Ionization of DNA Nucleotides in Explicit Solution
Abstract
1. Introduction
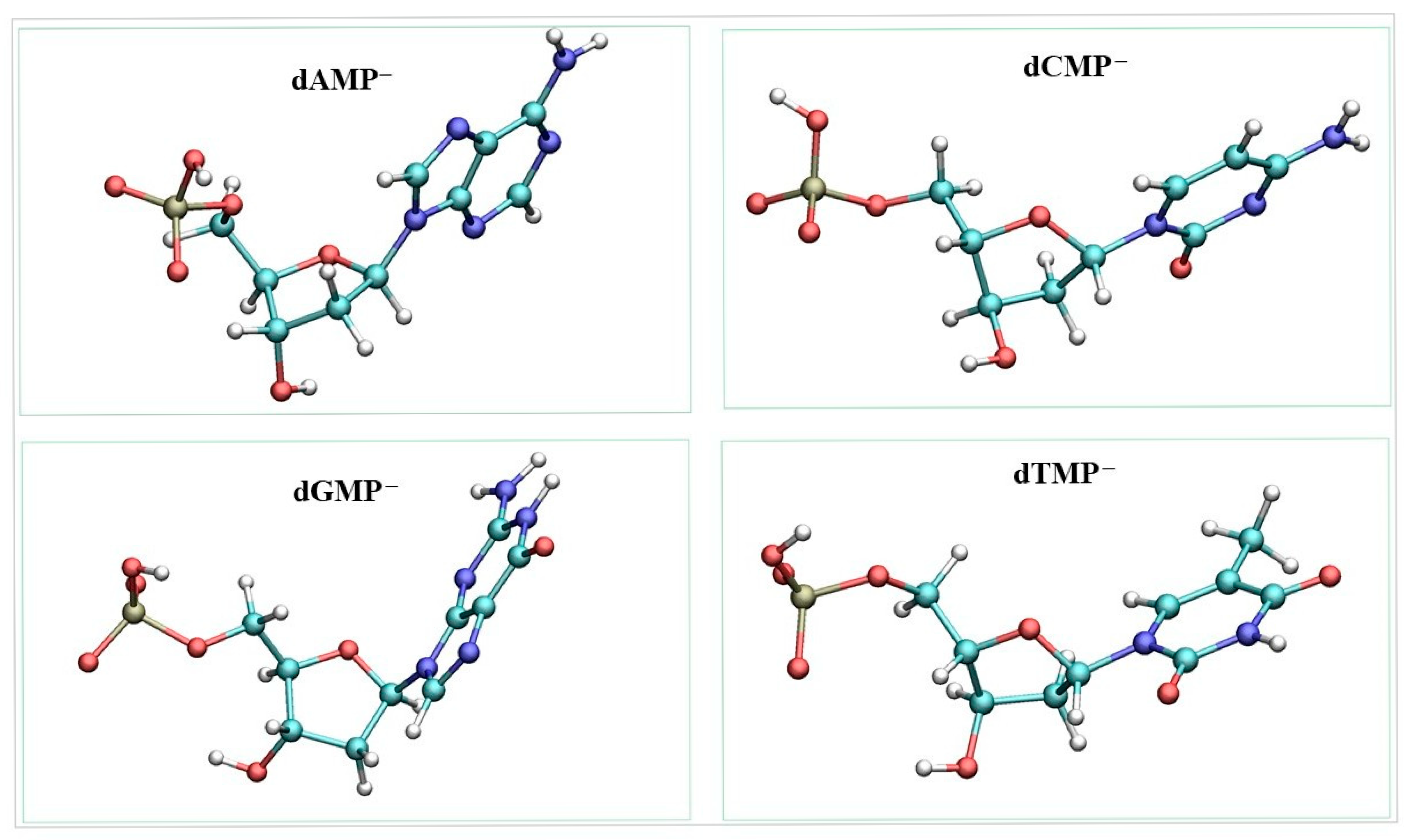
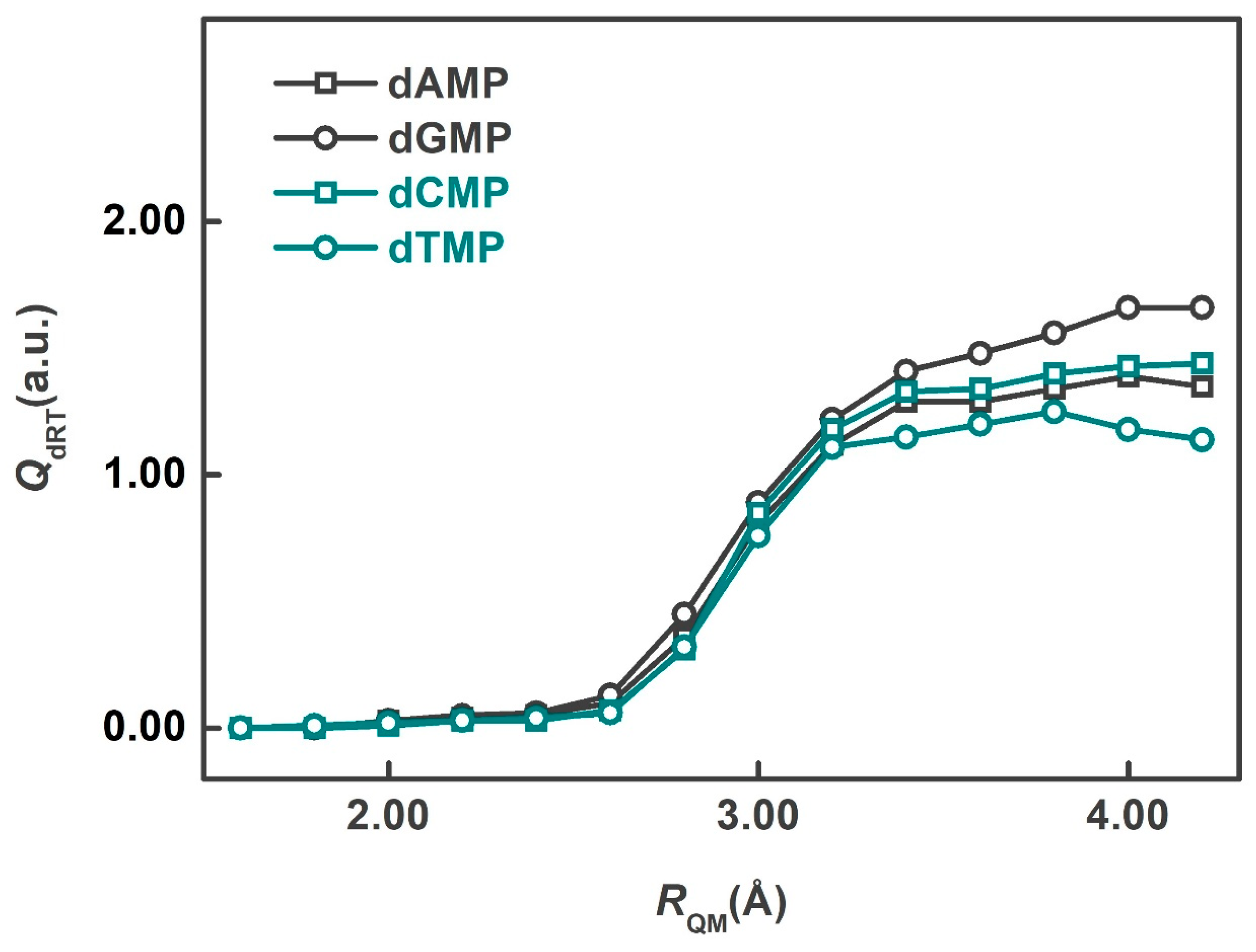
2. Results and Discussion
3. Computational Details
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ward, J.F. DNA damage produced by ionizing radiation in mammalian cells: Identities, mechanisms of formation, and reparability. Prog. Nucleic Acid Res. Mol. Biol. 1988, 35, 95–125. [Google Scholar]
- Måsson, E.P.; Camillis, S.D.; Castrovilli, M.C.; Galli, M.; Nisoli, M.; Calegari, F.; Greenwood, J.B. Ultrafast dynamics in the DNA building blocks thymidine and thymine initiated by ionizing radiation. Phys. Chem. Chem. Phys. 2017, 19, 19815–19821. [Google Scholar] [CrossRef] [PubMed]
- Pluhařová, E.; Slavíček, P.; Jungwirth, P. Modeling photoionization of aqueous DNA and its components. Acc. Chem. Res. 2015, 48, 1209–1217. [Google Scholar] [CrossRef] [PubMed]
- Russo, N.; Toscano, M.; Grand, A. Theoretical determination of electron affinity and ionization potential of DNA and RNA bases. J. Comput. Chem. 2000, 21, 1243–1250. [Google Scholar] [CrossRef]
- Kostko, O.; Bravaya, K.; Krylov, A.; Ahmed, M. Ionization of cytosine monomer and dimer studied by VUV photoionization and electronic structure calculations. Phys. Chem. Chem. Phys. 2010, 12, 2860–2872. [Google Scholar] [CrossRef]
- Chakraborty, R.; Ghosh, D. The effect of sequence on the ionization of guanine in DNA. Phys. Chem. Chem. Phys. 2016, 18, 6526–6533. [Google Scholar] [CrossRef]
- Lin, J.; Yu, C.; Peng, S.; Akiyama, I.; Li, K.; Lee, L.K.; LeBreton, P.R. Ultraviolet photoelectron studies of the ground-state electronic structure and gas-phase tautomerism of purine and adenine. J. Am. Chem. Soc. 1980, 102, 4627–4631. [Google Scholar] [CrossRef]
- Choi, K.-W.; Lee, J.-H.; Kim, S.K. Ionization spectroscopy of a DNA base: Vacuum-ultraviolet mass-analyzed threshold ionization spectroscopy of jet-cooled thymine. J. Am. Chem. Soc. 2005, 127, 15674–15675. [Google Scholar] [CrossRef]
- Bravaya, K.B.; Kostko, O.; Dolgikh, S.; Landau, A.; Ahmed, M.; Krylov, A.I. Electronic structure and spectroscopy of nucleic acid bases: Ionization energies, ionization-induced structural changes, and photoelectron spectra. J. Phys. Chem. A 2010, 114, 12305–12317. [Google Scholar] [CrossRef]
