In Silico Exploration of Natural Antioxidants for Sepsis Drug Discovery
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of Receptor for Docking Studies
2.2. Selection of Ligands for Docking Studies
2.3. Molecular Docking
3. Results and Discussion
3.1. Docking Validation Using Re-Docking
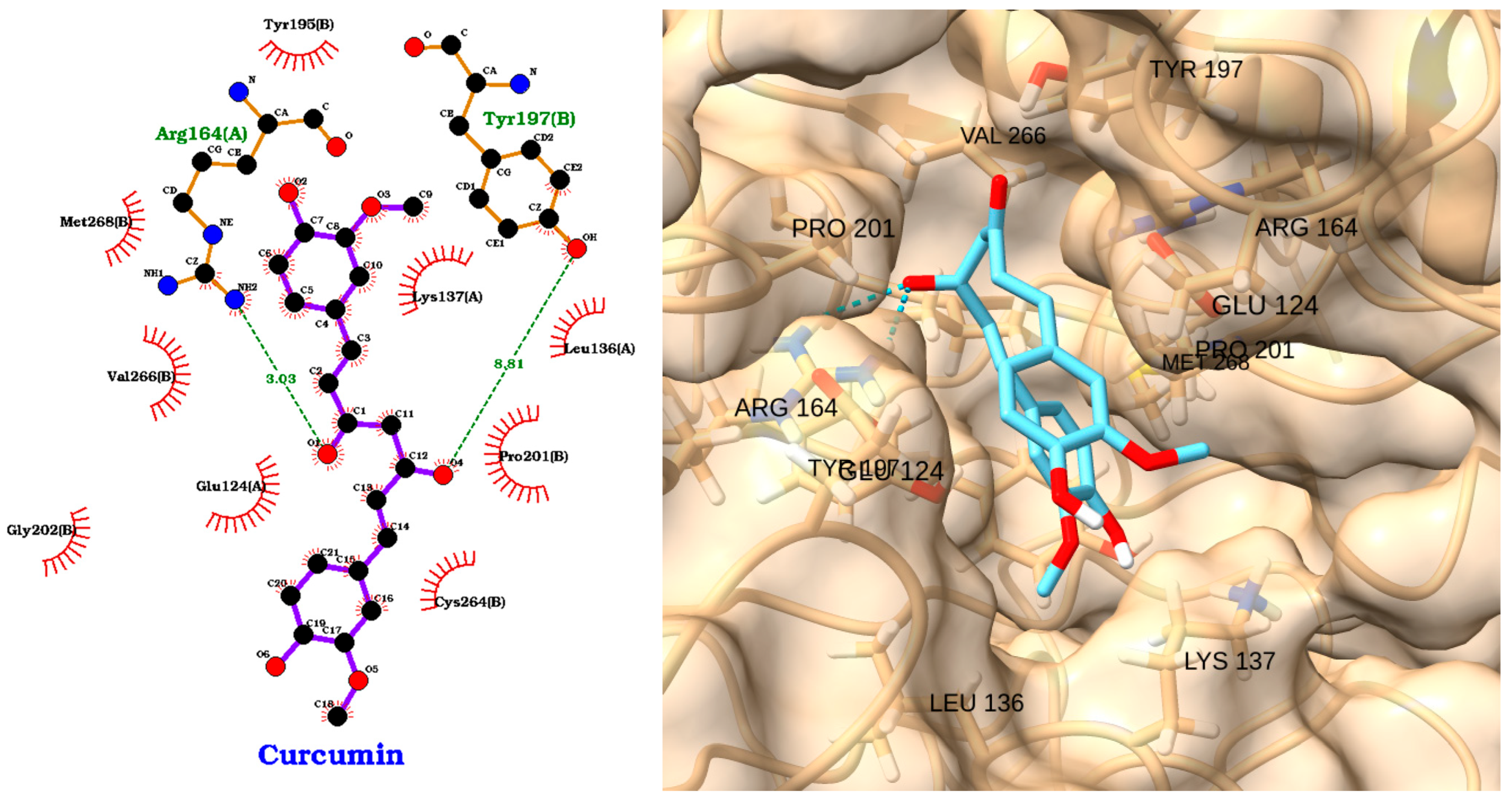
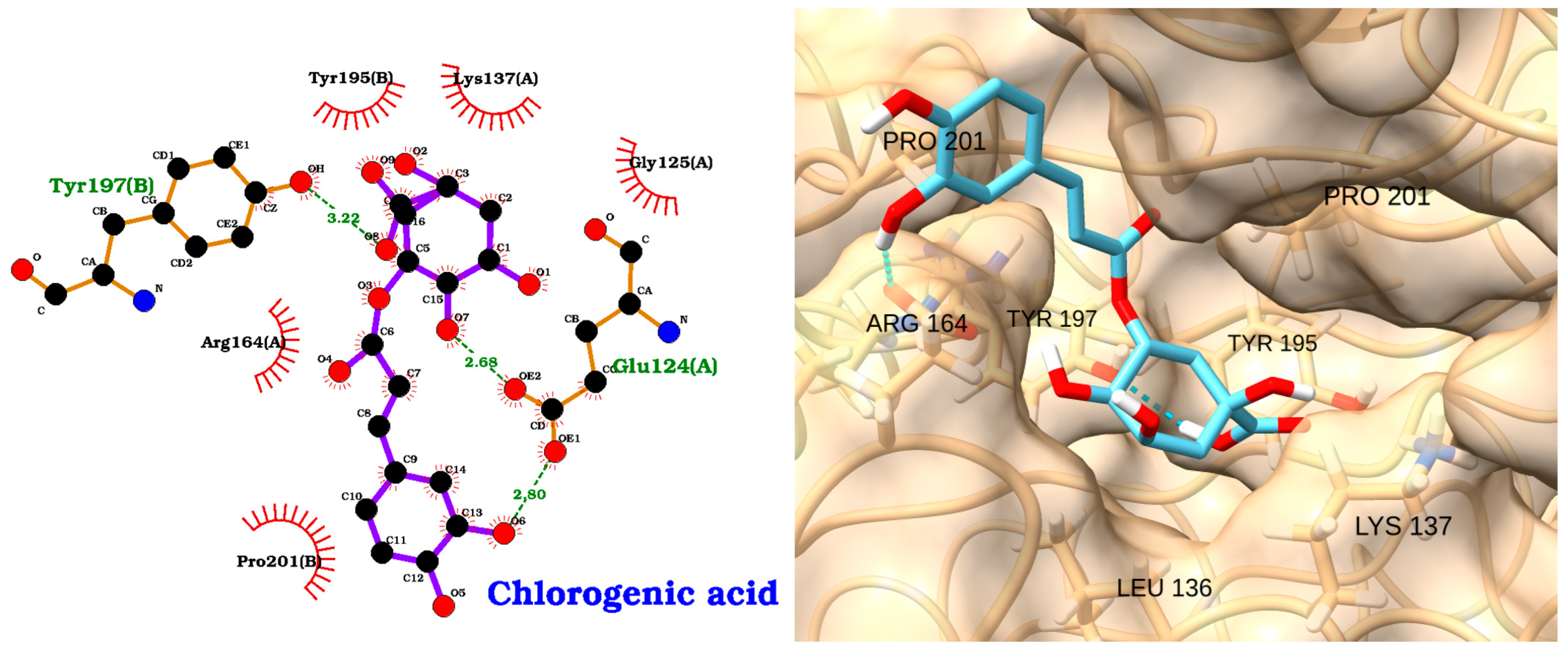
3.2. Docking of NAOs with TLR4
3.3. Docking of NAOs with IRAK1
3.4. Docking of NAOs with Caspase-3
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wiersinga, W.J.; van der Poll, T. Immunopathophysiology of human sepsis. EBioMedicine 2022, 86, 104363. [Google Scholar] [CrossRef] [PubMed]
- La Via, L.; Sangiorgio, G.; Stefani, S.; Marino, A.; Nunnari, G.; Cocuzza, S.; La Mantia, I.; Cacopardo, B.; Stracquadanio, S.; Spampinato, S. The global burden of sepsis and septic shock. Epidemiologia 2024, 5, 456–478. [Google Scholar] [CrossRef] [PubMed]
- Miliaraki, M.; Briassoulis, P.; Ilia, S.; Michalakakou, K.; Karakonstantakis, T.; Polonifi, A.; Bastaki, K.; Briassouli, E.; Vardas, K.; Pistiki, A. Oxidant/antioxidant status is impaired in sepsis and is related to anti-apoptotic, inflammatory, and innate immunity alterations. Antioxidants 2022, 11, 231. [Google Scholar] [CrossRef] [PubMed]
- Nedeva, C. Inflammation and cell death of the innate and adaptive immune system during sepsis. Biomolecules 2021, 11, 1011. [Google Scholar] [CrossRef]
- Luan, Y.-Y.; Dong, N.; Xie, M.; Xiao, X.-Z.; Yao, Y.-M. The significance and regulatory mechanisms of innate immune cells in the development of sepsis. J. Interferon Cytokine Res. 2014, 34, 2–15. [Google Scholar] [CrossRef]
- Wittebole, X.; Castanares-Zapatero, D.; Laterre, P.-F. Toll-like receptor 4 modulation as a strategy to treat sepsis. Mediat. Inflamm. 2010, 2010, 568396. [Google Scholar] [CrossRef]
- Roger, T.; Froidevaux, C.; Le Roy, D.; Reymond, M.K.; Chanson, A.-L.; Mauri, D.; Burns, K.; Riederer, B.M.; Akira, S.; Calandra, T. Protection from lethal gram-negative bacterial sepsis by targeting Toll-like receptor 4. Proc. Natl. Acad. Sci. USA 2009, 106, 2348–2352. [Google Scholar] [CrossRef] [PubMed]
- Gabarin, R.S.; Li, M.; Zimmel, P.A.; Marshall, J.C.; Li, Y.; Zhang, H. Intracellular and extracellular lipopolysaccharide signaling in sepsis: Avenues for novel therapeutic strategies. J. Innate Immun. 2021, 13, 323–332. [Google Scholar] [CrossRef]
- Abraham, E. Nuclear factor—κB and its role in sepsis-associated organ failure. J. Infect. Dis. 2003, 187, S364–S369. [Google Scholar] [CrossRef]
- Chandra, R.; Federici, S.; Bishwas, T.; Németh, Z.H.; Deitch, E.A.; Thomas, J.A.; Spolarics, Z. IRAK1-dependent signaling mediates mortality in polymicrobial sepsis. Inflammation 2013, 36, 1503–1512. [Google Scholar] [CrossRef]
- Chang, C.; Hu, L.; Sun, S.; Song, Y.; Liu, S.; Wang, J.; Li, P. Regulatory role of the TLR4/JNK signaling pathway in sepsis-induced myocardial dysfunction. Mol. Med. Rep. 2021, 23, 334. [Google Scholar] [CrossRef] [PubMed]
- Hossen, M.J.; Yang, W.S.; Kim, D.; Aravinthan, A.; Kim, J.-H.; Cho, J.Y. Thymoquinone: An IRAK1 inhibitor with in vivo and in vitro anti-inflammatory activities. Sci. Rep. 2017, 7, 42995. [Google Scholar] [CrossRef]
- Zhang, S.-y.; Xu, Q.-p.; Shi, L.-n.; Li, S.-w.; Wang, W.-h.; Wang, Q.-q.; Lu, L.-x.; Xiao, H.; Wang, J.-h.; Li, F.-y. Soluble CD4 effectively prevents excessive TLR activation of resident macrophages in the onset of sepsis. Signal Transduct. Target. Ther. 2023, 8, 236. [Google Scholar] [CrossRef] [PubMed]
- Kuzmich, N.N.; Sivak, K.V.; Chubarev, V.N.; Porozov, Y.B.; Savateeva-Lyubimova, T.N.; Peri, F. TLR4 signaling pathway modulators as potential therapeutics in inflammation and sepsis. Vaccines 2017, 5, 34. [Google Scholar] [CrossRef]
