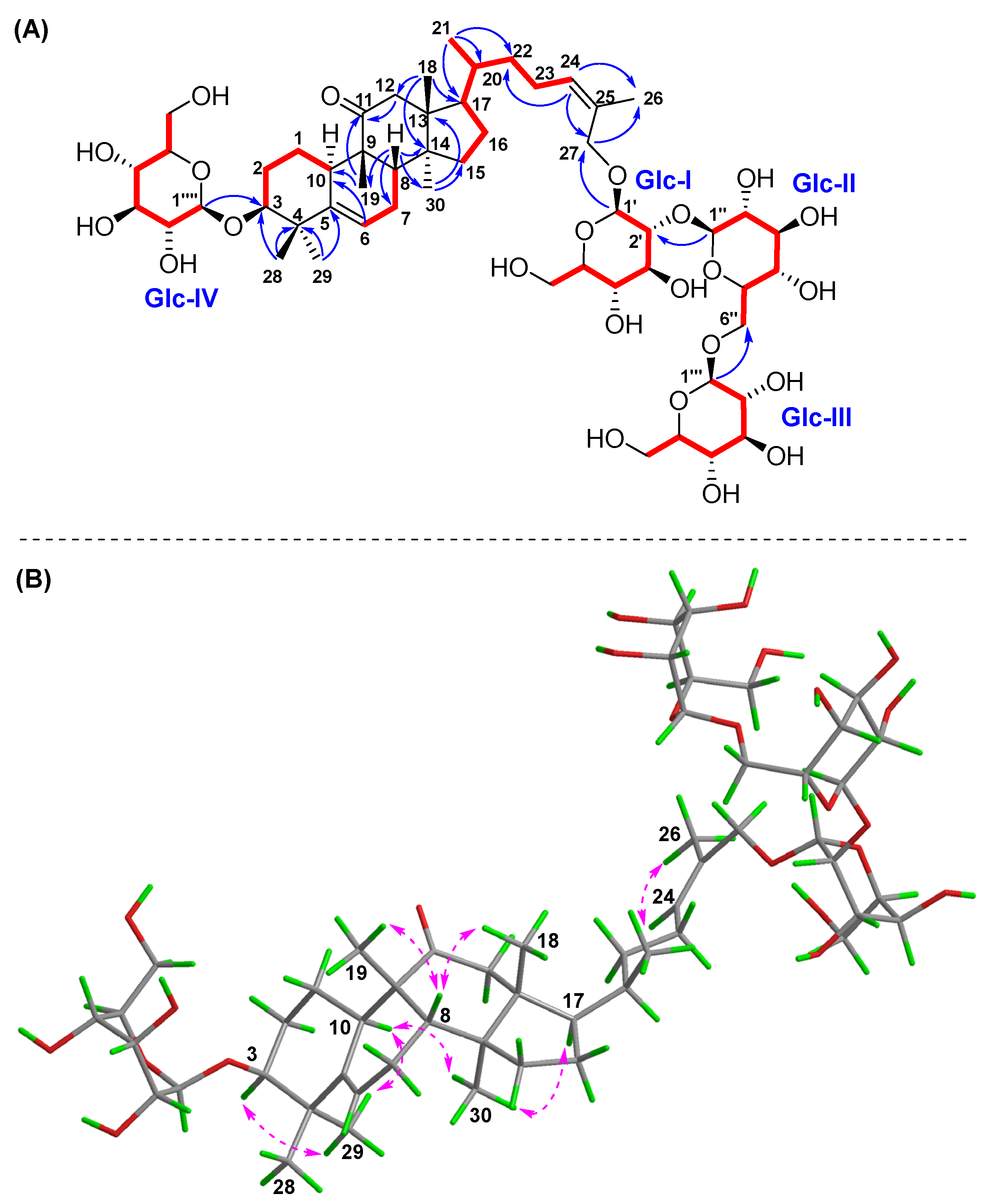

| NO. | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δH | δC | δH | δC | δH | δC | |
| 1 | 1.49 (m) 1.54 (m) | 23.2 | 1.50 (m) 1.54(m) | 23.3 | 1.93 (m) 2.14 (m) | 25.5 |
| 2 | 1.82 (m) 2.08 (m) | 29.4 | 1.91 (m) 2.07 (m) | 29.4 | 2.00 (m) 2.37 (m) | 24.9 |
| 3 | 3.40 (brs) | 88.0 | 3.42 (brs) | 88.0 | 3.40 (brs) | 88.0 |
| 4 | - | 42.5 | - | 42.6 | - | 42.9 |
| 5 | - | 144.0 | - | 141.0 | - | 141.2 |
| 6 | 5.51 (m) | 120.0 | 5.52 (d, 5.7) | 119.0 | 5.61 (d, 5.6) | 119.0 |
| 7 | 1.77 (m) 2.36 (m) | 25.3 | 1.80 (m) 2.37 (m) | 25.4 | 1.33 (m) 2.00 (m) | 29.2 |
| 8 | 2.29 (m) | 39.6 | 2.28 (m) | 39.6 | 1.93 (m) | 45.4 |
| 9 | - | 50.5 | 49.6 | - | 50.0 | |
| 10 | 1.75 (m) | 45.1 | 1.75 (m) | 45.2 | 2.40 (m) | 36.9 |
| 11 | 1.52 (m) 1.71 (m) | 31.6 | 1.44 (m) 1.69 (m) | 33.4 | - | 217.0 |
| 12 | 1.45 (m) 1.69 (m) | 33.4 | 1.15 (m) 1.25 (m) | 35.5 | 2.37 (d, 14.3) 3.05 (d, 14.3) | 50.1 |
| 13 | - | 50.5 | - | 50.5 | - | 50.5 |
| 14 | - | 48.1 | - | 47.5 | -- | 48.1 |
| 15 | 1.15 (m) 1.25 (m) | 35.8 | 1.15 (m) 1.23 (m) | 36.0 | 1.30 (m) 1.37 (m) | 35.4 |
| 16 | 1.29 (m) 1.94 (m) | 29.0 | 1.52 (m) 1.71 (m) | 31.6 | 1.06 (m) 1.45 (m) | 37.6 |
| 17 | 1.53 (m) | 51.9 | 1.49 (m) | 52.1 | 1.75 (m) | 50.5 |
| 18 | 0.89 (s) | 16.0 | 0.88 (s) | 16.1 | 0.73 (s) | 17.2 |
| 19 | 0.88 (s) | 28.7 | 0.87 (s) | 28.4 | 1.05 (s) | 20.0 |
| 20 | 1.34 (m) | 37.1 | 1.45 (m) | 37.3 | 1.43 (m) | 37.2 |
| 21 | 0.93 (d, 6.0) | 19.4 | 0.93 (d, 6.0) | 19.5 | 0.93 (d, 6.4) | 18.9 |
| 22 | 1.48 (m) 1.51 (m) | 37.2 | 1.04 (m) 1.46 (m) | 37.8 | 1.44 (m) 2.40 (m) | 37.1 |
| 23 | 1.95 (m) 2.10 (m) | 25.5 | 1.96 (m) 2.10 (m) | 25.8 | 1.97 (m) 2.14 (m) | 26.0 |