- Chen, Z.; Lau, K.-C.; Garcia, G.A.; Nahon, L.; Bozanić, D.K.; Poisson, L.; Al-Mogren, M.M.; Schwell, M.; Francisco, J.S.; Bellili, A.; et al. Identifying cytosine-specific isomers via high-accuracy single photon ionization. J. Am. Chem. Soc. 2016, 138, 16596–16599. [Google Scholar] [CrossRef]
- Cauët, E.; Valiev, M.; Weare, J.H. Vertical ionization potentials of nucleobases in a fully solvated dna environment. J. Phys. Chem. B 2010, 114, 5886–5894. [Google Scholar] [CrossRef]
- Ghosh, D.; Isayev, O.; Slipchenko, L.V.; Krylov, A.I. Effect of solvation on the vertical ionization energy of thymine: From microhydration to bulk. J. Phys. Chem. A 2011, 115, 6028–6038. [Google Scholar] [CrossRef]
- Muñoz-Losa, A.; Markovitsi, D.; Improta, R. A state-specific PCM–DFT method to include dynamic solvent effects in the calculation of ionization energies: Application to DNA bases. Chem. Phys. Lett. 2015, 634, 20–24. [Google Scholar] [CrossRef]
- Zhang, Y.; Xie, P.; Yang, S.; Han, K. Ionization and electron attachment for the nucleobases in water. J. Phys. Chem. B 2019, 123, 1237–11247. [Google Scholar] [CrossRef]
- Sun, L.; Bu, Y. Oxidative damage to DNA: Theoretical determination of ionization potential of deoxyriboguanosine (dG)–deoxyribocytidine (dC) and proton transfer in its cation. J. Mol. Struc. THEOCHEM 2009, 909, 25–32. [Google Scholar] [CrossRef]
- Pluhařová, E.; Jungwirth, P.; Bradforth, S.E.; Slavíček, P. Ionization of purine tautomers in nucleobases, nucleosides, and nucleotides: From the gas phase to the aqueous environment. J. Phys. Chem. B 2011, 115, 1294–1305. [Google Scholar] [CrossRef]
- Palivec, V.; Pluhařová, E.; Unger, I.; Winter, B.; Jungwirth, P. DNA lesion can facilitate base ionization: Vertical ionization energies of aqueous 8-oxoguanine and its nucleoside and nucleotide. J. Phys. Chem. B 2014, 118, 13833–13837. [Google Scholar] [CrossRef]
- Chakraborty, R.; Bose, S.; Ghosh, D. Effect of solvation on the ionization of guanine nucleotide: A hybrid QM/EFP study. J. Comput. Chem. 2017, 38, 2528–2537. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Fernandez, L.; Muñoz-Losa, A.; Esposito, L.; Improta, R. The optical properties of adenine cation in different oligonucleotides: A PCM/TD-DFT study. Theor. Chem. Acc. 2018, 137, 39. [Google Scholar] [CrossRef]