- Chousterman, B.G.; Swirski, F.K.; Weber, G.F. Cytokine storm and sepsis disease pathogenesis. In Seminars in Immunopathology; Springer: Berlin/Heidelberg, Germany, 2017; pp. 517–528. [Google Scholar]
- Reddy, H.; Javvaji, C.K.; Malali, S.; Kumar, S.; Acharya, S.; Toshniwal, S.; Malali, S., Jr.; Toshniwal, S.S. Navigating the cytokine storm: A comprehensive review of chemokines and cytokines in sepsis. Cureus 2024, 16, e54275. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Saxena, J.; Srivastava, V.K.; Kaushik, S.; Singh, H.; Abo-EL-Sooud, K.; Abdel-Daim, M.M.; Jyoti, A.; Saluja, R. The interplay of oxidative stress and ROS scavenging: Antioxidants as a therapeutic potential in sepsis. Vaccines 2022, 10, 1575. [Google Scholar] [CrossRef]
- Hotchkiss, R.S.; Nicholson, D.W. Apoptosis and caspases regulate death and inflammation in sepsis. Nat. Rev. Immunol. 2006, 6, 813–822. [Google Scholar] [CrossRef]
- Mahidhara, R.; Billiar, T.R. Apoptosis in sepsis. Crit. Care Med. 2000, 28, N105–N113. [Google Scholar] [CrossRef]
- Lorente, L.; Martín, M.M.; Ferreres, J.; Solé-Violán, J.; Labarta, L.; Díaz, C.; Jiménez, A.; Borreguero-León, J.M. Serum caspase 3 levels are associated with early mortality in severe septic patients. J. Crit. Care 2016, 34, 103–106. [Google Scholar] [CrossRef]
- Hotchkiss, R.S.; Swanson, P.E.; Freeman, B.D.; Tinsley, K.W.; Cobb, J.P.; Matuschak, G.M.; Buchman, T.G.; Karl, I.E. Apoptotic cell death in patients with sepsis, shock, and multiple organ dysfunction. Crit. Care Med. 1999, 27, 1230–1251. [Google Scholar] [CrossRef]
- Cao, C.; Yu, M.; Chai, Y. Pathological alteration and therapeutic implications of sepsis-induced immune cell apoptosis. Cell Death Dis. 2019, 10, 782. [Google Scholar] [CrossRef] [PubMed]
- Patil, N.K.; Guo, Y.; Luan, L.; Sherwood, E.R. Targeting immune cell checkpoints during sepsis. Int. J. Mol. Sci. 2017, 18, 2413. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Zhou, F.; Wu, S.; Tong, Z. Research progress on natural small-molecule compounds for the prevention and treatment of sepsis. Int. J. Mol. Sci. 2023, 24, 12732. [Google Scholar] [CrossRef]
- Zhang, C.; Singla, R.K.; Tang, M.; Shen, B. Natural Products Act as Game-Changer Potentially in Treatment and Management of Sepsis-Mediated Inflammation: A Clinical Perspective. Phytomedicine 2024, 130, 155710. [Google Scholar] [CrossRef]
- Usmani, J.; Khan, T.; Ahmad, R.; Sharma, M. Potential role of herbal medicines as a novel approach in sepsis treatment. Biomed. Pharmacother. 2021, 144, 112337. [Google Scholar] [CrossRef]
- Ho, T.T.; Tran, Q.T.; Chai, C.L. The polypharmacology of natural products. Future Med. Chem. 2018, 10, 1361–1368. [Google Scholar] [CrossRef]
- Topliss, J.; Clark, A.; Ernst, E.; Hufford, C.; Johnston, G.; Rimoldi, J.; Weimann, B. Natural and synthetic substances related to human health (IUPAC Technical Report). Pure Appl. Chem. 2002, 74, 1957–1985. [Google Scholar] [CrossRef]
- Ghosh, D.; Mukherjee, P.K. Natural medicines: Clinical Efficacy, Safety and Quality; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Mandal, M.K.; Domb, A.J. Antimicrobial activities of natural bioactive polyphenols. Pharmaceutics 2024, 16, 718. [Google Scholar] [CrossRef]
- Veres, B. Anti-inflammatory role of natural polyphenols and their degradation products. In Severe Sepsis and Septic Shock-Understanding a Serious Killer; IntechOpen: London, UK, 2012. [Google Scholar]
- Zhang, Y.; Mu, T.; Deng, X.; Guo, R.; Xia, B.; Jiang, L.; Wu, Z.; Liu, M. New insights of biological functions of natural polyphenols in inflammatory intestinal diseases. Int. J. Mol. Sci. 2023, 24, 9581. [Google Scholar] [CrossRef]
- Zamanian, M.Y.; Alsaab, H.O.; Golmohammadi, M.; Yumashev, A.; Jabba, A.M.; Abid, M.K.; Joshi, A.; Alawadi, A.H.; Jafer, N.S.; Kianifar, F. Nf-κb pathway as a molecular target for curcumin in diabetes Mellitus treatment: Focusing on oxidative stress and inflammation. Cell Biochem. Funct. 2024, 42, e4030. [Google Scholar] [CrossRef]
