Antimicrobial Activity of Copper(II), Nickel(II) and Zinc(II) Complexes with Semicarbazone and Thiosemicarbazone Ligands Derived from Substituted Salicylaldehydes
Abstract
1. Introduction
2. Results and Discussion
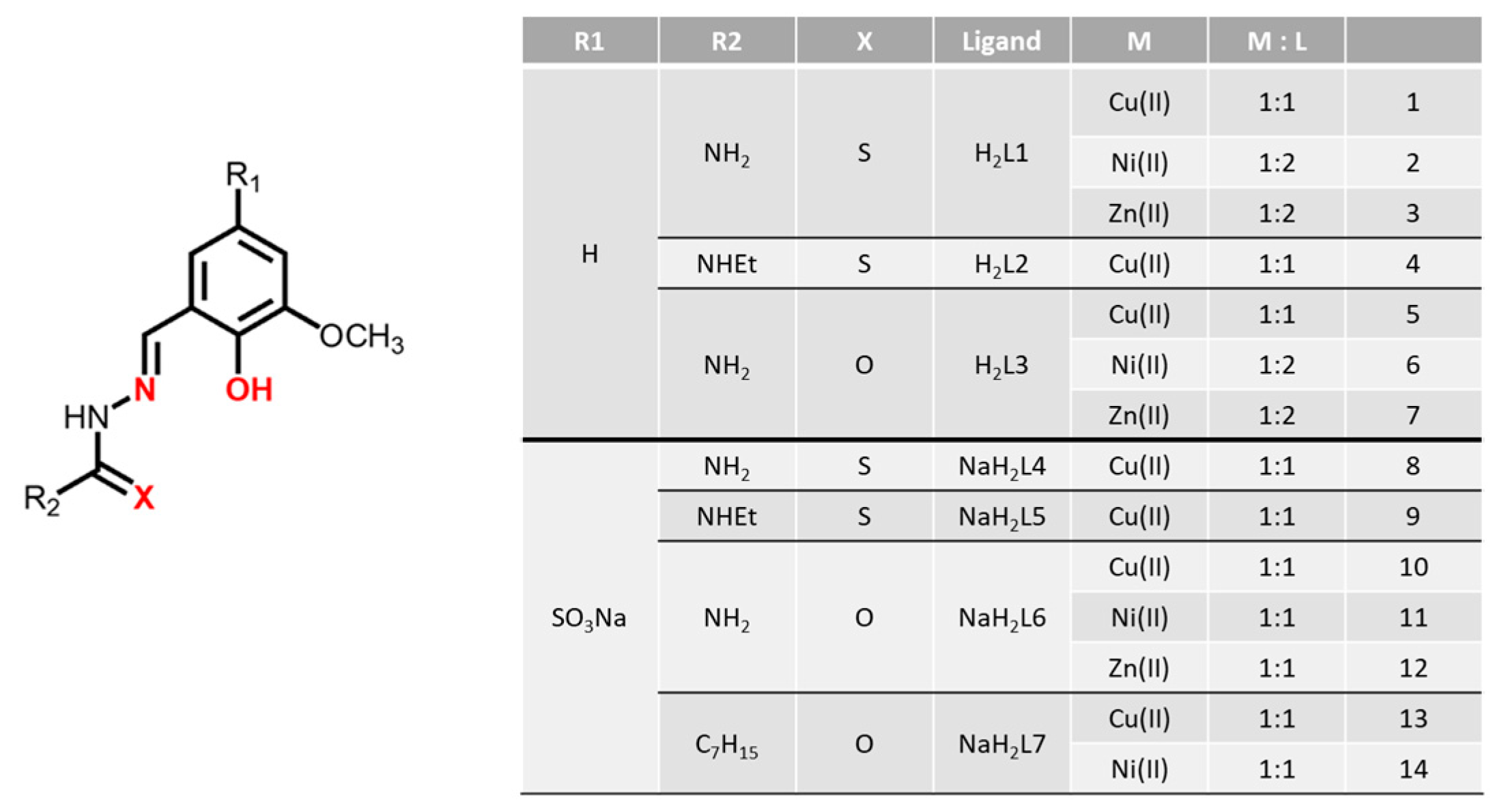
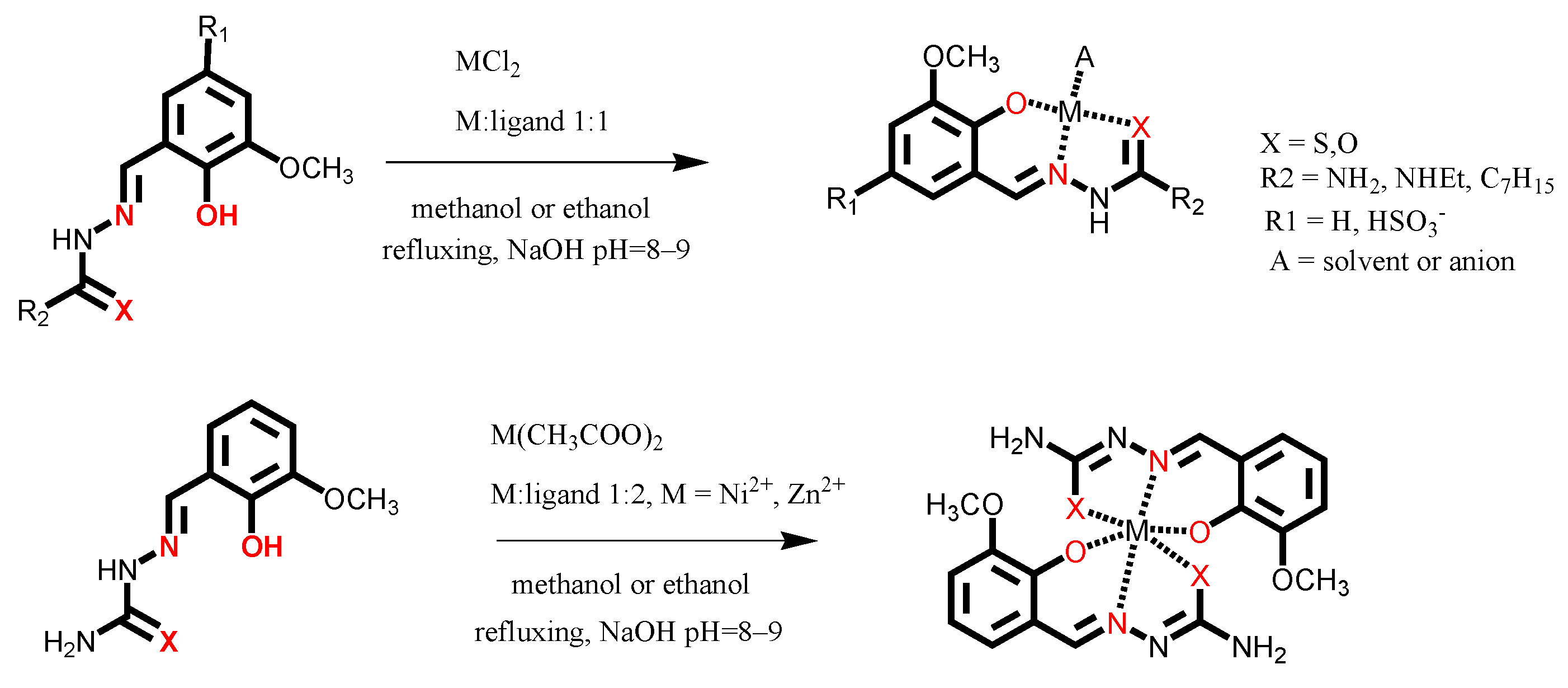
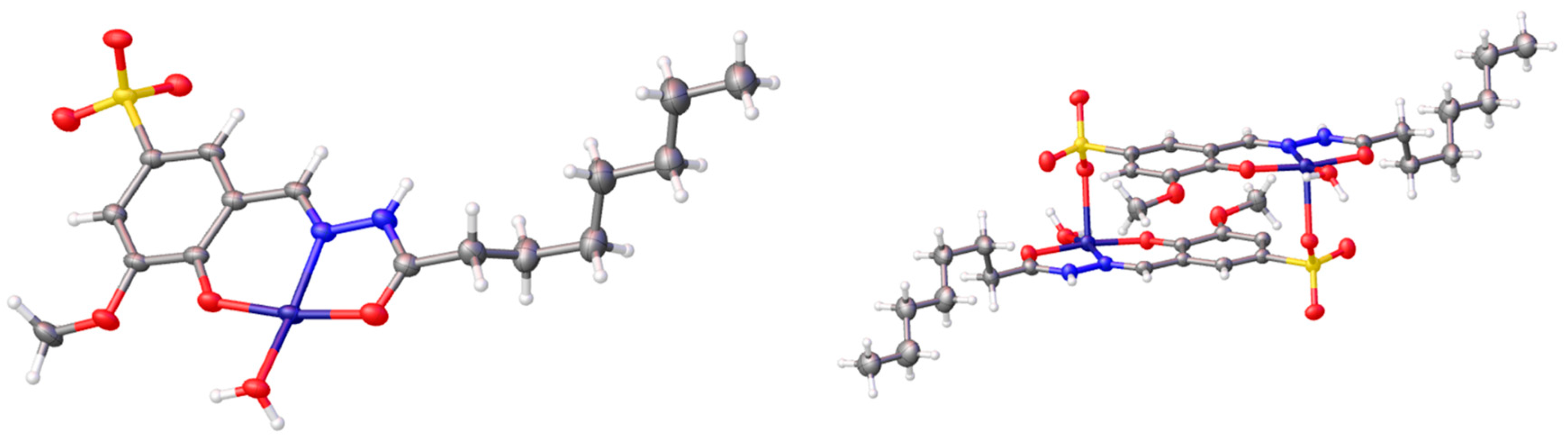
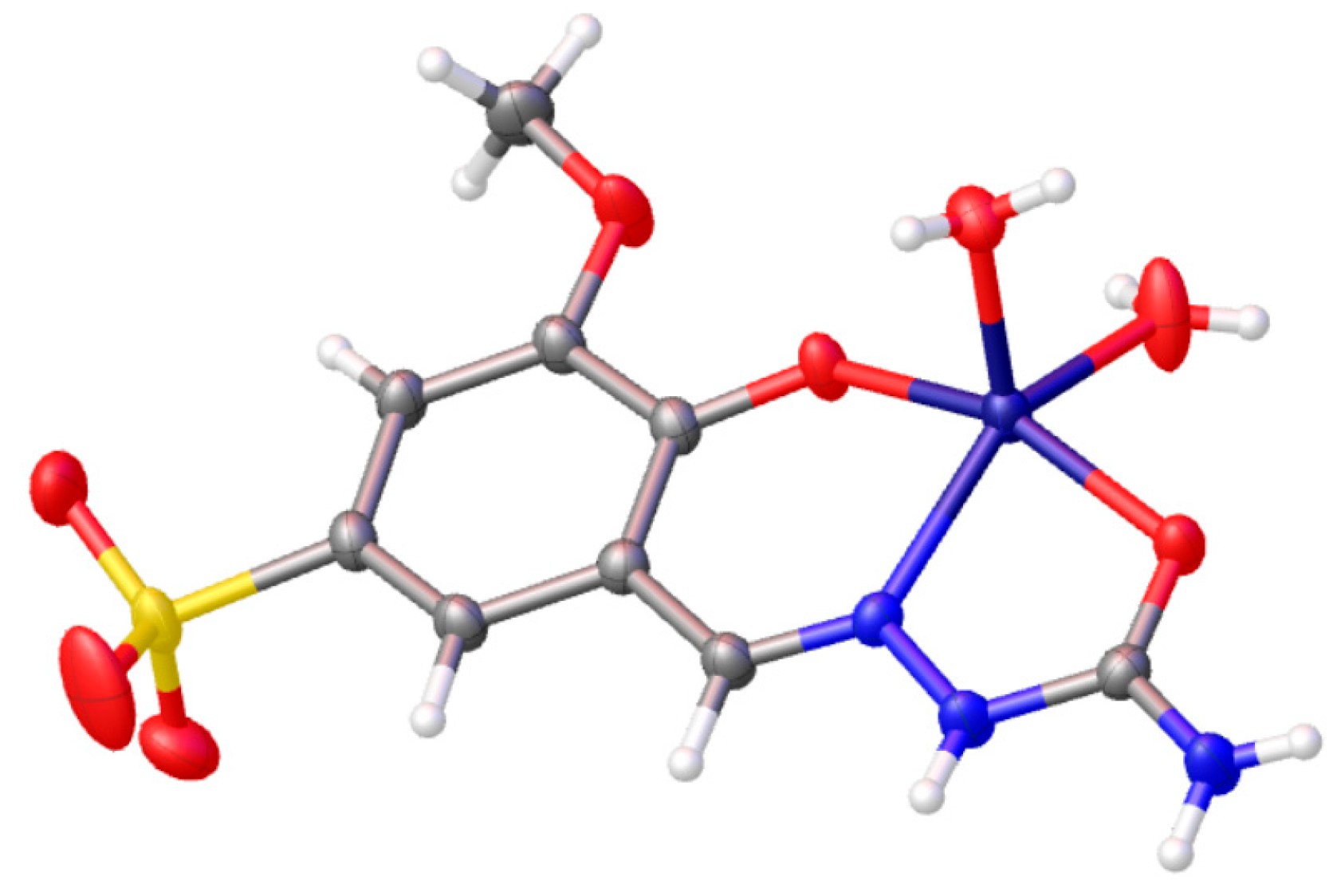
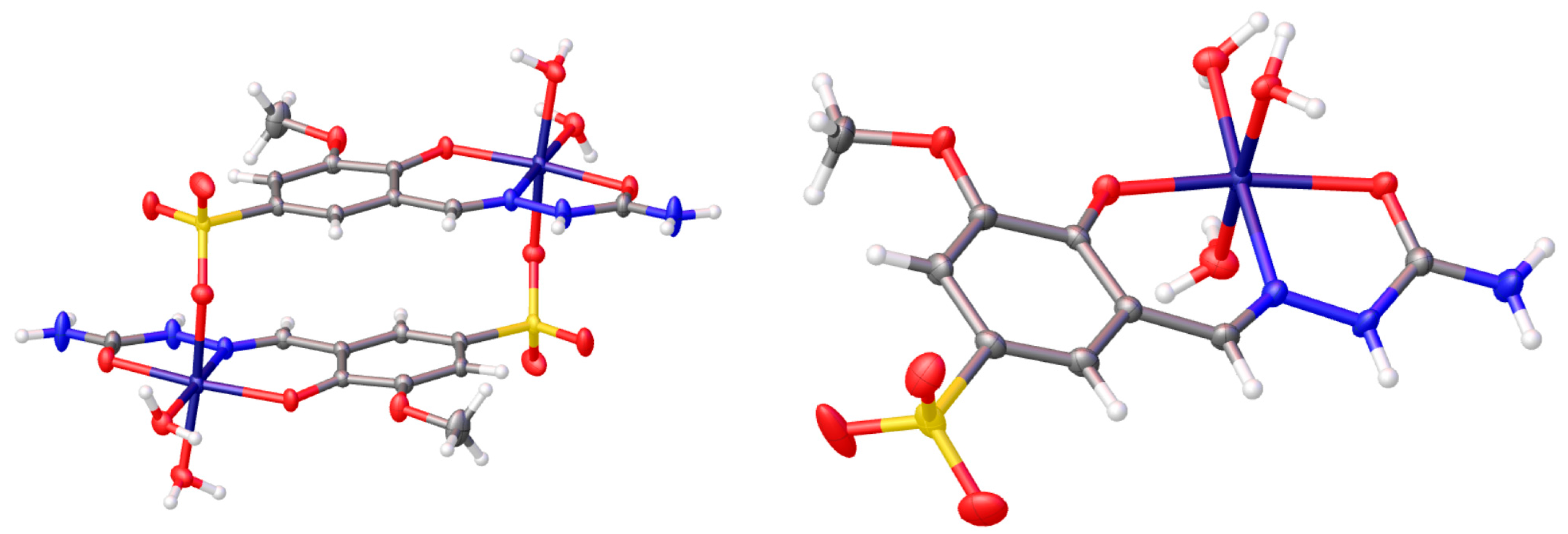
2.1. Synthesis
2.2. Biological Evaluations
2.2.1. In Vitro Antimicrobial Activity
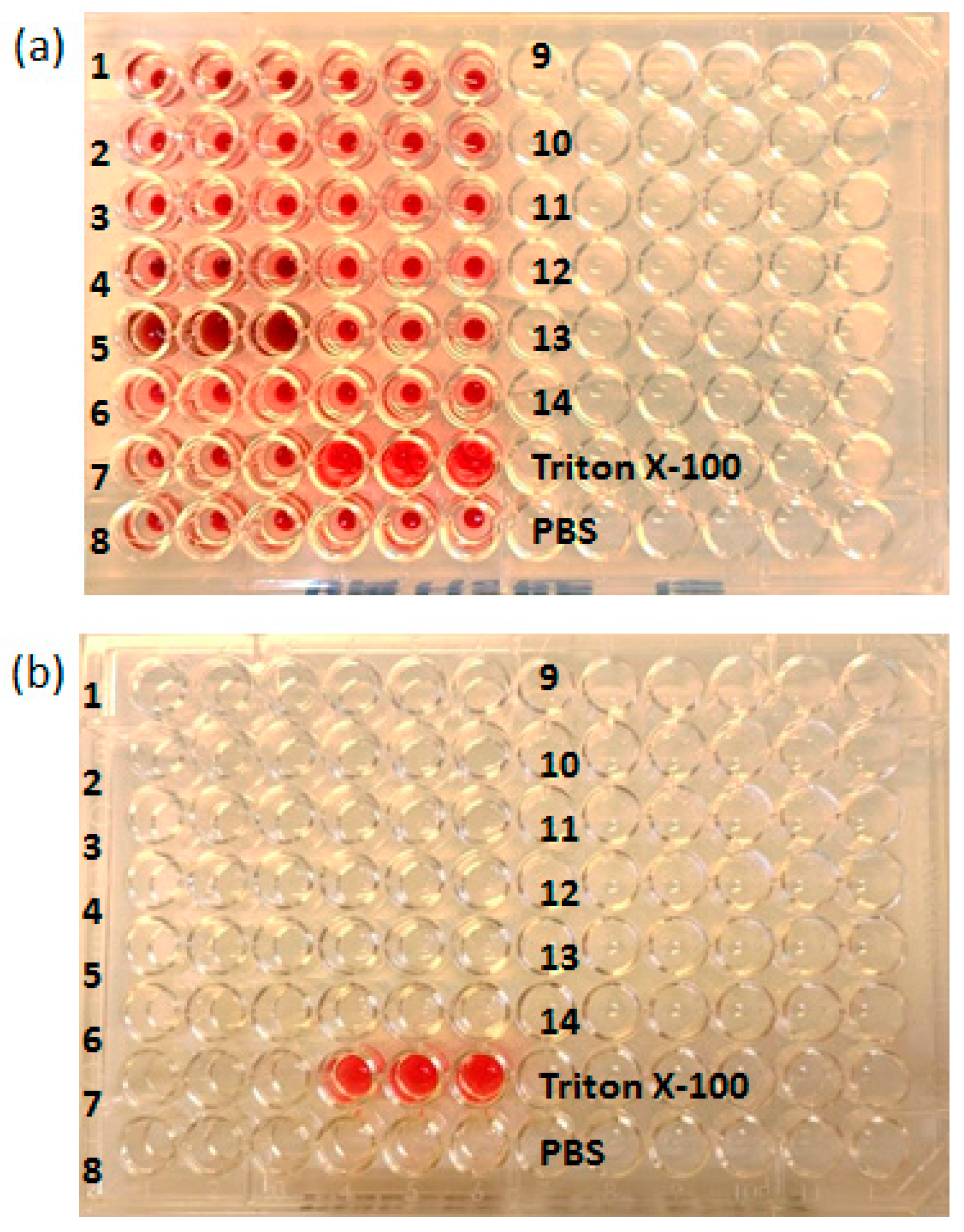
2.2.2. Cytotoxicity and Hemolytic Activity
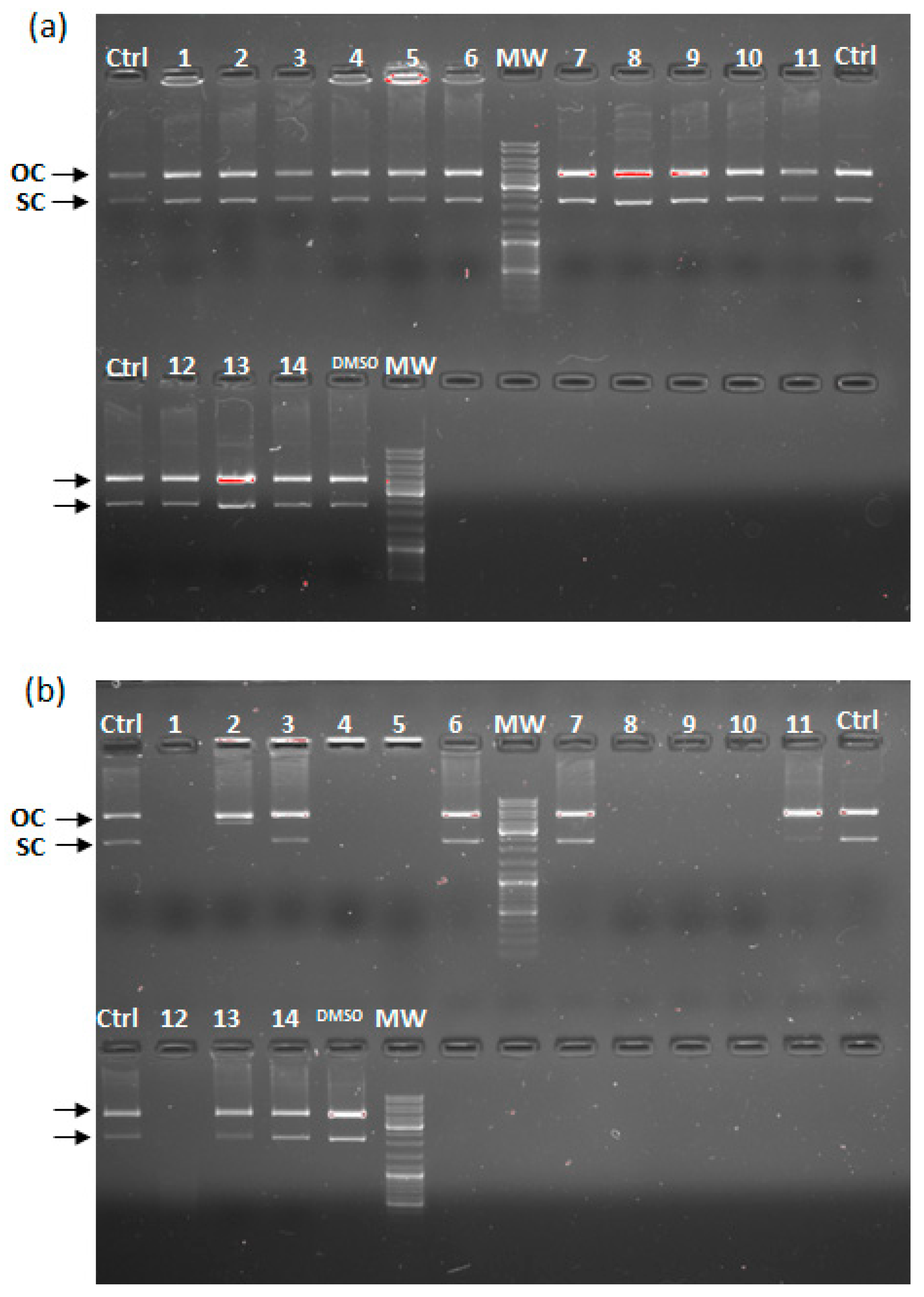
2.3. In Vitro DNA Cleavage Assay
2.4. Cell-Selective Activity
3. Materials and Methods
3.1. Synthesis
3.2. Metal Complexes and Reference Drugs
3.3. Biological Evaluation
3.3.1. Microbial Strains and Growth Conditions
3.3.2. MIC and IC50 Determination
3.3.3. Cell Viability and Proliferation Assay
3.3.4. Hemolytic Activity Assay
3.4. In Vitro DNA Cleavage/Mobility Shift Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Turner, R.J. The Good, the Bad, and the Ugly of Metals as Antimicrobials. BioMetals 2024, 37, 545–559. [Google Scholar] [CrossRef] [PubMed]
- Waters, J.E.; Stevens-Cullinane, L.; Siebenmann, L.; Hess, J. Recent Advances in the Development of Metal Complexes as Antibacterial Agents with Metal-Specific Modes of Action. Curr. Opin. Microbiol. 2023, 75, 102347. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014; ISBN 978-92-4-156474-8. [Google Scholar]