| 24 | 5.48 (m) | 130.0 | 5.34 (t, 5.2) | 132.1 | 5.35 (m) | 132.7 |
| 25 | - | 132.0 | - | 130.0 | - | 129.0 |
| 26 | 4.01 (d, 11.7) 4.21 (d, 11.7) | 76.4 | 1.79 (s) | 22.1 | 1.79 (s) | 22.0 |
| 27 | 1.67 (s) | 14.1 | 4.26 (m) | 68.5 | 4.27 (m) | 68.1 |
| 28 | 1.19 (s) | 26.1 | 1.19 (s) | 26.1 | 1.22 (s) | 26.1 |
| 29 | 1.02 (s) | 28.8 | 1.02 (s) | 28.8 | 1.06 (s) | 28.9 |
| 30 | 0.84 (s) | 18.9 | 0.84 (s) | 18.5 | 1.08 (s) | 19.1 |
| NO. | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δH | δC | δH | δC | δH | δC | |
| Glc-I | ||||||
| 1 | 4.39 (d, 7.7) | 101.4 | 4.28 (d, 7.8) | 101.6 | 4.35 (d, 7.5) | 101.2 |
| 2 | 3.48 (m) | 82.0 | 3.17 (m) | 82.0 | 3.57 (m) | 82.0 |
| 3 | 3.40 (m) | 76.7 | 3.20 (m) | 76.7 | 3.37 (m) | 71.4 |
| 4 | 3.25 (m) | 71.6 | 3.23 (dt, 8.6, 3.0) | 71.4 | 3.24 (m) | 75.5 |
| 5 | 3.54 (t, 6.2) | 77.9 | 3.24 (dt, 8.6, 3.0) | 78.1 | 3.21 (m) | 75.3 |
| 6 | 3.77 (m) 4.13 (d, 11.3) | 69.6 | 3.68 (dt, 11.6, 5.5) 3.85 (m) | 69.7 | 3.64 (m) 3.82 (d, 2.3) | 62.5 |
| Glc-II | ||||||
| 1 | 4.61 (d, 7.8) | 104.9 | 4.61 (d, 7.5) | 104.7 | 4.59 (d, 7.9) | 104.6 |
| 2 | 3.21 (m) | 75.5 | 3.23 (m) | 75.9 | 3.26 (m) | 77.9 |
| 3 | 3.27 (m) | 78.0 | 3.52 (m) | 78.0 | 3.19 (m) | 77.8 |
| 4 | 3.42 (m) | 71.2 | 3.41 (q, 3.6) | 71.6 | 3.34 (m) | 78.3 |
| 5 | 3.36 (m) | 78.0 | 3.37 (m) | 77.8 | 3.45 (m) | 77.5 |
| 6 | 3.62 (m) 3.75 (m) | 62.7 | 3.67 (dt, 11.6, 5.4) 3.82 (m) | 62.8 | 3.77 (d, 5.8) 4.09 (d, 2.1) | 69.6 |
| Glc-III | ||||||
| 1 | 4.37 (d, 7.7) | 104.7 | 4.34 (d, 7.9) | 104.9 | 4.47 (d, 7.8) | 104.6 |
| 2 | 3.22 (m) | 78.0 | 3.21 (d, 1.4) | 75.2 | 3.18 (m) | 75.1 |
| 3 | 3.54 (t,6.2) | 77.7 | 3.35 (dd, 8.6, 1.9) | 77.7 | 3.23 (dd, 7.1, 1.8) | 78.0 |