- Tošović, J.; Marković, S.; Marković, J.M.D.; Mojović, M.; Milenković, D. Antioxidative mechanisms in chlorogenic acid. Food Chem. 2017, 237, 390–398. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.A.; Mandal, A.K.A.; Khan, Z.A. Potential neuroprotective properties of epigallocatechin-3-gallate (EGCG). Nutr. J. 2015, 15, 1–17. [Google Scholar] [CrossRef]
- Cho, S.; Namkoong, K.; Shin, M.; Park, J.; Yang, E.; Ihm, J.; Thu, V.T.; Kim, H.K.; Han, J. Cardiovascular protective effects and clinical applications of resveratrol. J. Med. Food 2017, 20, 323–334. [Google Scholar] [CrossRef]
- Costa, L.G.; Garrick, J.M.; Roquè, P.J.; Pellacani, C. Mechanisms of neuroprotection by quercetin: Counteracting oxidative stress and more. Oxidative Med. Cell. Longev. 2016, 2016, 2986796. [Google Scholar] [CrossRef]
- Fukutomi, R.; Ohishi, T.; Koyama, Y.; Pervin, M.; Nakamura, Y.; Isemura, M. Beneficial effects of epigallocatechin-3-O-gallate, chlorogenic acid, resveratrol, and curcumin on neurodegenerative diseases. Molecules 2021, 26, 415. [Google Scholar] [CrossRef]
- Liu, K.; Chen, Y.-Y.; Pan, L.-H.; Li, Q.-M.; Luo, J.-P.; Zha, X.-Q. Co-encapsulation systems for delivery of bioactive ingredients. Food Res. Int. 2022, 155, 111073. [Google Scholar] [CrossRef]
- Shahidi, F.; Dissanayaka, C.S. Phenolic-protein interactions: Insight from in-silico analyses–a review. Food Prod. Process. Nutr. 2023, 5, 2. [Google Scholar] [CrossRef]
- Surana, K.R.; Ahire, E.D.; Sonawane, V.N.; Talele, S.G. Biomolecular and molecular docking: A modern tool in drug discovery and virtual screening of natural products. In Applied Pharmaceutical Practice and Nutraceuticals; Apple Academic Press: Cambridge, MA, USA, 2021; pp. 209–223. [Google Scholar]
- Agu, P.C.; Afiukwa, C.A.; Orji, O.; Ezeh, E.; Ofoke, I.; Ogbu, C.; Ugwuja, E.I.; Aja, P. Molecular docking as a tool for the discovery of molecular targets of nutraceuticals in diseases management. Sci. Rep. 2023, 13, 13398. [Google Scholar] [CrossRef]
- Shapiro, H.; Lev, S.; Cohen, J.; Singer, P. Polyphenols in the prevention and treatment of sepsis syndromes: Rationale and pre-clinical evidence. Nutrition 2009, 25, 981–997. [Google Scholar] [CrossRef]
- Bauer, J.A.; Bauerová-Hlinková, V. Extracting the dynamic motion of proteins using normal mode analysis. In Data Mining Techniques for the Life Sciences; Springer: Berlin/Heidelberg, Germany, 2022; pp. 213–231. [Google Scholar]
- Jastrzębski, M.K. Computational Methods to Target Protein-Protein Interactions. In Protein-Protein Docking: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2024; pp. 327–343. [Google Scholar]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Hill, A.D.; Reilly, P.J. Scoring functions for AutoDock. Methods Mol. Biol. 2015, 1273, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Mendez, D.; Gaulton, A.; Bento, A.P.; Chambers, J.; De Veij, M.; Félix, E.; Magariños, M.P.; Mosquera, J.F.; Mutowo, P.; Nowotka, M. ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Res. 2019, 47, D930–D940. [Google Scholar] [CrossRef]
- Zhang, Y.; Liang, X.; Bao, X.; Xiao, W.; Chen, G. Toll-like receptor 4 (TLR4) inhibitors: Current research and prospective. Eur. J. Med. Chem. 2022, 235, 114291. [Google Scholar] [CrossRef]
- Mustafa, A.M.; Shaheen, A.M.; Zaki, H.F.; Rabie, M.A. Nicorandil and carvedilol mitigates motor deficits in experimental autoimmune encephalomyelitis-induced multiple sclerosis: Role of TLR4/TRAF6/MAPK/NF-κB signalling cascade. Int. Immunopharmacol. 2024, 127, 111387. [Google Scholar] [CrossRef]
- Jiang, Y.; Hansen, T.V. Isatin 1,2,3-triazoles as potent inhibitors against caspase-3. Bioorg. Med. Chem. Lett. 2011, 21, 1626–1629. [Google Scholar] [CrossRef]