- Frei, A.; Verderosa, A.D.; Elliott, A.G.; Zuegg, J.; Blaskovich, M.A.T. Metals to Combat Antimicrobial Resistance. Nat. Rev. Chem. 2023, 7, 202–224. [Google Scholar] [CrossRef]
- Fabra, D.; Amariei, G.; Ruiz-Camino, D.; Matesanz, A.I.; Rosal, R.; Quiroga, A.G.; Horcajada, P.; Hidalgo, T. Proving the Antimicrobial Therapeutic Activity on a New Copper–Thiosemicarbazone Complex. Mol. Pharm. 2024, 21, 1987–1997. [Google Scholar] [CrossRef] [PubMed]
- Bisceglie, F.; Bacci, C.; Vismarra, A.; Barilli, E.; Pioli, M.; Orsoni, N.; Pelosi, G. Antibacterial Activity of Metal Complexes Based on Cinnamaldehyde Thiosemicarbazone Analogues. J. Inorg. Biochem. 2020, 203, 110888. [Google Scholar] [CrossRef]
- Low, M.L.; Maigre, L.; Dorlet, P.; Guillot, R.; Pagès, J.-M.; Crouse, K.A.; Policar, C.; Delsuc, N. Conjugation of a New Series of Dithiocarbazate Schiff Base Copper(II) Complexes with Vectors Selected to Enhance Antibacterial Activity. Bioconjug. Chem. 2014, 25, 2269–2284. [Google Scholar] [CrossRef]
- Miglioli, F.; De Franco, M.; Bartoli, J.; Scaccaglia, M.; Pelosi, G.; Marzano, C.; Rogolino, D.; Gandin, V.; Carcelli, M. Anticancer Activity of New Water-Soluble Sulfonated Thiosemicarbazone Copper(II) Complexes Targeting Disulfide Isomerase. Eur. J. Med. Chem. 2024, 276, 116697. [Google Scholar] [CrossRef]