- Diamantis, P.; Tavernelli, I.; Rothlisberger, U. Vertical ionization energies and electron affinities of native and damaged DNA bases, nucleotides, and pairs from density functional theory calculations: Model assessment and implications for DNA damage recognition and repair. J. Chem. Theory Comput. 2019, 15, 2042–2052. [Google Scholar] [CrossRef]
- Uddin, I.A.; Stec, E.; Papadantonakis, G.A. Ionization patterns and chemical reactivity of cytosine-guanine watson-crick pairs. ChemPhysChem 2024, 25, e202300946. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Adhikary, A.; Sevilla, M.D.; Close, D.M. One-electron oxidation of ds(5′-GGG-3′) and ds(5′-G(8OG)G-3′) and the nature of hole distribution: A density functional theory (DFT) study. Phys. Chem. Chem. Phys. 2020, 22, 5078–5089. [Google Scholar] [CrossRef]
- Lucia-Tamudo, J.; Díaz-Tendero, S.; Nogueira, J.J. Intramolecular and intermolecular hole delocalization rules the reducer character of isolated nucleobases and homogeneous single-stranded DNA. Phys. Chem. Chem. Phys. 2023, 25, 14578–14589. [Google Scholar] [CrossRef] [PubMed]
- Diamantis, P.; Tavernelli, I.; Rothlisberger, U. Redox properties of native and damaged DNA from mixed quantum mechanical/molecular mechanics molecular dynamics simulations. J. Chem. Theory Comput. 2020, 16, 6690–6701. [Google Scholar] [CrossRef]
- Mukherjee, M.; Haldar, S.; Dutta, A.K. Solvation effect on the vertical ionization energy of adenine-thymine base pair: From microhydration to bulk. Int. J. Quantum Chem. 2019, 22, e26127. [Google Scholar] [CrossRef]
- Tóth, Z.; Kubečka1, J.; Muchová, E.; Slavíček, P. Ionization energies in solutions with QM:QM approach. Phys. Chem. Chem. Phys. 2020, 22, 10550–10560. [Google Scholar] [CrossRef]
- Slavíček, P.; Winter, B.; Faubel, M.; Bradforth, S.E.; Jungwirth, P. Ionization energies of aqueous nucleic acids: Photoelectron spectroscopy of pyrimidine nucleosides and ab initio calculations. J. Am. Chem. Soc. 2009, 131, 6460–6467. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, C.A.; Pluhařová, E.; Seidel, R.; Schroeder, W.P.; Faubel, M.; Slavíček, P.; Winter, B.; Jungwirth, P.; Bradforth, S.E. Oxidation half-reaction of aqueous nucleosides and nucleotides via photoelectron spectroscopy augmented by ab initio calculations. J. Am. Chem. Soc. 2015, 137, 201–209. [Google Scholar] [CrossRef]