- Newton, A.S.; Glória, P.M.; Gonçalves, L.M.; dos Santos, D.J.; Moreira, R.; Guedes, R.C.; Santos, M.M. Synthesis and evaluation of vinyl sulfones as caspase-3 inhibitors. A structure-activity study. Eur. J. Med. Chem. 2010, 45, 3858–3863. [Google Scholar] [CrossRef]
- Asim, A. Approaches to Backbone Flexibility in Protein–Protein Docking. In Protein-Protein Docking: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2024; pp. 45–68. [Google Scholar]
- Talevi, A. Computer-aided drug discovery and design: Recent advances and future prospects. In Computational Drug Discovery and Design; Springer: Berlin/Heidelberg, Germany, 2023; pp. 1–20. [Google Scholar]
- Akash, S.; Kumer, A.; Rahman, M.M.; Emran, T.B.; Sharma, R.; Singla, R.K.; Alhumaydhi, F.A.; Khandaker, M.U.; Park, M.N.; Idris, A.M. Development of new bioactive molecules to treat breast and lung cancer with natural myricetin and its derivatives: A computational and SAR approach. Front. Cell. Infect. Microbiol. 2022, 12, 952297. [Google Scholar] [CrossRef]
- Fatriansyah, J.F.; Rizqillah, R.K.; Yandi, M.Y. Molecular docking and molecular dynamics simulation of fisetin, galangin, hesperetin, hesperidin, myricetin, and naringenin against polymerase of dengue virus. J. Trop. Med. 2022, 2022, 7254990. [Google Scholar] [CrossRef]
- Malathi, R.; Dsouza, V.; Rithika, R.; Sneha, P. Molecular Docking of Fisetin as a Multi-target drug in the treatment of Alzheimer’s disease. Res. J. Pharm. Technol. 2023, 16, 5813–5817. [Google Scholar]
- Cosconati, S.; Forli, S.; Perryman, A.L.; Harris, R.; Goodsell, D.S.; Olson, A.J. Virtual screening with AutoDock: Theory and practice. Expert Opin. Drug Discov. 2010, 5, 597–607. [Google Scholar] [CrossRef]
- Khan, A.; Khan, S.U.; Khan, A.; Shal, B.; Rehman, S.U.; Rehman, S.U.; Htar, T.T.; Khan, S.; Anwar, S.; Alafnan, A.; et al. Anti-Inflammatory and Anti-Rheumatic Potential of Selective Plant Compounds by Targeting TLR-4/AP-1 Signaling: A Comprehensive Molecular Docking and Simulation Approaches. Molecules 2022, 27, 4319. [Google Scholar] [CrossRef]
- Kingsley, M.K.; Rao, G.K.; Bhat, B.V. Effectiveness of Narciclasine in Suppressing the Inflammatory Response in Sepsis: Molecular Docking and In Silico Studies. Bioinform. Biol. Insights 2024, 18, 11779322241233436. [Google Scholar] [CrossRef]
- Kang, C.; Li, X.; Liu, P.; Liu, Y.; Niu, Y.; Zeng, X.; Liu, J.; Zhao, H.; Qiu, S. Quercetin inhibits the activity and function of dendritic cells through the TLR4/IRAK4/NF-κB signalling pathway. Contemp. Oncol. 2023, 27, 182–189. [Google Scholar] [CrossRef]
- Hong Byun, E.; Fujimura, Y.; Yamada, K.; Tachibana, H. TLR4 Signaling Inhibitory Pathway Induced by Green Tea Polyphenol Epigallocatechin-3-Gallate through 67-kDa Laminin Receptor. J. Immunol. 2010, 185, 33–45. [Google Scholar] [CrossRef]
- Gao, Y.; Zhuang, Z.; Lu, Y.; Tao, T.; Zhou, Y.; Liu, G.; Wang, H.; Zhang, D.; Wu, L.; Dai, H.; et al. Curcumin Mitigates Neuro-Inflammation by Modulating Microglia Polarization Through Inhibiting TLR4 Axis Signaling Pathway Following Experimental Subarachnoid Hemorrhage. Front. Neurosci. 2019, 13, 1223. [Google Scholar] [CrossRef]
- Wu, Y.; Guo, F.; Ma, Y.; Chai, W.; Li, J.; Shi, W.; Liu, J. Curcumin’s Protective Role in Heatstroke-Induced Acute Liver Injury: Targeting Pyroptosis and Enhancing SIRT1 Expression. Glob. Chall. 2024, 8, 2400178. [Google Scholar] [CrossRef]
- Zhang, L.; Tao, X.; Fu, Q.; Ge, C.; Li, R.; Li, Z.; Zhu, Y.; Tian, H.; Li, Q.; Liu, M.; et al. Curcumin inhibits cell proliferation and migration in NSCLC through a synergistic effect on the TLR4/MyD88 and EGFR pathways. Oncol. Rep. 2019, 42, 1843–1855. [Google Scholar] [CrossRef]
- Chen, D.; Pan, D.; Tang, S.; Tan, Z.; Zhang, Y.; Fu, Y.; Lü, G.; Huang, Q. Administration of chlorogenic acid alleviates spinal cord injury via TLR4/NF-κB and p38 signaling pathway anti-inflammatory activity. Mol. Med. Rep. 2018, 17, 1340–1346. [Google Scholar] [CrossRef]