- Carcelli, M.; Tegoni, M.; Bartoli, J.; Marzano, C.; Pelosi, G.; Salvalaio, M.; Rogolino, D.; Gandin, V. In Vitro and in Vivo Anticancer Activity of Tridentate Thiosemicarbazone Copper Complexes: Unravelling an Unexplored Pharmacological Target. Eur. J. Med. Chem. 2020, 194, 112266. [Google Scholar] [CrossRef]
- Carcelli, M.; Rogolino, D.; Gatti, A.; De Luca, L.; Sechi, M.; Kumar, G.; White, S.W.; Stevaert, A.; Naesens, L. N-Acylhydrazone Inhibitors of Influenza Virus PA Endonuclease with Versatile Metal Binding Modes. Sci. Rep. 2016, 6, 31500. [Google Scholar] [CrossRef]
- Elmusa, S.; Elmusa, M.; Elmusa, F.; Karaer Ozmen, F.; Kasimogullari, R. Synthesis of New Structural Thiosemicarbazones and Investigation of Their Antibacterial Activities Against E. Coli. Chem. 2024, 9, e202401981. [Google Scholar] [CrossRef]
- Chylewska, A.; Biedulska, M.; Sumczynski, P.; Makowski, M. Metallopharmaceuticals in Therapy—A New Horizon for Scientific Research. Curr. Med. Chem. 2018, 25, 1729–1791. [Google Scholar] [CrossRef] [PubMed]
- Peana, M.; Pelucelli, A.; Medici, S.; Cappai, R.; Nurchi, V.M.; Zoroddu, M.A. Metal Toxicity and Speciation: A Review. Curr. Med. Chem. 2021, 28, 7190–7208. [Google Scholar] [CrossRef]
- Carcelli, M.; Montalbano, S.; Rogolino, D.; Gandin, V.; Miglioli, F.; Pelosi, G.; Buschini, A. Antiproliferative Activity of Nickel(II), Palladium(II) and Zinc(II) Thiosemicarbazone Complexes. Inorganica Chim. Acta 2022, 533, 120779. [Google Scholar] [CrossRef]
- Zimmer, M.; Schulte, G.; Luo, X.-L.; Crabtree, R.H. Functional Modeling of Ni,Fe Hydrogenases: A Nickel Complex in an N,O,S Environment. Angew. Chem. Int. Ed. Engl. 1991, 30, 193–194. [Google Scholar]