- Wang, J.; Yang, S.; Zhang, Y. One-electron oxidation and redox potential of nucleobases and deoxyribonucleosides computed by QM/MM simulations. Chem. Phys. Lett. 2020, 739, 136948. [Google Scholar] [CrossRef]
- Rubio, M.; Roca-Sanjuán, D.; Merchán, M.; Serrano-Andrés, L. Determination of the lowest-energy oxidation site in nucleotides: 2′-deoxythymidine 5′-monophosphate anion. J. Phys. Chem. B 2006, 110, 10234–10235. [Google Scholar] [CrossRef]
- Rubio, M.; Roca-Sanjuán, D.; Serrano-Andrés, L.; Merchán, M. Determination of the electron-detachment energies of 2′-deoxyguanosine 5′-monophosphate anion: Influence of the conformation. J. Phys. Chem. B 2009, 113, 2451–2457. [Google Scholar] [CrossRef]
- Ma, J.; Denisov, S.A.; Marignier, J.-L.; Pernot, P.; Adhikary, A.; Seki, S.; Mostafavi, M. Ultrafast electron attachment and hole transfer following ionizing radiation of aqueous uridine monophosphate. J. Phys. Chem. Lett. 2018, 9, 5105–5109. [Google Scholar] [CrossRef] [PubMed]
- D’Annibale, V.; Nardi, A.N.; Amadei, A.; D’Abramo, M. Theoretical characterization of the reduction potentials of nucleic acids in solution. J. Chem. Theory Comput. 2021, 17, 1301–1307. [Google Scholar] [CrossRef] [PubMed]
- Sarangi, R.; Maity, S.; Acharya, A. Machine learning approach to vertical energy gap in redox processes. J. Chem. Theory Comput. 2024, 20, 6747–6755. [Google Scholar] [CrossRef] [PubMed]
- Brunk, E.; Rothlisberger, U. Mixed quantum mechanical/molecular mechanical molecular dynamics simulations of biological systems in ground and electronically excited states. Chem. Rev. 2015, 115, 6217–6263. [Google Scholar] [CrossRef]
- Close, D.M.; Crespo-Hernández, C.E.; Gorb, L.; Leszczynski, J. Ionization energy thresholds of microhydrated adenine and its tautomers. J. Phys. Chem. A 2008, 112, 12702–12706. [Google Scholar] [CrossRef]
- Khistyaev, K.; Bravaya, K.B.; Kamarchik, E.; Kostko, O.; Ahmed, M.; Krylov, A.I. The effect of microhydration on ionization energies of thymine. Faraday Discuss. 2011, 150, 313–330. [Google Scholar] [CrossRef]