- Zheng, Y.; Gao, Y.; Zhu, W.; Bai, X.-g.; Qi, J. Advances in molecular agents targeting toll-like receptor 4 signaling pathways for potential treatment of sepsis. Eur. J. Med. Chem. 2024, 268, 116300. [Google Scholar] [CrossRef] [PubMed]
- Arcaroli, J.; Silva, E.; Maloney, J.P.; He, Q.; Svetkauskaite, D.; Murphy, J.R.; Abraham, E. Variant IRAK-1 haplotype is associated with increased nuclear factor-kappaB activation and worse outcomes in sepsis. Am. J. Respir. Crit. Care. Med. 2006, 173, 1335–1341. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, I.S.; Hatmal, M.m.M.; Abuarqoub, D.; Esawi, E.; Zalloum, H.; Wehaibi, S.; Nsairat, H.; Alshaer, W. 1,4-Naphthoquinone Is a Potent Inhibitor of IRAK1 Kinases and the Production of Inflammatory Cytokines in THP-1 Differentiated Macrophages. ACS Omega 2021, 6, 25299–25310. [Google Scholar] [CrossRef]
- Kim, K.M.; Hwang, N.-H.; Hyun, J.-S.; Shin, D. Recent Advances in IRAK1: Pharmacological and Therapeutic Aspects. Molecules 2024, 29, 2226. [Google Scholar] [CrossRef]
- Pan, B.; Gao, J.; Chen, W.; Liu, C.; Shang, L.; Xu, M.; Fu, C.; Zhu, S.; Niu, M.; Xu, K. Selective inhibition of interleukin-1 receptor-associated kinase 1 ameliorates lipopolysaccharide-induced sepsis in mice. Int. Immunopharmacol. 2020, 85, 106597. [Google Scholar] [CrossRef]
- Li, W.; Wang, Y.; Tang, Y.; Lu, H.; Qi, Y.; Li, G.; He, H.; Lu, F.; Yang, Y.; Sun, H. Quercetin Alleviates Osteoarthritis Progression in Rats by Suppressing Inflammation and Apoptosis via Inhibition of IRAK1/NLRP3 Signaling. J. Inflamm. Res. 2021, 14, 3393–3403. [Google Scholar] [CrossRef]
- Hellal, D.; Abdelaziz Emam, W. Effect of Quercetin on Inflammatory Pathways in Animal Model of Osteoarthritis. Al-Azhar Med. J. 2023, 52, 230–240. [Google Scholar]
- Xu, J.; Li, Y.; Yang, X.; Li, H.; Xiao, X.; You, J.; Li, H.; Zheng, L.; Yi, C.; Li, Z. Quercetin inhibited LPS-induced cytokine storm by interacting with the AKT1-FoxO1 and Keap1-Nrf2 signaling pathway in macrophages. Sci. Rep. 2024, 14, 20913. [Google Scholar]
- Singh, A.K.; Umar, S.; Riegsecker, S.; Chourasia, M.; Ahmed, S. Regulation of Transforming Growth Factor β-Activated Kinase Activation by Epigallocatechin-3-Gallate in Rheumatoid Arthritis Synovial Fibroblasts: Suppression of K(63) -Linked Autoubiquitination of Tumor Necrosis Factor Receptor-Associated Factor 6. Arthritis Rheumatol. 2016, 68, 347–358. [Google Scholar] [CrossRef]
- Singh, A.; Riegsecker, S.; Umar, S.; Ahmed, S. Epigallocatechin-3-Gallate (EGCG) Suppresses IL-1β-Induced IL-6 and IL-8 Synthesis By Selectively Inhibiting TAK1 Activation in Human Rheumatoid Arthritis Synovial Fibroblasts: 2879. Arthritis Rheumatol. 2014, 66, S1257–S1258. [Google Scholar]
- Malaguarnera, L. Influence of Resveratrol on the Immune Response. Nutrients 2019, 11, 946. [Google Scholar] [CrossRef] [PubMed]
- Li, D.-y.; Cui, Y.-L.; Li, H.; Huang, S.-L.; Fan, Y. Resveratrol attenuates inflammation by regulating macrophage polarization via inhibition of toll-like receptor 4/MyD88 signaling pathway. Pharmacogn. Mag. 2021, 17, 321–326. [Google Scholar]
- Li, H.; Sun, H.; Xu, Y.; Xing, G.; Wang, X. Curcumin plays a protective role against septic acute kidney injury by regulating the TLR9 signaling pathway. Transl. Androl. Urol. 2021, 10, 2103–2112. [Google Scholar] [CrossRef]
- Karimi, A.; Ghodsi, R.; Kooshki, F.; Karimi, M.; Asghariazar, V.; Tarighat-Esfanjani, A. Therapeutic effects of curcumin on sepsis and mechanisms of action: A systematic review of preclinical studies. Phytother. Res. 2019, 33, 2798–2820. [Google Scholar] [CrossRef]
- Chen, D.; Wang, H.; Cai, X. Curcumin interferes with sepsis-induced cardiomyocyte apoptosis via TLR1 inhibition. Rev. Port. De Cardiol. 2023, 42, 209–221. [Google Scholar] [CrossRef]