- Hosseini-Yazdi, S.A.; Mirzaahmadi, A.; Khandar, A.A.; Mahdavi, M.; Rahimian, A.; Eigner, V.; Dušek, M.; Zarrini, G. Copper, Nickel and Zinc Complexes of a New Water-Soluble Thiosemicarbazone Ligand: Synthesis, Characterization, Stability and Biological Evaluation. J. Mol. Liq. 2017, 248, 658–667. [Google Scholar] [CrossRef]
- Sîrbu, A.; Palamarciuc, O.; Babak, M.V.; Lim, J.M.; Ohui, K.; Enyedy, E.A.; Shova, S.; Darvasiová, D.; Rapta, P.; Ang, W.H.; et al. Copper(ii) Thiosemicarbazone Complexes Induce Marked ROS Accumulation and Promote Nrf2-Mediated Antioxidant Response in Highly Resistant Breast Cancer Cells. Dalton Trans. 2017, 46, 3833–3847. [Google Scholar] [CrossRef]
- Mahmudov, K.T.; Sutradhar, M.; Martins, L.M.D.R.S.; Guedes Da Silva, M.F.C.; Ribera, A.; Nunes, A.V.M.; Gahramanova, S.I.; Marchetti, F.; Pombeiro, A.J.L. MnII and CuII Complexes with Arylhydrazones of Active Methylene Compounds as Effective Heterogeneous Catalysts for Solvent- and Additive-Free Microwave-Assisted Peroxidative Oxidation of Alcohols. RSC Adv. 2015, 5, 25979–25987. [Google Scholar] [CrossRef]
- Jlassi, R.; Ribeiro, A.P.C.; Alegria, E.C.B.A.; Naïli, H.; Tiago, G.A.O.; Rüffer, T.; Lang, H.; Zubkov, F.I.; Pombeiro, A.J.L.; Rekik, W. Copper(II) Complexes with an Arylhydrazone of Methyl 2-Cyanoacetate as Effective Catalysts in the Microwave-Assisted Oxidation of Cyclohexane. Inorganica Chim. Acta 2018, 471, 658–663. [Google Scholar] [CrossRef]