- Brooks, B.R.; Brooks, C.L., III; Mackerell, A.D.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1615. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Sherwood, P.; de Vries, A.H.; Guest, M.F.; Schreckenbach, G.; Catlow, C.R.A.; French, S.A.; Sokol, A.A.; Bromley, S.T.; Thiel, W.; Turner, A.J.; et al. QUASI: A general purpose implementation of the QM/MM approach and its application to problems in catalysis. J. Mol. Struct.-THEOCHEM 2003, 632, 1–28. [Google Scholar] [CrossRef]
- TURBOMOLE V6.4; a Development of University of Karlsruhe and Forschungszentrum Karlsruhe GmbH.; TURBOMOLE GmbH; 2012. Available online: http://www.turbomole.com (accessed on 8 May 2025).
- Roca-Sanjuán, D.; Rubio, M.; Merchán, M.; Serrano-Andrés, L. Ab initio determination of the ionization potentials of DNA and RNA nucleobases. J. Chem. Phys. 2006, 125, 084302. [Google Scholar] [CrossRef] [PubMed]
- Singh, U.C.; Kollman, P.A. An approach to computing electrostatic charges for molecules. J. Comput. Chem. 1984, 5, 129–145. [Google Scholar] [CrossRef]
- Martin, F.; Zipse, H. Charge distribution in the water molecule—A comparison of methods. J. Comput. Chem. 2005, 26, 97–105. [Google Scholar] [CrossRef]
- Jacquemin, D.; Bahers, T.L.; Adamobc, C.; Ciofini, I. What is the “best” atomic charge model to describe through-space charge-transfer excitations? Phys. Chem. Chem. Phys. 2012, 14, 5383–5388. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xie, P.; He, X.; Han, K. High-efficiency microiterative optimization in QM/MM simulations of large flexible systems. J. Chem. Theory Comput. 2016, 12, 4632–4643. [Google Scholar] [CrossRef]
- Kästner, J.; Carr, J.M.; Keal, T.W.; Thiel, W.; Wander, A.; Sherwood, P. DL-FIND: An open-source geometry optimizer for atomistic simulations. J. Phys. Chem. A 2009, 113, 11856–11865. [Google Scholar] [CrossRef]
| Nucleotides | Nucleosides | ||||
|---|---|---|---|---|---|
| QM/MM | Gas-QM | NEPCM 1 | Exp. 2 | QM/MM 3 | |
| dAMP− | 7.26 ± 0.08 | 5.68 ± 0.04 | 7.53 | 7.7 | 7.99 |
| dGMP− | 6.92 ± 0.07 | 5.51 ± 0.04 | 7.23 | n/a | 7.67 |
| dCMP− | 7.45 ± 0.07 | 5.69 ± 0.04 | 7.82 | n/a | 8.34 |
| dTMP− | 7.63 ± 0.06 | 5.95 ± 0.04 | 7.77 | n/a | 8.38 |
| dRT | Bases | Ribose | Phosphate | |||||