- Rana, M.; Maurya, P.; Reddy, S.S.; Singh, V.; Ahmad, H.; Dwivedi, A.K.; Dikshit, M.; Barthwal, M.K. A Standardized Chemically Modified Curcuma longa Extract Modulates IRAK-MAPK Signaling in Inflammation and Potentiates Cytotoxicity. Front. Pharmacol. 2016, 7, 223. [Google Scholar] [CrossRef]
- Huang, J.; Xie, M.; He, L.; Song, X.; Cao, T. Chlorogenic acid: A review on its mechanisms of anti-inflammation, disease treatment, and related delivery systems. Front. Pharmacol. 2023, 14, 1218015. [Google Scholar] [CrossRef]
- Park, S.H.; Baek, S.-I.; Yun, J.; Lee, S.; Yoon, D.Y.; Jung, J.-K.; Jung, S.-H.; Hwang, B.Y.; Hong, J.T.; Han, S.-B. IRAK4 as a molecular target in the amelioration of innate immunity–related endotoxic shock and acute liver injury by chlorogenic acid. J. Immunol. 2015, 194, 1122–1130. [Google Scholar] [CrossRef]
- Choi, J.H.; Park, S.H.; Jung, J.-K.; Cho, W.-J.; Ahn, B.; Yun, C.-Y.; Choi, Y.P.; Yeo, J.H.; Lee, H.; Hong, J.T. Caffeic acid cyclohexylamide rescues lethal inflammation in septic mice through inhibition of IκB kinase in innate immune process. Sci. Rep. 2017, 7, 41180. [Google Scholar] [CrossRef]
- Qin, H.; Lu, N.; Chen, K.; Huang, Y.; Rui, Y.; Huang, L.; Gao, Q.; Hu, J. Inhibiting caspase-3/GSDME-mediated pyroptosis ameliorates septic lung injury in mice model. Mol. Immunol. 2024, 172, 96–104. [Google Scholar] [CrossRef]
- Mokhtari, B.; Yavari, R.; Badalzadeh, R.; Mahmoodpoor, A. An overview on mitochondrial-based therapies in sepsis-related myocardial dysfunction: Mitochondrial transplantation as a promising approach. Can. J. Infect. Dis. Med. Microbiol. 2022, 2022, 3277274. [Google Scholar] [CrossRef] [PubMed]
- Wan Chik, W.D.; Abdullah, Z.L. Molecular docking studies of selected natural compounds as caspase-3 enzyme inhibitors. ESTEEM Acad. J. 2022, 18, 112–119. [Google Scholar]
- Dhani, S.; Zhao, Y.; Zhivotovsky, B. A long way to go: Caspase inhibitors in clinical use. Cell Death Dis. 2021, 12, 949. [Google Scholar] [CrossRef]
- Wang, D.; Li, X.; Li, Y.; Wang, R.; Wang, C.; Li, Y. New molecular mechanisms of quercetin in improving recurrent spontaneous abortion based on in-depth network pharmacology and molecular docking. Front. Chem. 2024, 12, 1407667. [Google Scholar] [CrossRef]
- Unsal, V.; Keskin, C.; Oner, E. Can quercetin reduce arsenic induced toxicity in mouse BALB/c 3T3 fibroblast cells? A study involving in vitro, molecular docking, and ADME predictions. BMC Pharmacol. Toxicol. 2025, 26, 68. [Google Scholar] [CrossRef]
- Eleyan, M.; Ibrahim, K.A.; Mohamed, R.A.; Hussien, M.; Zughbur, M.R.; Aldalou, A.R.; Masad, A.; El-Rahman, H.A.A.; Abdelgaid, H.A. Quercetin diminishes the apoptotic pathway of magnetite nanoparticles in rats’ ovary: Antioxidant status and hormonal profiles. Env. Anal. Health. Toxicol. 2024, 39, e2024025-2024020. [Google Scholar] [CrossRef]
- Seo, H.-S.; Ku, J.M.; Choi, H.-S.; Choi, Y.K.; Woo, J.-K.; Kim, M.; Kim, I.; Na, C.H.; Hur, H.; Jang, B.H.; et al. Quercetin induces caspase-dependent extrinsic apoptosis through inhibition of signal transducer and activator of transcription 3 signaling in HER2-overexpressing BT-474 breast cancer cells. Oncol. Rep. 2016, 36, 31–42. [Google Scholar] [CrossRef]
- Li, X.; Guo, S.; Xiong, X.K.; Peng, B.Y.; Huang, J.M.; Chen, M.F.; Wang, F.Y.; Wang, J.N. Combination of quercetin and cisplatin enhances apoptosis in OSCC cells by downregulating xIAP through the NF-κB pathway. J. Cancer 2019, 10, 4509–4521. [Google Scholar] [CrossRef]
- Chen, T.; Zhang, X.; Zhu, G.; Liu, H.; Chen, J.; Wang, Y.; He, X. Quercetin inhibits TNF-α induced HUVECs apoptosis and inflammation via downregulating NF-kB and AP-1 signaling pathway in vitro. Medicine 2020, 99, e22241. [Google Scholar] [CrossRef]
- Cai, X.; Bao, L.; Ding, Y.; Dai, X.; Zhang, Z.; Li, Y. Quercetin alleviates cell apoptosis and inflammation via the ER stress pathway in vascular endothelial cells cultured in high concentrations of glucosamine. Mol. Med. Rep. 2017, 15, 825–832. [Google Scholar] [CrossRef]