- Morigi, R.; Esposito, D.; Calvaresi, M.; Marforio, T.D.; Gentilomi, G.A.; Bonvicini, F.; Locatelli, A. Isatin Bis-Imidathiazole Hybrids Identified as FtsZ Inhibitors with On-Target Activity Against Staphylococcus Aureus. Antibiotics 2024, 13, 992. [Google Scholar] [CrossRef]
- Bonvicini, F.; Manet, I.; Belluti, F.; Gobbi, S.; Rampa, A.; Gentilomi, G.A.; Bisi, A. Targeting the Bacterial Membrane with a New Polycyclic Privileged Structure: A Powerful Tool To Face Staphylococcus aureus Infections. ACS Infect. Dis. 2019, 5, 1524–1534. [Google Scholar] [CrossRef]
- Bonvicini, F.; Locatelli, A.; Morigi, R.; Leoni, A.; Gentilomi, G.A. Isatin Bis-Indole and Bis-Imidazothiazole Hybrids: Synthesis and Antimicrobial Activity. Molecules 2022, 27, 5781. [Google Scholar] [CrossRef] [PubMed]
- Cox, G.; Wright, G.D. Intrinsic Antibiotic Resistance: Mechanisms, Origins, Challenges and Solutions. Int. J. Med. Microbiol. 2013, 303, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Bruker (2016) APEX3 and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA. Available online: https://www.bruker.com/en/products-and-solutions/diffractometers-and-x-ray-microscopes/single-crystal-x-ray-diffractometers/sc-xrd-software/apex.html (accessed on 10 April 2025).
