Chalkophomycin Biosynthesis Revealing Unique Enzyme Architecture for a Hybrid Nonribosomal Peptide Synthetase and Polyketide Synthase
Abstract
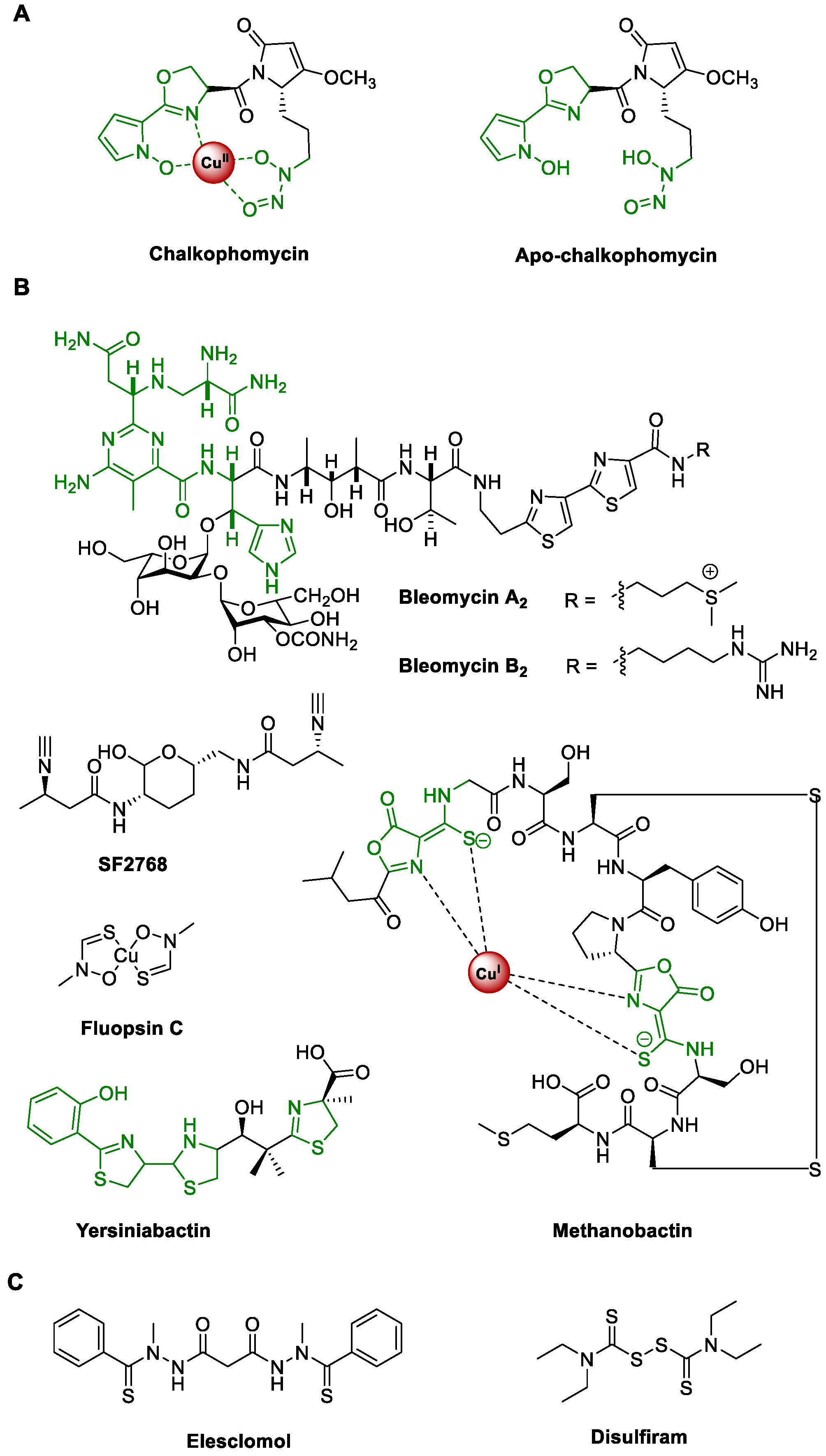
1. Introduction
2. Results and Discussion
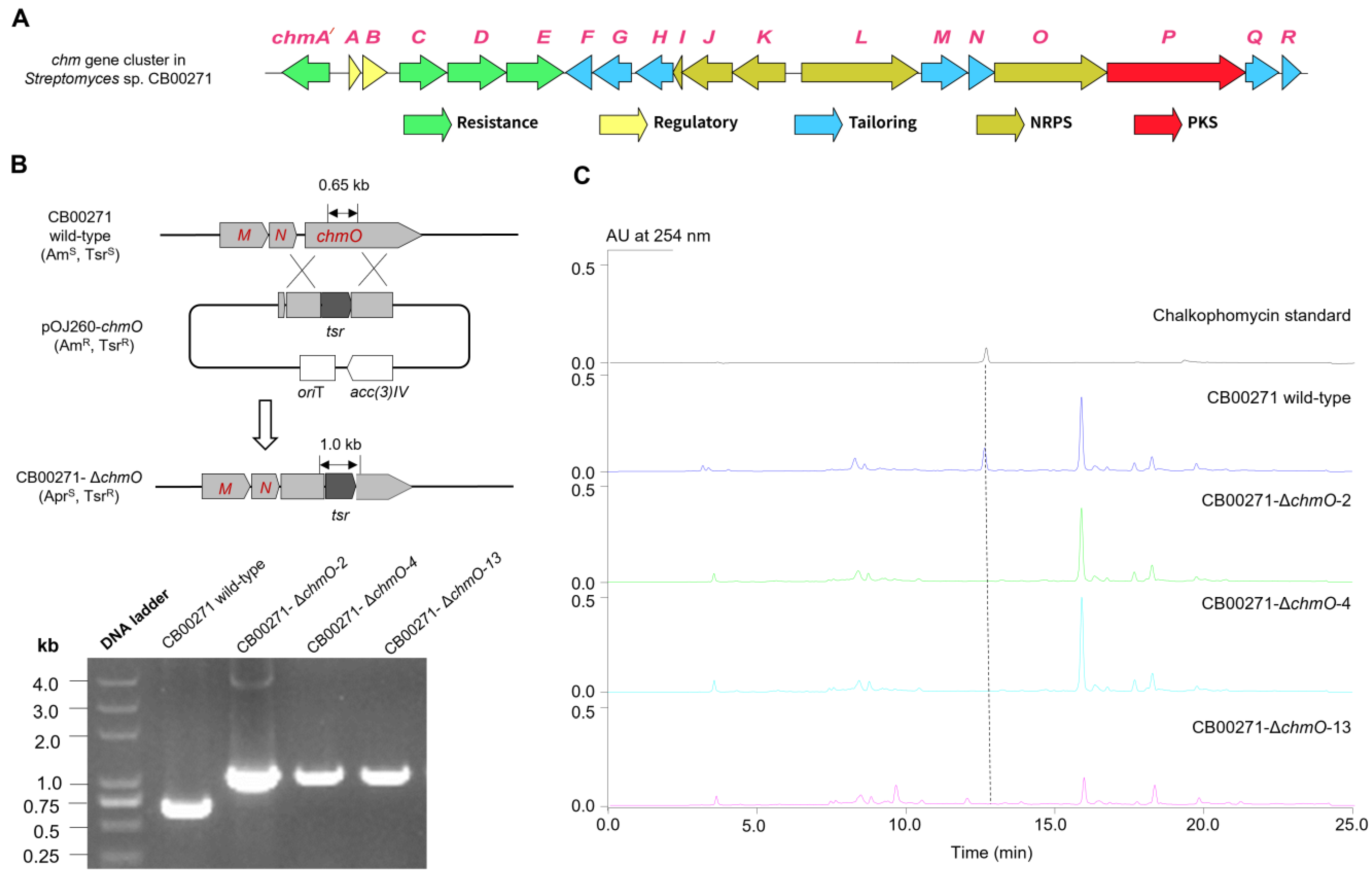
2.1. Discovery and Genetic Characterization of the chm Gene Cluster in S. sp. CB00271
2.2. Bioinformatics Analysis of the chm Cluster in S. sp. CB00271 Revealed a Hybrid NRPS/PKS for Chalkophomycin Biosynthesis
2.2.1. Overview of the chm Gene Cluster
2.2.2. Biosynthesis of NRPS Precursors N-Hydroxylpyrrole and l-Graminine
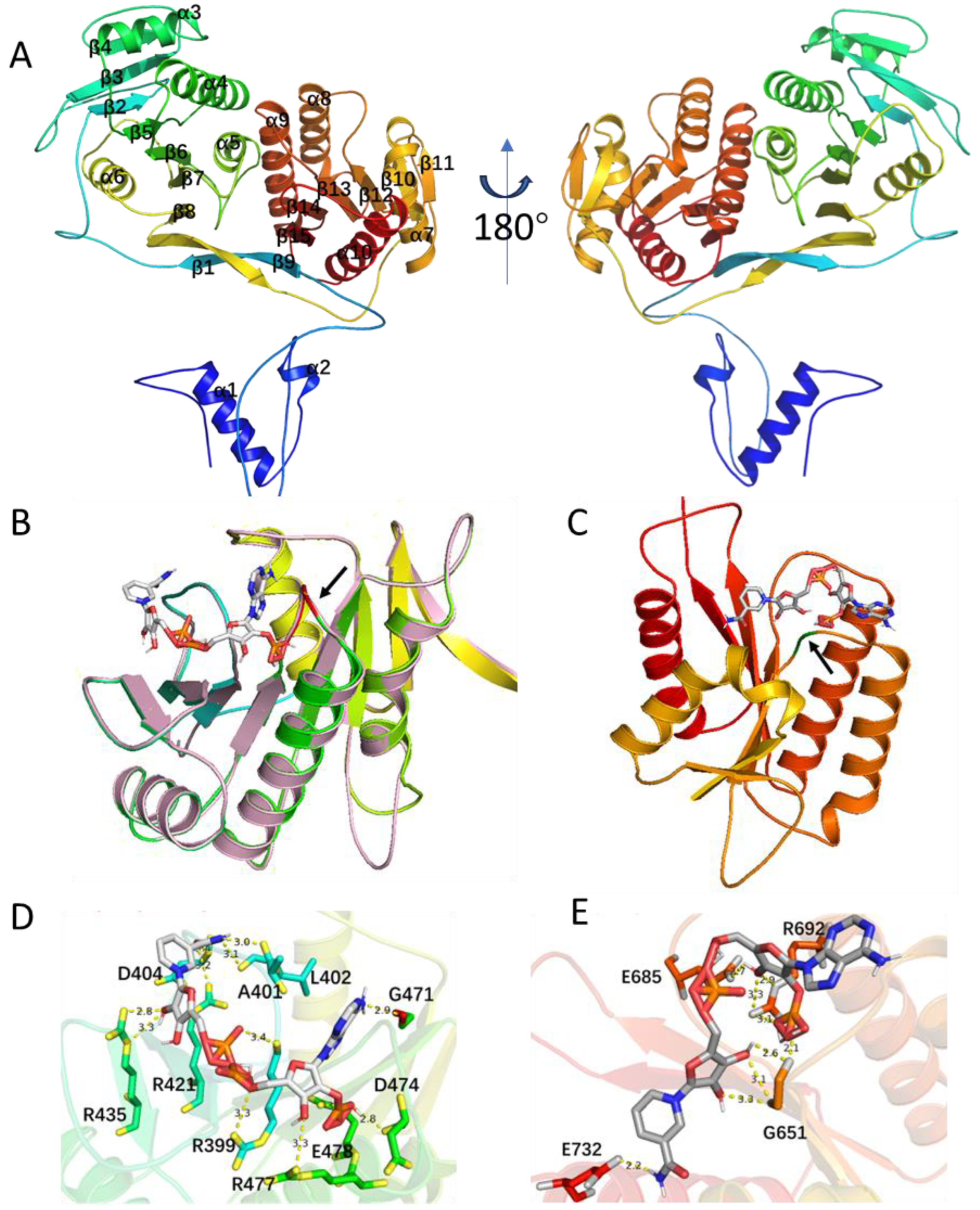
2.2.3. Chalkophomycin Biosynthesis by a Hybrid NRPS/PKS
2.2.4. Regulatory and Resistance Genes for Chalkophomycin Biosynthesis
2.3. Chalkophomycin-Type Gene Clusters Are Wide-Spread among Diverse Bacterial Strains
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Strains, Plasmids, and Culture Conditions
3.3. Fermentation Production and HPLC Analysis of Chalkophomycin
3.4. Gene Replacement of chmO in S. sp. CB00271
3.5. Structural Analysis of the ChmP_R0 Domains in S. sp. CB00271
3.6. Gene Cluster Similarity Network Analysis of chm Genes in Public Databases