- Ghasemi-Pirbaluti, M.; Pourgheysari, B.; Shirzad, H.; Sourani, Z.; Beshkar, P. The Inhibitory Effect of Epigallocatechin Gallate on the Viability of T Lymphoblastic Leukemia Cells is Associated with Increase of Caspase-3 Level and Fas Expression. Indian J. Hematol. Blood. Transfus. 2018, 34, 253–260. [Google Scholar] [CrossRef]
- Liu, L.; Hou, L.; Gu, S.; Zuo, X.; Meng, D.; Luo, M.; Zhang, X.; Huang, S.; Zhao, X. Molecular mechanism of epigallocatechin-3-gallate in human esophageal squamous cell carcinoma in vitro and in vivo. Oncol. Rep. 2015, 33, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.R.; Rauf, A.; Akter, S.; Akter, H.; Al-Imran, M.I.K.; Islam, S.; Nessa, M.; Shompa, C.J.; Shuvo, M.N.R.; Khan, I.; et al. Epigallocatechin 3-gallate-induced neuroprotection in neurodegenerative diseases: Molecular mechanisms and clinical insights. Mol. Cell. Biochem. 2025, 1–21. [Google Scholar] [CrossRef]
- Li, J.; Zeng, X.; Yang, F.; Wang, L.; Luo, X.; Liu, R.; Zeng, F.; Lu, S.; Huang, X.; Lei, Y. Resveratrol: Potential application in sepsis. Front. Pharmacol. 2022, 13, 821358. [Google Scholar] [CrossRef]
- Chen, Y.; Ye, X.; Jin, S.; Huang, W.; Chen, C.; Chen, X. Resveratrol protects against sepsis induced acute kidney injury in mice by inducing Klotho mediated apoptosis inhibition. Trop. J. Pharm. Res. 2022, 21, 1615–1623. [Google Scholar] [CrossRef]
- Silswal, N.; Reddy, N.S.; Qureshi, A.A.; Qureshi, N. Resveratrol downregulates biomarkers of sepsis via inhibition of proteasome’s proteases. Shock 2018, 50, 579–588. [Google Scholar] [CrossRef]
- Vajdi, M.; Sefidmooye Azar, P.; Mahmoodpoor, A.; Dashti, F.; Sanaie, S.; Kiani Chalmardi, F.; Karimi, A. A comprehensive insight into the molecular and cellular mechanisms of action of resveratrol on complications of sepsis a systematic review. Phytother. Res. 2023, 37, 3780–3808. [Google Scholar] [CrossRef]
- Manica, D.; Silva, G.B.d.; Silva, A.P.d.; Marafon, F.; Maciel, S.F.V.d.O.; Bagatini, M.D.; Moreno, M. Curcumin promotes apoptosis of human melanoma cells by caspase 3. Cell Biochem. Funct. 2023, 41, 1295–1304. [Google Scholar] [CrossRef]
- Yavuz Türel, G.; Şahin Calapoğlu, N.; Bayram, D.; Özgöçmen, M.; Toğay, V.A.; Evgen Tülüceoğlu, E. Curcumin induces apoptosis through caspase dependent pathway in human colon carcinoma cells. Mol. Biol. Rep. 2022, 49, 1351–1360. [Google Scholar] [CrossRef]
- Tao, H.; Shen, L. Research progress of curcumin in the treatment of sepsis. Shock 2024, 61, 805–816. [Google Scholar] [CrossRef]
- Elengoe, A.; Sundramoorthy, N.D. Molecular docking of curcumin with breast cancer cell line proteins. Pharm. Biomed. Res. 2020, 6, 27–36. [Google Scholar] [CrossRef]
- Moslehi, A.; Komeili-Movahhed, T.; Ahmadian, M.; Ghoddoosi, M.; Heidari, F. Chlorogenic acid attenuates liver apoptosis and inflammation in endoplasmic reticulum stress-induced mice. Iran. J. Basic. Med. Sci. 2023, 26, 478–485. [Google Scholar] [CrossRef]
- Althagafi, H.A. Neuroprotective role of chlorogenic acid against hippocampal neuroinflammation, oxidative stress, and apoptosis following acute seizures induced by pentylenetetrazole. Metab. Brain Dis. 2024, 39, 1307–1321. [Google Scholar] [CrossRef]
- Zhao, Q.-Y.; Cheng, X.; Li, Q. Protective Impacts and Involved Mechanisms of Chlorogenic Acid on Sepsis-Associated Cognitive Deficits in Rats. Pak. Vet. J. 2023, 43, 585–590. [Google Scholar]
- Kumar, B.; Malik, J.K.; Singh, S.P.; Singh, G. Screening Hepatoprotective Effective Components of Leonotis nepetifolia Root Based on the Molecular Docking and its Mechanism Exploring. Saudi J. Med. Pharm. Sci. 2025, 11, 94–102. [Google Scholar] [CrossRef]