| 4 | 3.28 (m) | 71.5 | 3.27 (dt, 8.6, 3.0) | 71.6 | 3.28 (s) | 71.5 |
| 5 | 3.37 (m) | 77.5 | 3.41 (m) | 76.9 | 3.42 (d, 3.8) | 77.5 |
| 6 | 3.68 (m) 3.85 (m) | 62.7 | 3.65 (dt, 11.6, 5.4) 3.80 (m) | 62.8 | 3.69 (d, 5.5) 3.85 (s) | 62.7 |
| Glc-IV | ||||||
| 1 | 4.26 (d, 7.8) | 106.9 | 4.28 (d, 7.8) | 106.7 | 4.26 (d, 7.3) | 106.4 |
| 2 | 3.17 (d, 8.3) | 75.1 | 3.17 (m) | 75.1 | 3.16 (m) | 75.6 |
| 3 | 3.31 (m) | 78.1 | 3.20 (m) | 75.6 | 3.20 (m) | 78.2 |
| 4 | 3.29 (d) | 71.4 | 3.23 (dt, 8.6, 3.0) | 71.7 | 3.28 (m) | 71.6 |
| 5 | 3.34 (m) | 78.2 | 3.24 (dt, 8.6, 3.0) | 77.6 | 3.23 (m) | 77.8 |
| 6 | 3.69 (m) 3.89 (d, 2.0) | 62.6 | 3.68 (dt, 11.6, 5.5) 3.85 (m) | 62.9 | 3.66 (m) 3.89 (s) | 62.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chi, J.; Li, M.; Lian, F.; Li, Y.; Dai, L. Cucurbitane Glycosides and Their Potential Anti-Inflammatory Activities from Hemsleya chinensis Tubers. Molecules 2025, 30, 2349. https://doi.org/10.3390/molecules30112349
Chi J, Li M, Lian F, Li Y, Dai L. Cucurbitane Glycosides and Their Potential Anti-Inflammatory Activities from Hemsleya chinensis Tubers. Molecules. 2025; 30(11):2349. https://doi.org/10.3390/molecules30112349
Chicago/Turabian StyleChi, Jun, Miaomiao Li, Feihe Lian, Yixiao Li, and Liping Dai. 2025. "Cucurbitane Glycosides and Their Potential Anti-Inflammatory Activities from Hemsleya chinensis Tubers" Molecules 30, no. 11: 2349. https://doi.org/10.3390/molecules30112349
APA StyleChi, J., Li, M., Lian, F., Li, Y., & Dai, L. (2025). Cucurbitane Glycosides and Their Potential Anti-Inflammatory Activities from Hemsleya chinensis Tubers. Molecules, 30(11), 2349. https://doi.org/10.3390/molecules30112349