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gong, B.; Bai, E.; Feng, X.; Yi, L.; Wang, Y.; Chen, X.; Zhu, X.; Duan, Y.; Huang, Y. Characterization of chalkophomycin, a copper(II) metallophore with an unprecedented molecular architecture. J. Am. Chem. Soc. 2021, 143, 20579–20584. [Google Scholar] [CrossRef]
- Schönewolf, M.; Rohr, J. Biogenesis of the carbon skeleton of glycerinopyrin: New biosynthetic pathway for pyrroles. Angew. Chem. Int. Ed. Engl. 1991, 30, 183–185. [Google Scholar] [CrossRef]
- Li, X.; Shimaya, R.; Dairi, T.; Chang, W.C.; Ogasawara, Y. Identification of cyclopropane formation in the biosyntheses of hormaomycins and belactosins: Sequential nitration and cyclopropanation by metalloenzymes. Angew. Chem. Int. Ed. Engl. 2022, 61, e202113189. [Google Scholar] [CrossRef]
- Sugiyama, Y.; Watanabe, K.; Hirota, A. Surugapyrroles A and B, two new-hydroxypyrroles, as DPPH radical-scavengers from Streptomyces sp. USF-6280 strain. Biosci. Biotechnol. Biochem. 2009, 73, 230–232. [Google Scholar] [CrossRef]
- Kallifidas, D.; Pascoe, B.; Owen, G.A.; Strain-Damerell, C.M.; Hong, H.J.; Paget, M.S. The zinc-responsive regulator Zur controls expression of the coelibactin gene cluster in Streptomyces coelicolor. J. Bacteriol. 2010, 192, 608–611. [Google Scholar] [CrossRef]