- Sheldrick, G.M. A Short History of SHELX. Acta Crystallogr. A 2008, 64, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. Crystal Structure Refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A Complete Structure Solution, Refinement and Analysis Program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Burnett, M.N.; Johnson, C.K. ORTEP-III: Oak Ridge Thermal Ellipsoid Plot Program for Crystal Structure Illustrations; Citeseer: Princeton, NJ, USA, 1996. [Google Scholar]
- Macrae, C.F.; Sovago, I.; Cottrell, S.J.; Galek, P.T.A.; McCabe, P.; Pidcock, E.; Platings, M.; Shields, G.P.; Stevens, J.S.; Towler, M.; et al. Mercury 4.0: From Visualization to Analysis, Design and Prediction. J. Appl. Crystallogr. 2020, 53, 226–235. [Google Scholar] [CrossRef]
- Hager, E.B.; Makhubela, B.C.E.; Smith, G.S. Aqueous-Phase Hydroformylation of 1-Octene Using Hydrophilic Sulfonate Salicylaldimine Dendrimers. Dalton Trans. 2012, 41, 13927. [Google Scholar] [CrossRef]
- Liu, J.; Cheng, J.; Ma, X.; Zhou, X.; Xiang, H. Photophysical Properties and PH Sensing Applications of Luminescent Salicylaldehyde Derivatives. Res. Chem. Intermed. 2016, 42, 5027–5048. [Google Scholar] [CrossRef]
- Lloyd, D.R.; Phillips, D.H. Oxidative DNA Damage Mediated by Copper(II), Iron(II) and Nickel(II) Fenton Reactions: Evidence for Site-Specific Mechanisms in the Formation of Double-Strand Breaks, 8-Hydroxydeoxyguanosine and Putative Intrastrand Cross-Links. Mutat. Res./Fundam. Mol. Mech. Mutagen. 1999, 424, 23–36. [Google Scholar] [CrossRef]
| Compound | S. aureus | E. coli | C. albicans |
|---|---|---|---|
| 1 | 0.3 ± 0.5 | 88.9 ± 8.3 | 43.6 ± 6.1 |