- Kimsa-Dudek, M.; Synowiec-Wojtarowicz, A.; Krawczyk, A.; Kosowska, A.; Kimsa-Furdzik, M.; Francuz, T. The Apoptotic Effect of Caffeic or Chlorogenic Acid on the C32 Cells That Have Simultaneously Been Exposed to a Static Magnetic Field. Int. J. Mol. Sci. 2022, 23, 3859. [Google Scholar] [CrossRef]
- Agudelo-Quintero, M.L.; Varela, S.A.; Lopera-Rodríguez, J.A.; Llano-Ramirez, M.A. Molecular affinity between caffeic and chlorogenic acid with proteins associated with apoptosis pathways using docking molecular analysis. In Proceedings of the 2021 IEEE 2nd International Congress of Biomedical Engineering and Bioengineering, Kragujevac, Serbia, 25–27 October 2021; IEEE: New York, NY, USA, 2021; pp. 1–4. [Google Scholar]

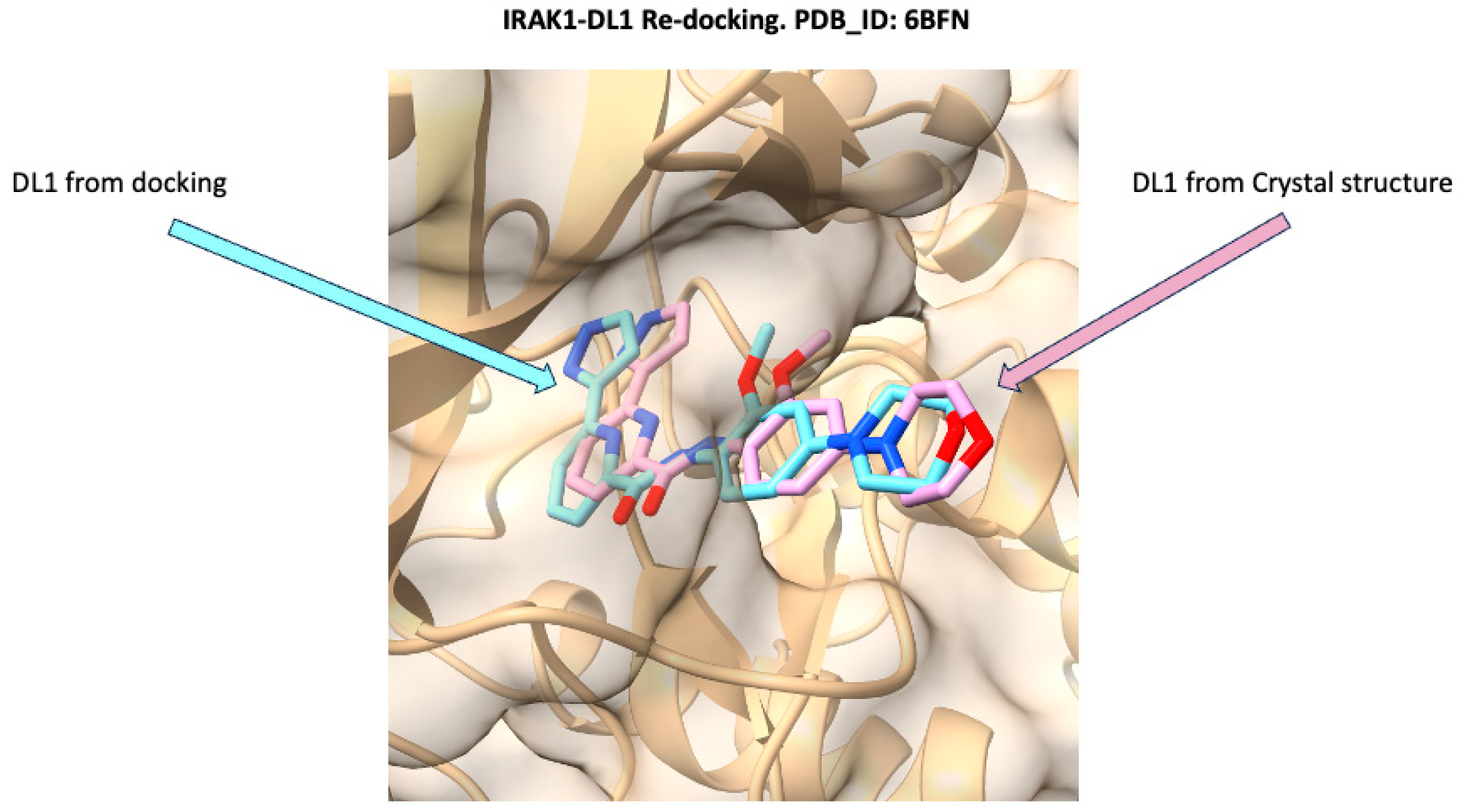
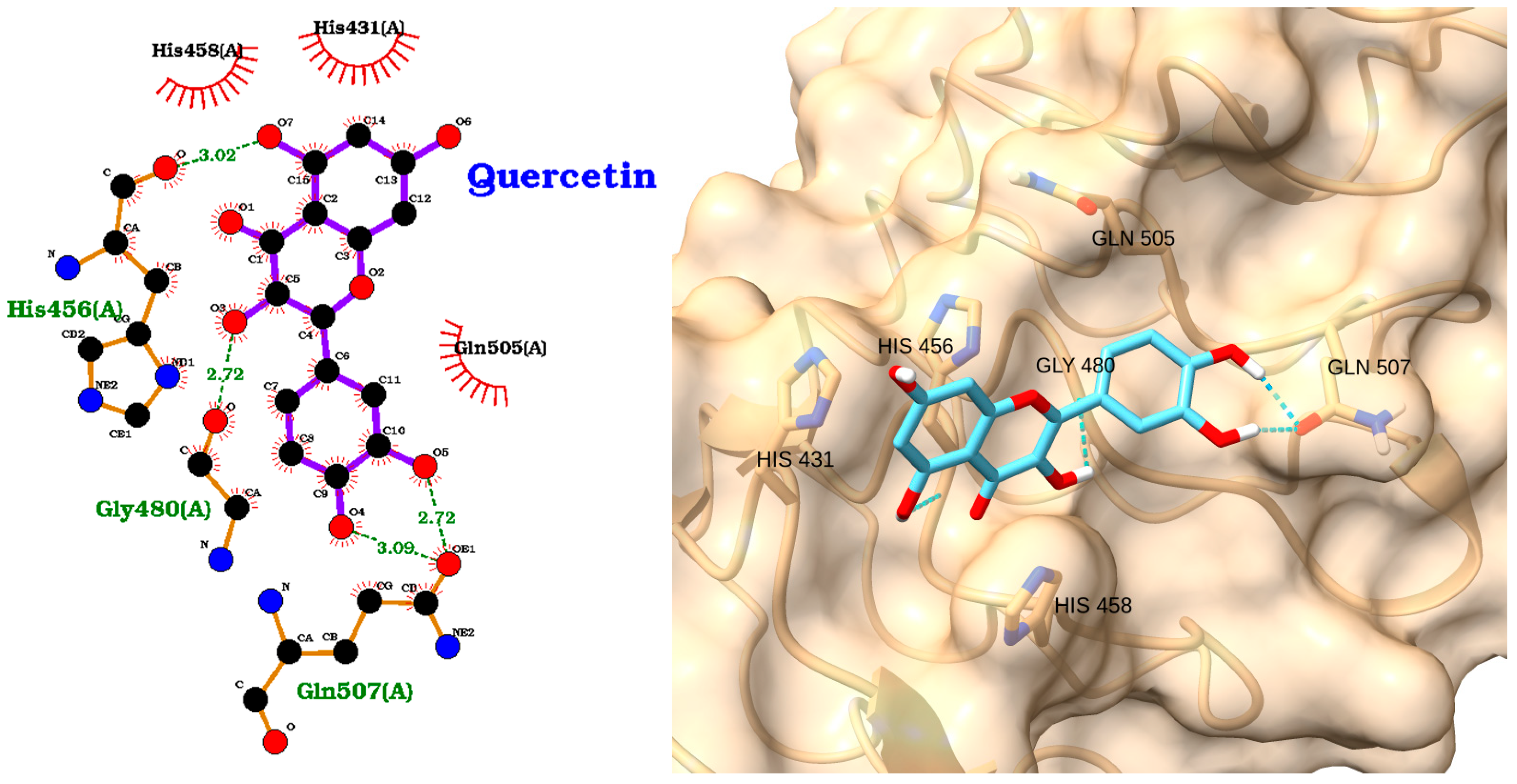
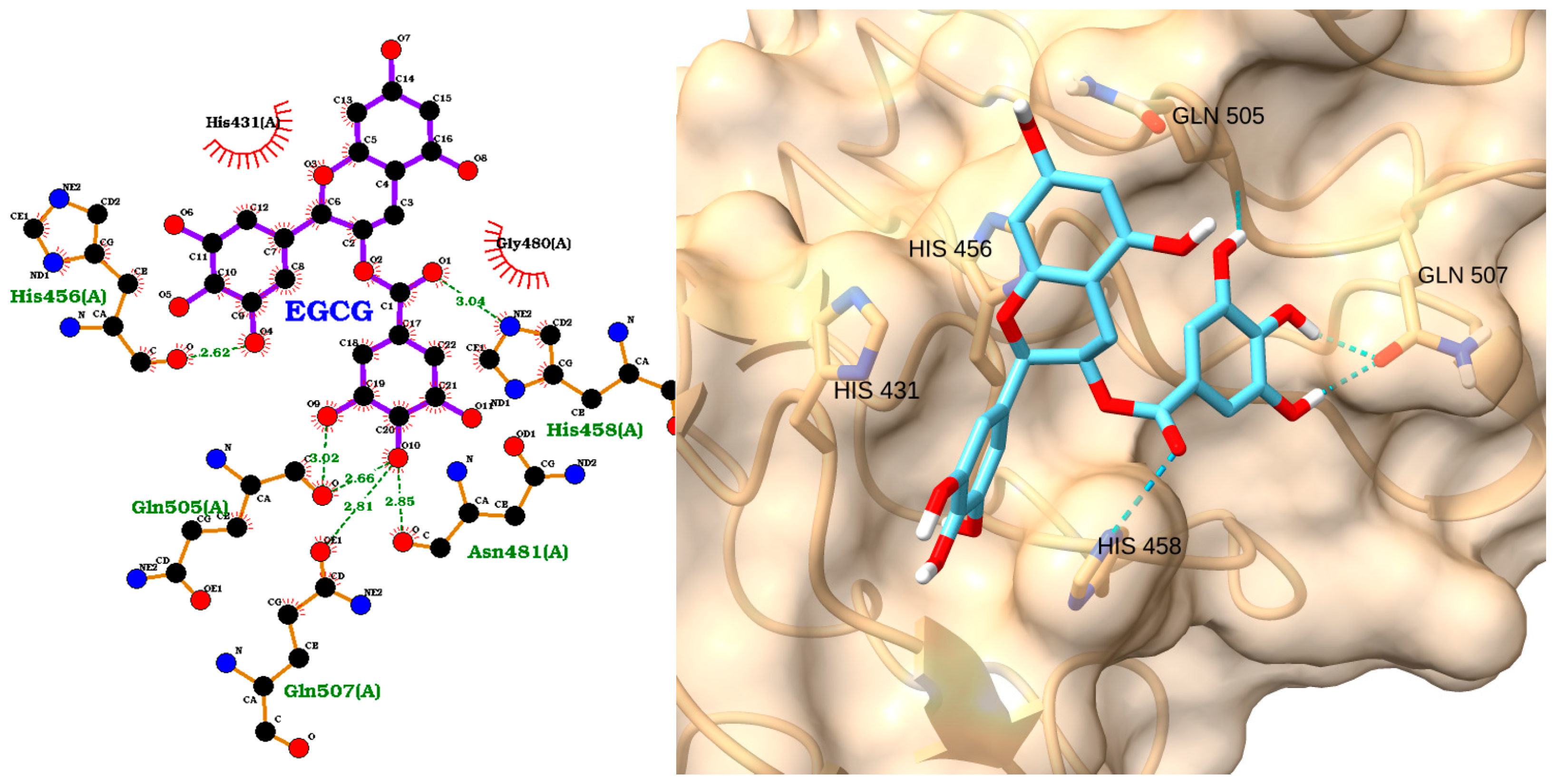
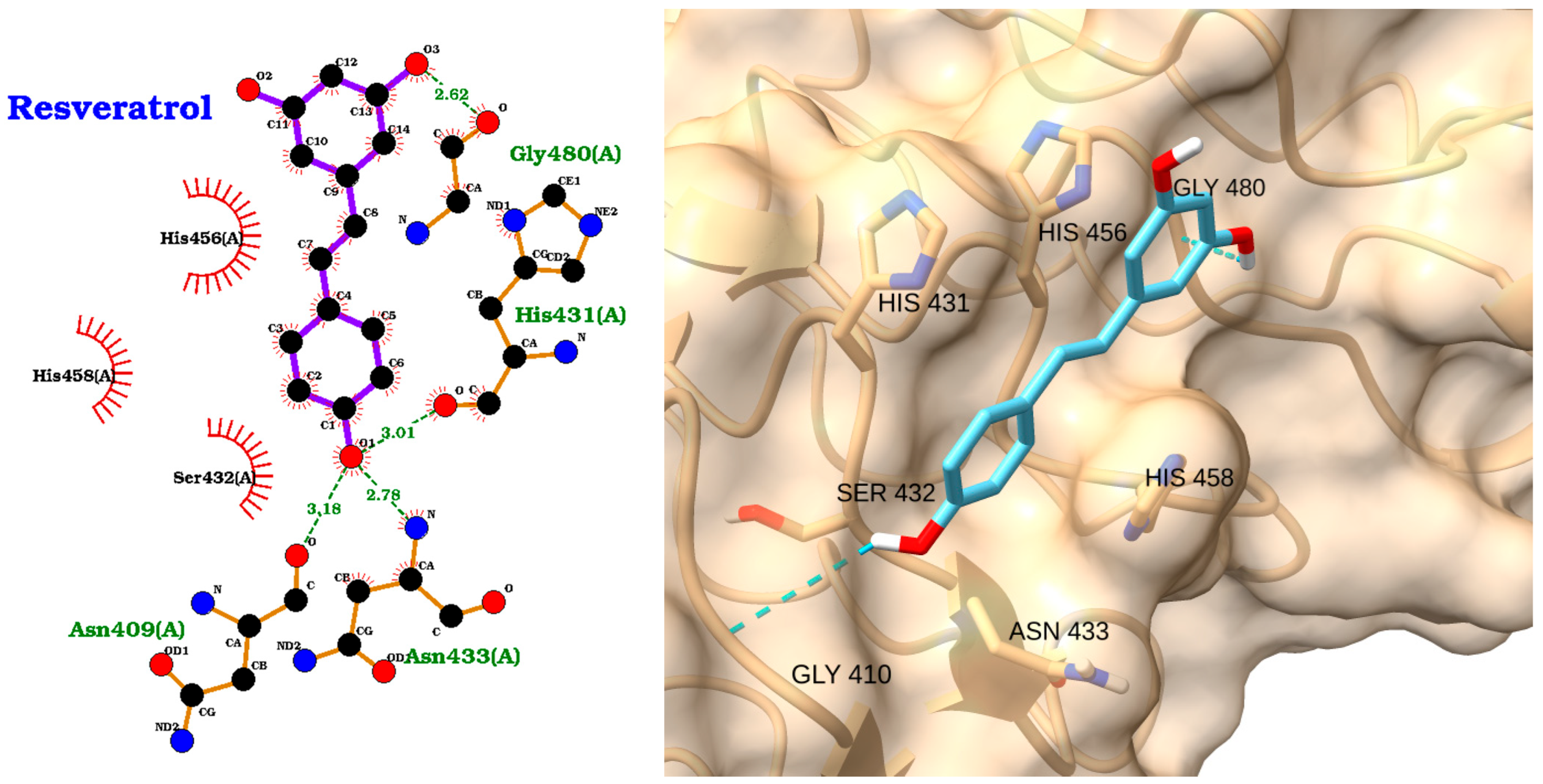
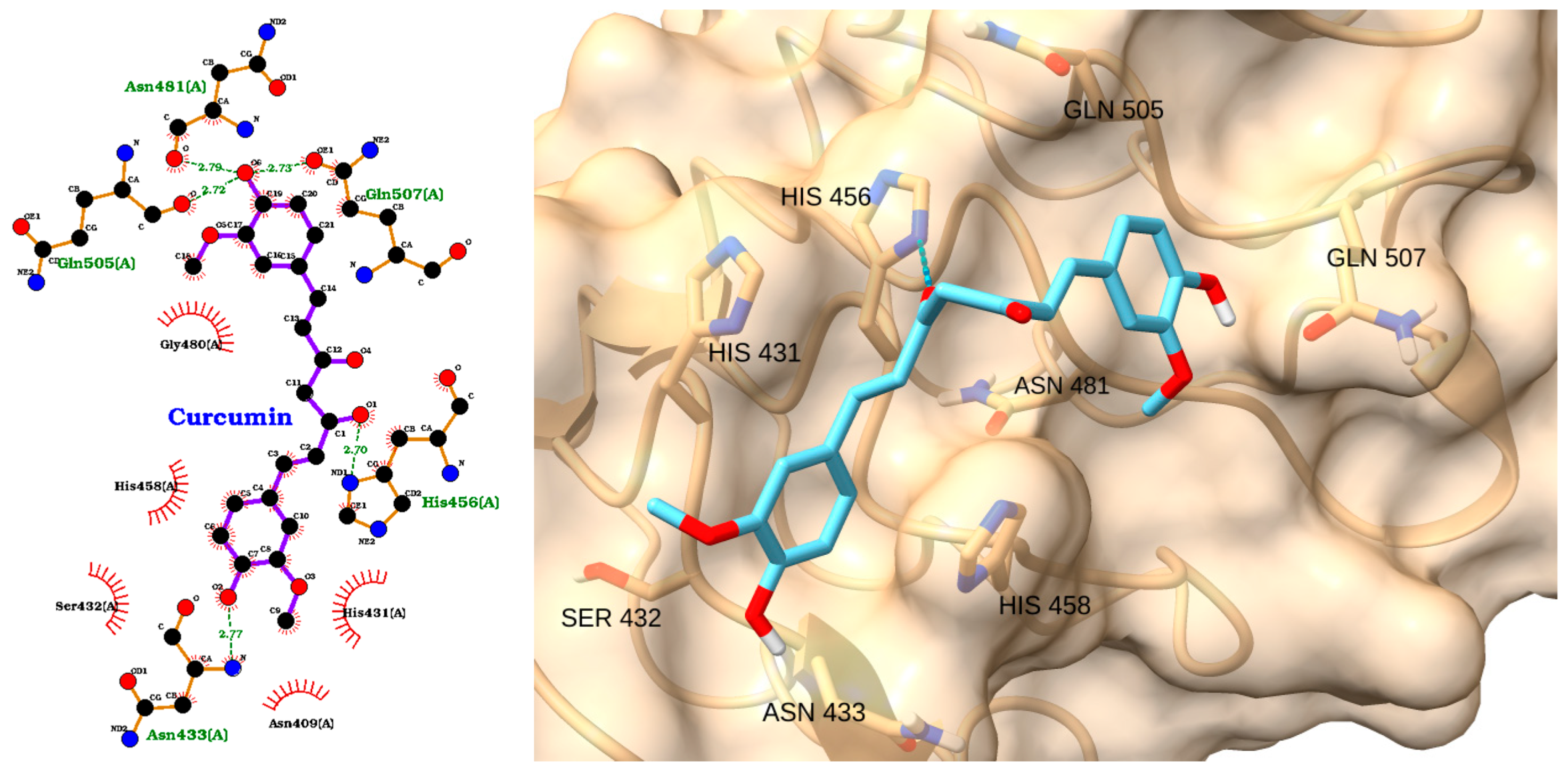
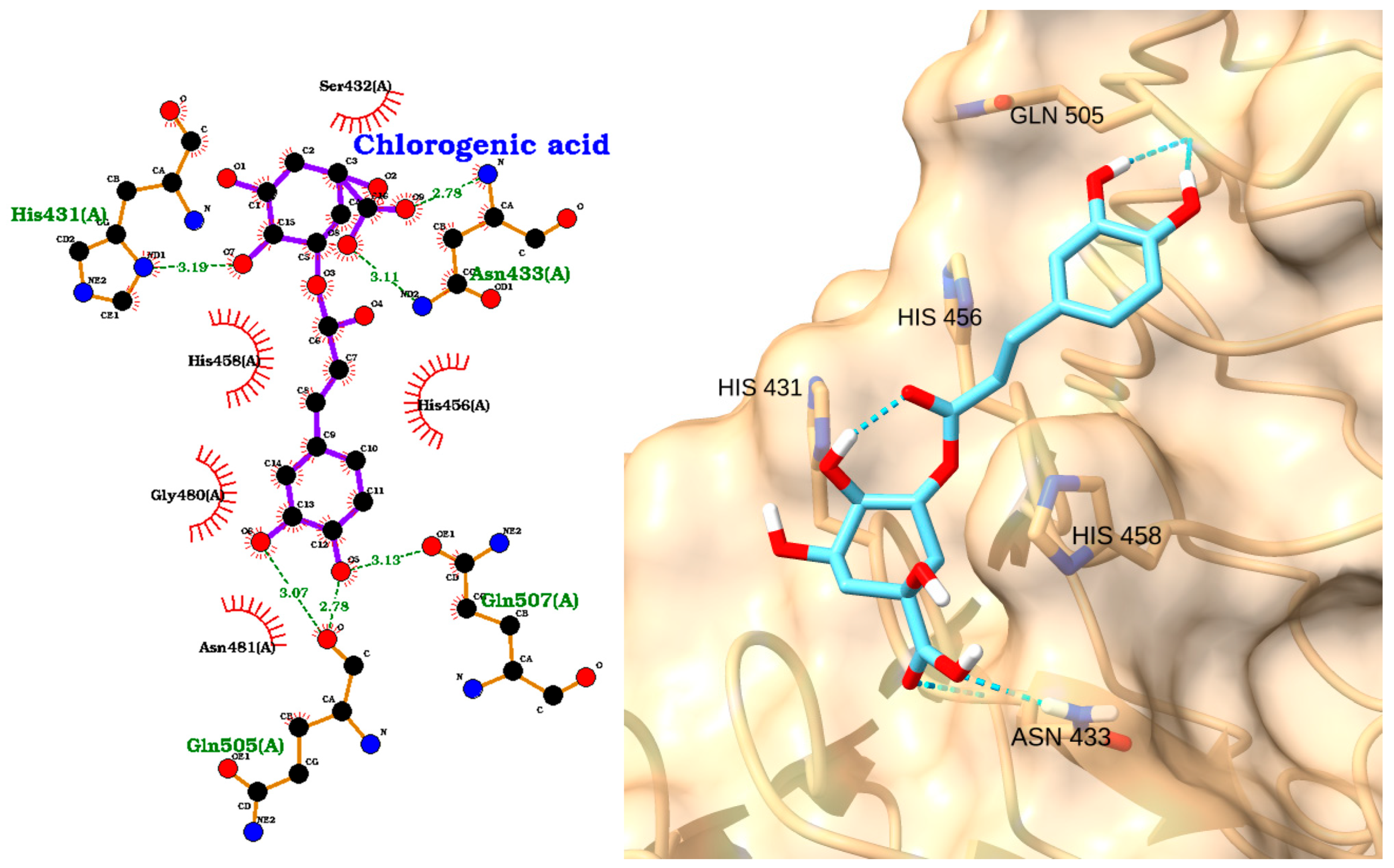
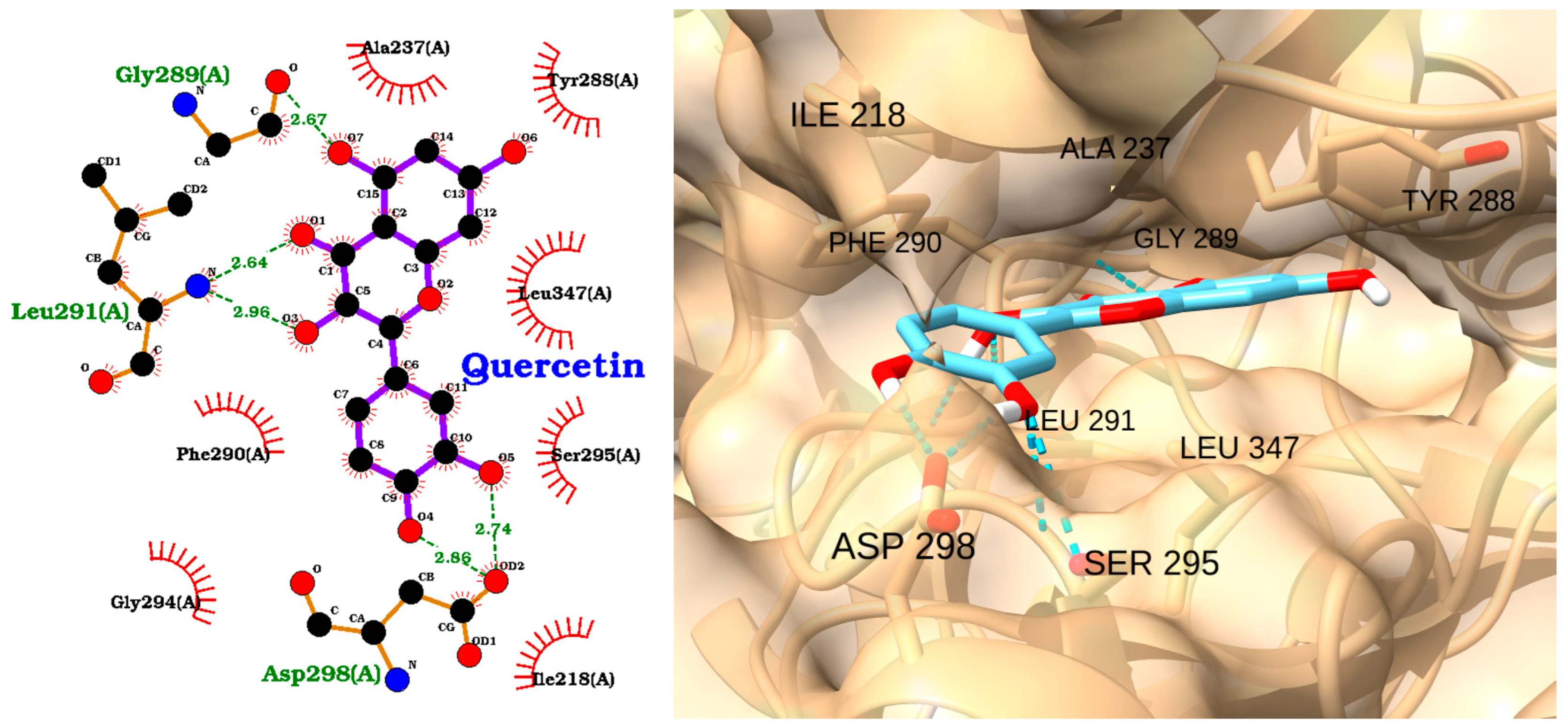

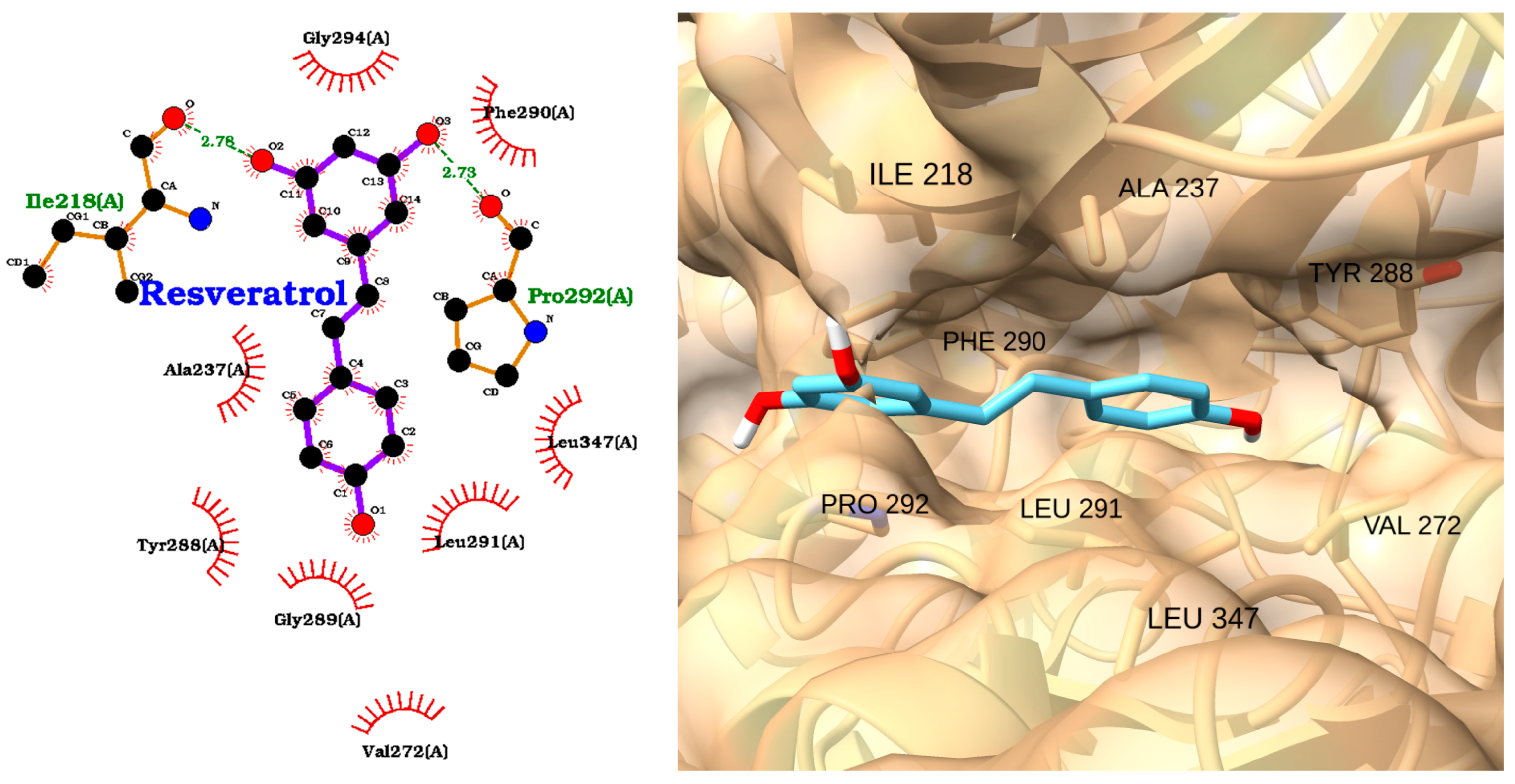
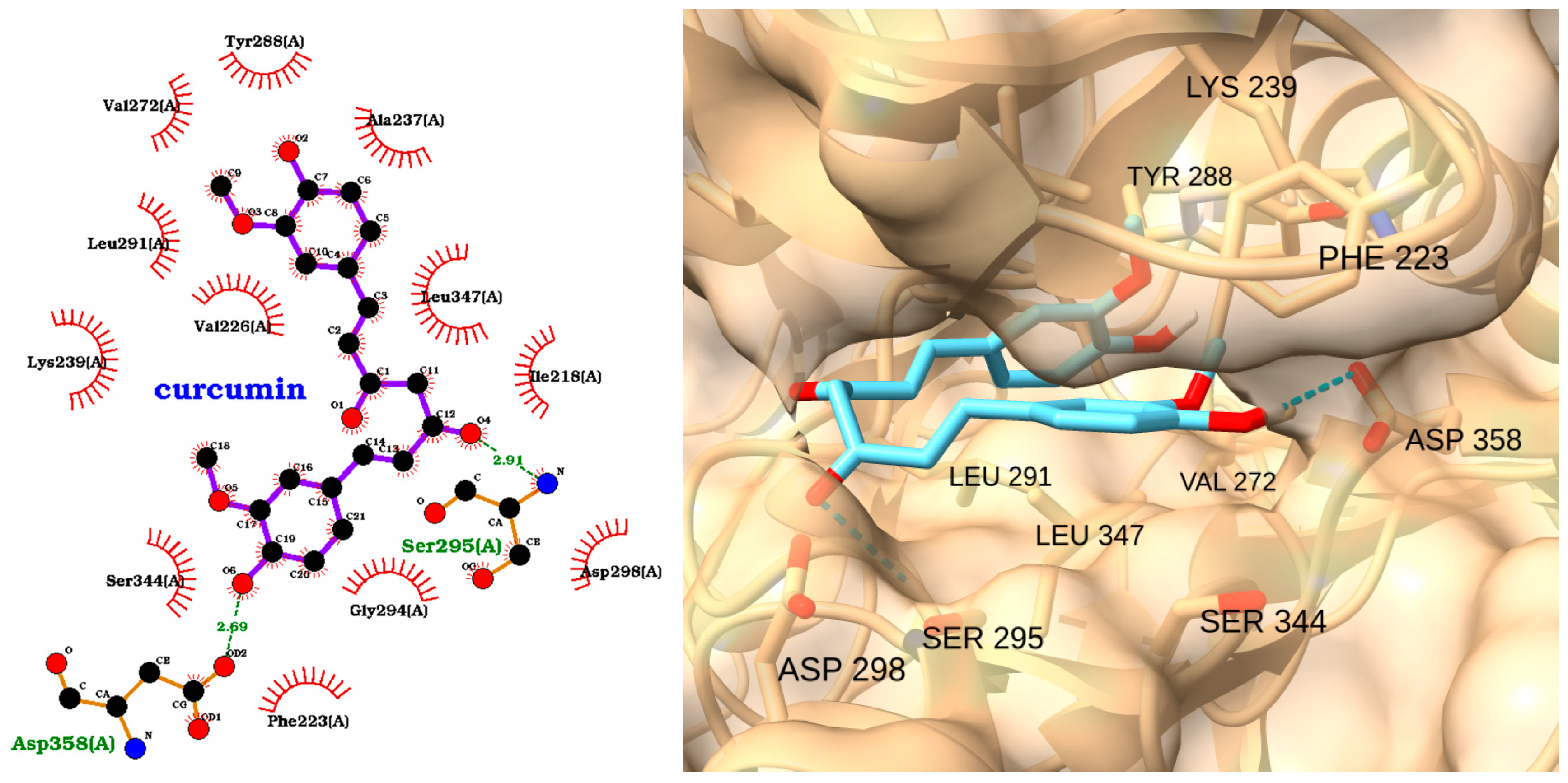
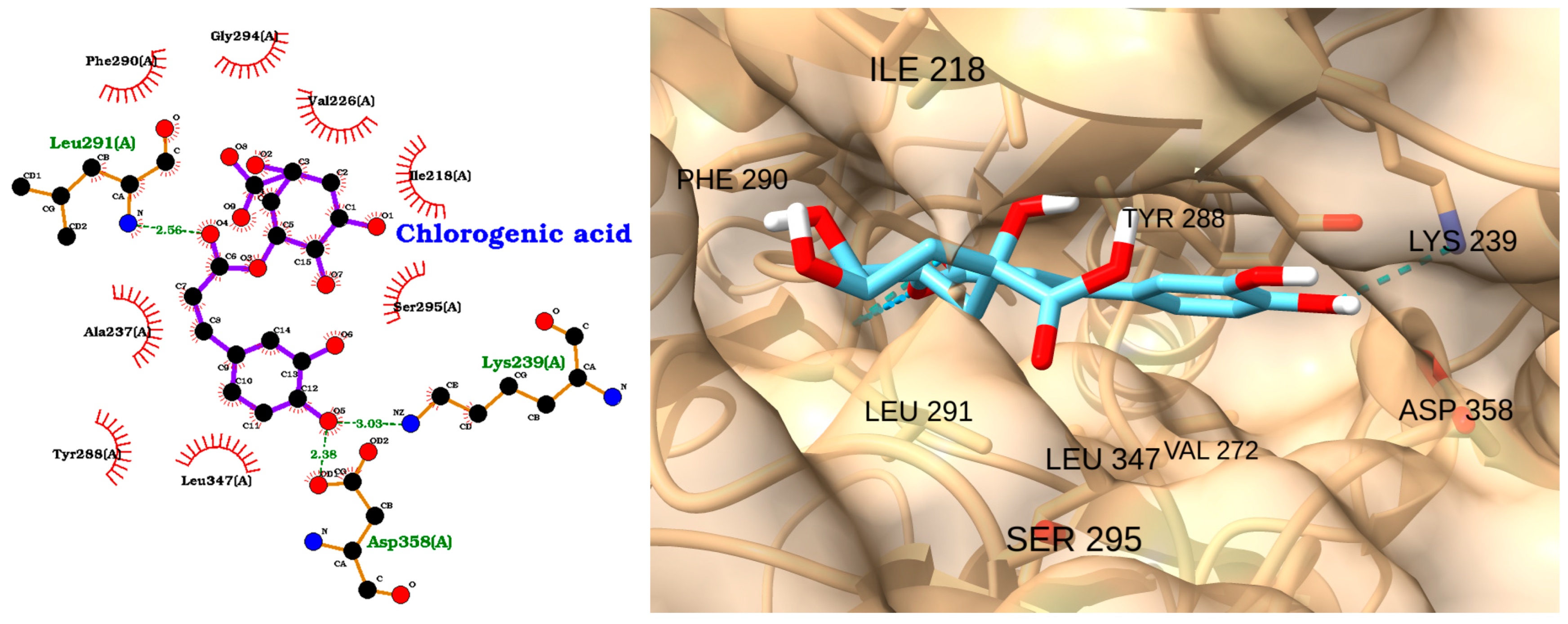
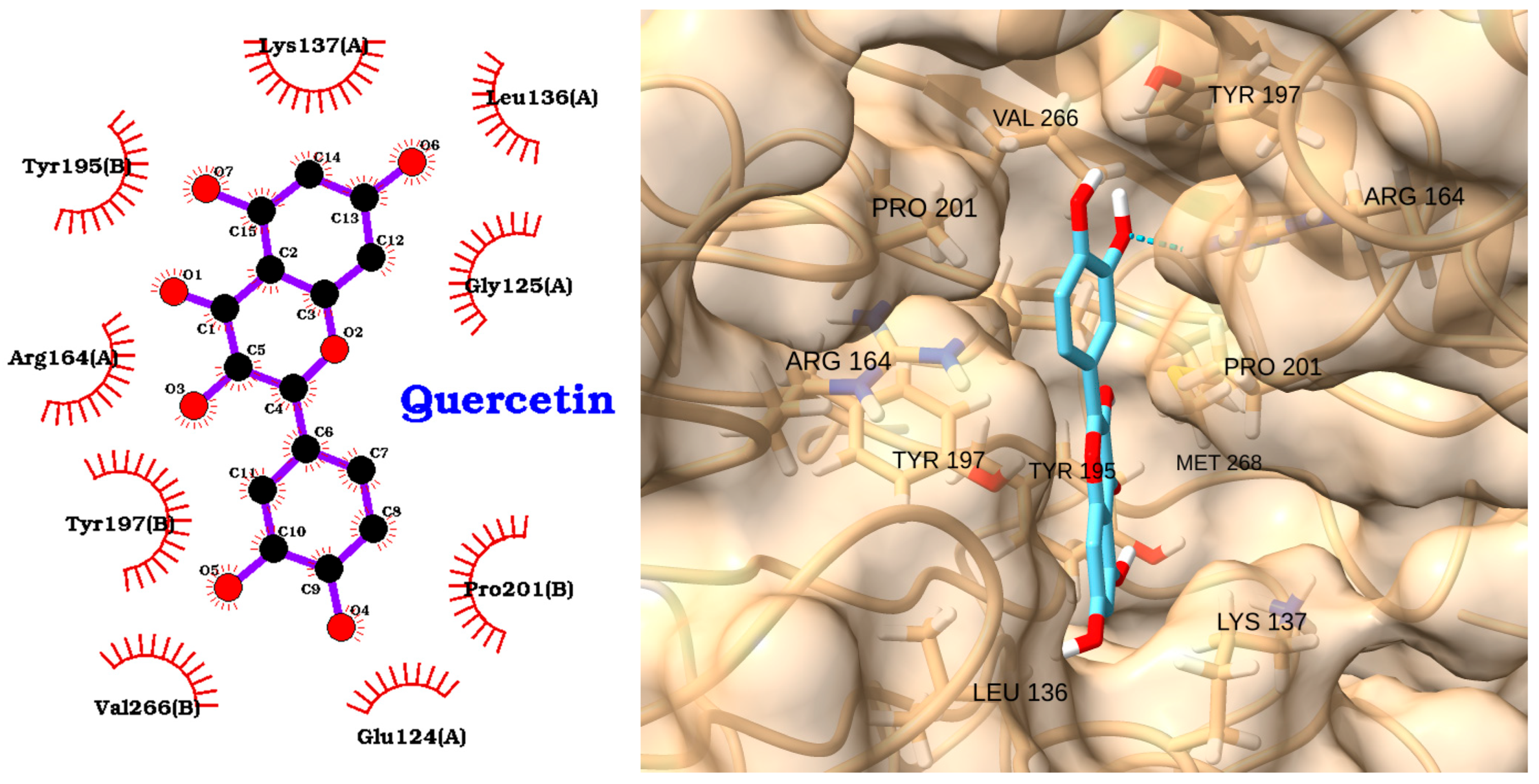

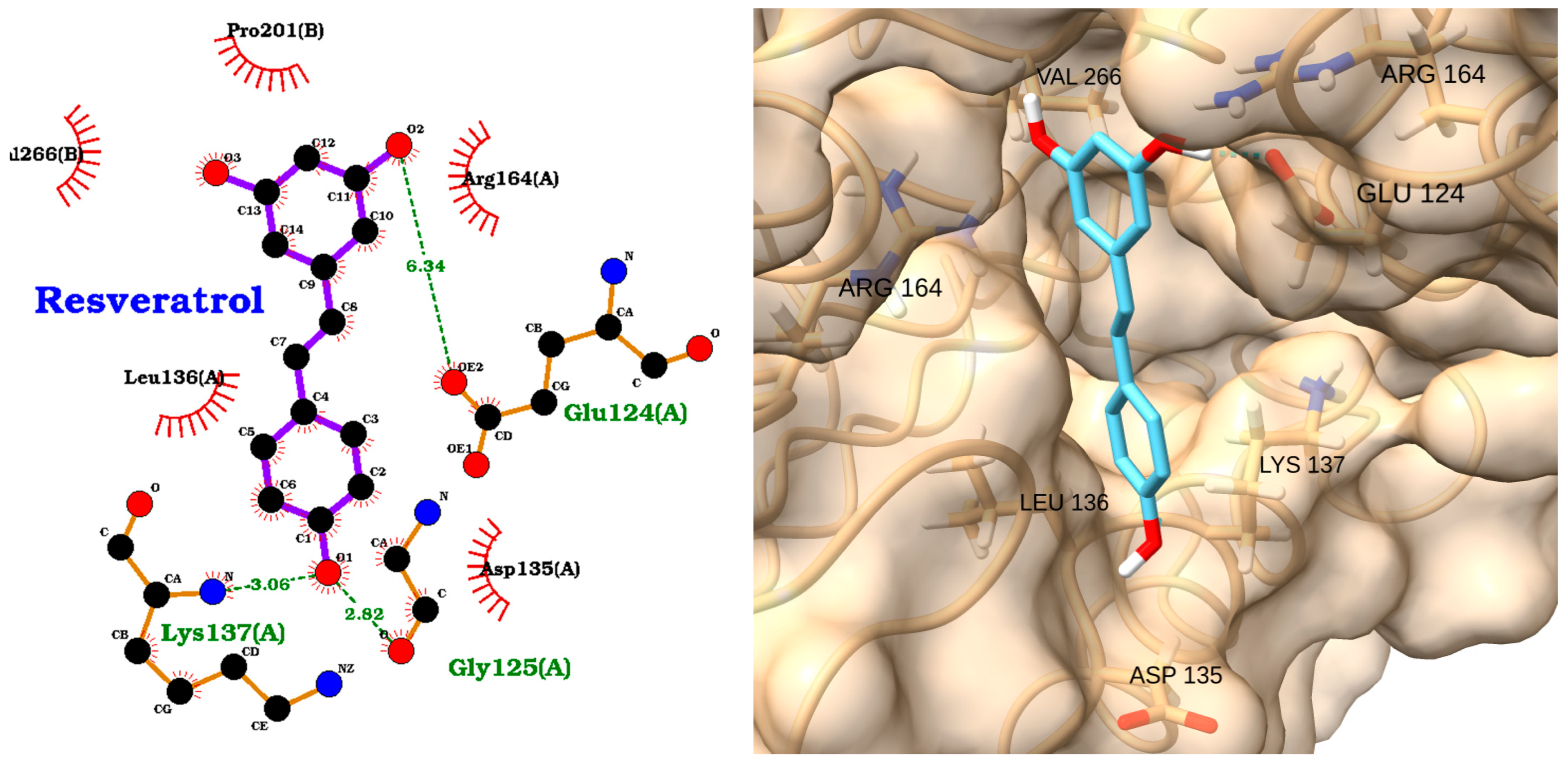
| Receptor | Uniprot ID | Role in Sepsis | Mechanism Represented |
|---|---|---|---|
| TLR4 | O00206 | Innate immunity | Recognition of LPS, initiating immune response |
| IRAK1 | P51617 | Inflammation | Involved in TLR and IL-1R signaling pathways |
| Caspase-3 | P42574 | Apoptosis | Key enzyme in programmed cell death |
| NAO | CID | Potential Role in Sepsis-Related Diseases | Clinical Trial |
|---|---|---|---|
| Quercetin | 5280343 | Chronic obstructive pulmonary disease | NCT01708278 |
| EGCG | 65064 | Prophylaxis of influenza infection | NCT01008020 |
| Resveratrol | 445154 | Inflammation and oxidative stress in chronic kidney disease | NCT02433925 |
| Curcumin | 969516 | Modulating gut microbiota, reducing endotoxemia | NCT03329781 |
| Chlorogenic acid | 1794427 | Renal insufficiency | NCT02524938 |
| Ligand | Score | RMSD |
|---|---|---|
| Quercetin | −5.20 | 2.279 |
| EGCG | −5.15 | 0.3575 |
| Resveratrol | −4.60 | 2.122 |
| Curcumin | −4.46 | 1.455 |
| Chlorogenic_acid | −3.76 | 2.655714 |
| CHEMBL5174883 (Positive) | −4.22 | 1.552 |
| Carvedilol (Negative) | −3.41 | 2.81 |
| Ligand | Score | RMSD |
|---|---|---|
| Quercetin | −8.18 | 0.269 |
| EGCG | −8.86 | 1.8475 |
| Resveratrol | −6.72 | 0.379 |
| Curcumin | −9.05 | 2 |
| Chlorogenic_acid | −7.14 | 2.1275 |
| DL1 ligand in PDB | −9.30 | 0.792 |
| Ligand | Score | RMSD |
|---|---|---|
| Quercetin | −5.66 | 1.35125 |
| EGCG | −6.22 | 1.786667 |
| Resveratrol | −5.49 | 0.156 |
| Curcumin | −6.23 | 2.71 |
| Chlorogenic_acid | −5.28 | 1.926667 |
| CHEMBL456799 (positive) | −7.39 | 2.908 |
| CHEMBL1242700 (negative) | −5.59 | 3.9875 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrés, C.M.C.; Munguira, E.B.; Juan, C.A.; Lobo, F.; Pérez-Lebeña, E.; Pérez de la Lastra, J.M. In Silico Exploration of Natural Antioxidants for Sepsis Drug Discovery. Molecules 2025, 30, 2288. https://doi.org/10.3390/molecules30112288
Andrés CMC, Munguira EB, Juan CA, Lobo F, Pérez-Lebeña E, Pérez de la Lastra JM. In Silico Exploration of Natural Antioxidants for Sepsis Drug Discovery. Molecules. 2025; 30(11):2288. https://doi.org/10.3390/molecules30112288
Chicago/Turabian StyleAndrés, Celia María Curieses, Elena Bustamante Munguira, Celia Andrés Juan, Fernando Lobo, Eduardo Pérez-Lebeña, and José Manuel Pérez de la Lastra. 2025. "In Silico Exploration of Natural Antioxidants for Sepsis Drug Discovery" Molecules 30, no. 11: 2288. https://doi.org/10.3390/molecules30112288
APA StyleAndrés, C. M. C., Munguira, E. B., Juan, C. A., Lobo, F., Pérez-Lebeña, E., & Pérez de la Lastra, J. M. (2025). In Silico Exploration of Natural Antioxidants for Sepsis Drug Discovery. Molecules, 30(11), 2288. https://doi.org/10.3390/molecules30112288