|---|---|---|---|---|---|---|---|---|
| QM/MM | Gas-QM | QM/MM | Gas-QM | QM/MM | Gas-QM | QM/MM | Gas-QM | |
| dAMP− | 0.73 ± 0.01 | 0.32 ± 0.02 | 0.62 ± 0.01 | 0.29 ± 0.02 | 0.08 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.01 ± 0.01 |
| dGMP− | 0.75 ± 0.01 | 0.40 ± 0.02 | 0.66 ± 0.01 | 0.37 ± 0.02 | 0.05 ± 0.01 | 0.02 ± 0.01 | 0.03 ± 0.01 | 0.02 ± 0.01 |
| dCMP− | 0.76 ± 0.01 | 0.24 ± 0.02 | 0.61 ± 0.01 | 0.20 ± 0.03 | 0.07 ± 0.01 | 0.04 ± 0.01 | 0.09 ± 0.01 | −0.01 ± 0.01 |
| dTMP− | 0.75 ± 0.01 | 0.25 ± 0.02 | 0.58 ± 0.01 | 0.21 ± 0.02 | 0.05 ± 0.01 | 0.02 ± 0.01 | 0.11 ± 0.01 | 0.02 ± 0.01 |
| QM/MM | pol-QM | gas-QM | PCM 1 | |
|---|---|---|---|---|
| dAMP− | 5.40 ± 0.08 | 5.78 ± 0.09 | 5.51 ± 0.06 | 6.19 |
| dGMP− | 4.94 ± 0.11 | 5.50 ± 0.09 | 5.18 ± 0.05 | 5.82 |
| dCMP− | 5.84 ± 0.06 | 6.25 ± 0.07 | 5.60 ± 0.05 | 6.51 |
| dTMP− | 5.86 ± 0.07 | 6.33 ± 0.09 | 5.82 ± 0.05 | 6.43 |
| dRT | Bases | Ribose | Phosphate | |
|---|---|---|---|---|
| dAMP− | 0.84 ± 0.02 | 0.71 ± 0.02 | 0.13 ± 0.03 | −0.00 ± 0.02 |
| dGMP− | 0.86 ± 0.03 | 0.77 ± 0.03 | 0.06 ± 0.03 | 0.04 ± 0.02 |
| dCMP− | 0.85 ± 0.03 | 0.76 ± 0.04 | 0.05 ± 0.04 | 0.04 ± 0.02 |
| dTMP− | 0.86 ± 0.02 | 0.80 ± 0.04 | 0.04 ± 0.03 | 0.01 ± 0.02 |
| QM/MM | Pol-QM | Gas-QM | |
|---|---|---|---|
| dAMP− | 1.86 ± 0.11 | 1.48 ± 0.10 | 0.17 ± 0.04 |
| dGMP− | 1.97 ± 0.15 | 1.42 ± 0.10 | 0.33 ± 0.04 |
| dCMP− | 1.62 ± 0.08 | 1.20 ± 0.08 | 0.09 ± 0.04 |
| dTMP− | 1.76 ± 0.09 | 1.29 ± 0.09 | 0.12 ± 0.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, J.; Zhang, Y.; Yin, S.; Che, L.; Yang, S. Ionization of DNA Nucleotides in Explicit Solution. Molecules 2025, 30, 2213. https://doi.org/10.3390/molecules30102213
Bai J, Zhang Y, Yin S, Che L, Yang S. Ionization of DNA Nucleotides in Explicit Solution. Molecules. 2025; 30(10):2213. https://doi.org/10.3390/molecules30102213
Chicago/Turabian StyleBai, Junhao, Yan Zhang, Shuhui Yin, Li Che, and Songqiu Yang. 2025. "Ionization of DNA Nucleotides in Explicit Solution" Molecules 30, no. 10: 2213. https://doi.org/10.3390/molecules30102213
APA StyleBai, J., Zhang, Y., Yin, S., Che, L., & Yang, S. (2025). Ionization of DNA Nucleotides in Explicit Solution. Molecules, 30(10), 2213. https://doi.org/10.3390/molecules30102213