| 2 | 3.1 ± 4.3 | 86.1 ± 24.4 | 77.1 ± 6.0 |
| 3 | 102.3 ± 6.7 | 106.1 ± 4.3 | 124.3 ± 3.6 |
| 4 | 0.3 ± 0 5 | 110.5 ± 4.8 | 15.6 ± 5.5 |
| 5 | 83.6 ± 1.4 | 55.8 ± 0.5 | 77.8 ± 2.0 |
| 6 | 97.7 ± 3.0 | 101.9 ± 1.8 | 130.8 ± 3.6 |
| 7 | 107.0 ± 2.7 | 82.0 ± 3.2 | 121.4 ± 4.4 |
| 8 | 106.3 ± 1.6 | 104.1 ± 9.3 | 89.9 ± 5.3 |
| 9 | 104.8 ± 0.7 | 96.6 ± 1.1 | 91.4 ± 2.5 |
| 10 | 97.8 ± 1.4 | 99.8 ± 1.8 | 71.6 ± 1.3 |
| 11 | 90.0 ± 1.1 | 99.4 ± 0.8 | 82.7 ± 2.7 |
| 12 | 94.2 ± 0.9 | 108.0 ± 3.9 | 89.1 ± 3.8 |
| 13 | 99.8 ± 3.6 | 105.2 ± 2.0 | 81.7 ± 5.4 |
| 14 | 97.7 ± 5.3 | 113.6 ± 10.0 | 85.8 ± 3.4 |
| CIP 1 | 0.7 ± 0.6 | 0.2 ± 0.3 | n.d. 2 |
| FCZ 3 | n.d. | n.d. | 10.0 ± 0.6 |
| DMSO 4 | 100 ± 2.5 | 100 ± 4.4 | 100 ± 1.5 |
| Compound | HEL 299 | hRBCs |
|---|---|---|
| 1 | 3.0 ± 3.3 | <5 |
| 2 | 26.5 ± 5.8 | <5 |
| 3 | 68.5 ± 7.1 | <5 |
| 4 | 9.1 ± 1.1 | <5 |
| 5 | 20.3 ± 0.6 | <5 |
| 6 | 54.1 ± 8.0 | <5 |
| 7 | 55.3 ± 6.2 | <5 |
| 8 | 90.4 ± 0.6 | <5 |
| 9 | 85.8 ± 1.7 | <5 |
| 10 | 63.5 ± 3.8 | <5 |
| 11 | 87.3 ± 2.3 | <5 |
| 12 | 88.2 ± 3.5 | <5 |
| 13 | 88.7 ± 0.9 | <5 |
| 14 | 83.9 ± 0.8 | <5 |
| CP 1 | 9.2 ± 3.8 | n.d. 2 |
| Triton X-100 3 | n.d. | 100 ± 2.6 |
| Compound | S. aureus, IC50 | C. albicans, IC50 | HEL 299, CC50 | SI 1 |
|---|---|---|---|---|
| 1 | 4.2 [3.7–4.7] | n.d. 2 | 2.0 [1.7–2.3] | 0.5 |
| 2 | 61.8 [50.9–75.1] | n.d. | 73.1 [58.1–92.0] | 1.2 |