- Shyam, M.; Shilkar, D.; Verma, H.; Dev, A.; Sinha, B.N.; Brucoli, F.; Bhakta, S.; Jayaprakash, V. The mycobactin biosynthesis pathway: A prospective therapeutic target in the battle against tuberculosis. J. Med. Chem. 2021, 64, 71–100. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Niikura, H.; He, H.Y.; Daniel-Ivad, P.; Ryan, K.S. Biosynthesis of the N-N-bond-containing compound l-alanosine. Angew. Chem. Int. Ed. Engl. 2020, 59, 3881–3885. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.L.; McCallum, M.E.; Zheng, C.R.; Wang, J.X.; Wu, K.J.Y.; Balskus, E.P. The l-Alanosine gene cluster encodes a pathway for diazeniumdiolate biosynthesis. ChemBioChem 2020, 21, 1155–1160. [Google Scholar] [CrossRef]
- Jenul, C.; Sieber, S.; Daeppen, C.; Mathew, A.; Lardi, M.; Pessi, G.; Hoepfner, D.; Neuburger, M.; Linden, A.; Gademann, K.; et al. Biosynthesis of fragin is controlled by a novel quorum sensing signal. Nat. Commun. 2018, 9, 1297. [Google Scholar] [CrossRef]
- Hermenau, R.; Ishida, K.; Gama, S.; Hoffmann, B.; Pfeifer-Leeg, M.; Plass, W.; Mohr, J.F.; Wichard, T.; Saluz, H.P.; Hertweck, C. Gramibactin is a bacterial siderophore with a diazeniumdiolate ligand system. Nat. Chem. Biol. 2018, 14, 841–843. [Google Scholar] [CrossRef]