| 4 | 3.5 [2.4–5.1] | 54.8 [31.8–74.3] | 1.4 [1.3–1.6] | 0.4 3/0.02 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zavaroni, A.; Rigamonti, L.; Bisceglie, F.; Carcelli, M.; Pelosi, G.; Gentilomi, G.A.; Rogolino, D.; Bonvicini, F. Antimicrobial Activity of Copper(II), Nickel(II) and Zinc(II) Complexes with Semicarbazone and Thiosemicarbazone Ligands Derived from Substituted Salicylaldehydes. Molecules 2025, 30, 2329. https://doi.org/10.3390/molecules30112329
Zavaroni A, Rigamonti L, Bisceglie F, Carcelli M, Pelosi G, Gentilomi GA, Rogolino D, Bonvicini F. Antimicrobial Activity of Copper(II), Nickel(II) and Zinc(II) Complexes with Semicarbazone and Thiosemicarbazone Ligands Derived from Substituted Salicylaldehydes. Molecules. 2025; 30(11):2329. https://doi.org/10.3390/molecules30112329
Chicago/Turabian StyleZavaroni, Alessio, Luca Rigamonti, Franco Bisceglie, Mauro Carcelli, Giorgio Pelosi, Giovanna Angela Gentilomi, Dominga Rogolino, and Francesca Bonvicini. 2025. "Antimicrobial Activity of Copper(II), Nickel(II) and Zinc(II) Complexes with Semicarbazone and Thiosemicarbazone Ligands Derived from Substituted Salicylaldehydes" Molecules 30, no. 11: 2329. https://doi.org/10.3390/molecules30112329
APA StyleZavaroni, A., Rigamonti, L., Bisceglie, F., Carcelli, M., Pelosi, G., Gentilomi, G. A., Rogolino, D., & Bonvicini, F. (2025). Antimicrobial Activity of Copper(II), Nickel(II) and Zinc(II) Complexes with Semicarbazone and Thiosemicarbazone Ligands Derived from Substituted Salicylaldehydes. Molecules, 30(11), 2329. https://doi.org/10.3390/molecules30112329