- Hermenau, R.; Mehl, J.L.; Ishida, K.; Dose, B.; Pidot, S.J.; Stinear, T.P.; Hertweck, C. Genomics-driven discovery of NO-donating diazeniumdiolate siderophores in diverse plant-associated bacteria. Angew. Chem. Int. Ed. Engl. 2019, 58, 13024–13029. [Google Scholar] [CrossRef] [PubMed]
- Makris, C.; Carmichael, J.R.; Zhou, H.; Butler, A. C-diazeniumdiolate graminine in the siderophore gramibactin is photoreactive and originates from arginine. ACS Chem. Biol. 2022, 17, 3140–3147. [Google Scholar] [CrossRef] [PubMed]
- Gerc, A.J.; Song, L.; Challis, G.L.; Stanley-Wall, N.R.; Coulthurst, S.J. The insect pathogen Serratia marcescens Db10 uses a hybrid non-ribosomal peptide synthetase-polyketide synthase to produce the antibiotic althiomycin. PLoS ONE 2012, 7, e44673. [Google Scholar] [CrossRef] [PubMed]
- Cortina, N.S.; Revermann, O.; Krug, D.; Müller, R. Identification and characterization of the althiomycin biosynthetic gene cluster in Myxococcus xanthus DK897. ChemBioChem 2011, 12, 1411–1416. [Google Scholar] [CrossRef] [PubMed]
- Pettit, G.R.; Yoshiaki, K.; Claude, D.; Cerny, R.L.; Herald, C.L.; Schmidt, J.M. Isolation and structure of the cytostatic linear depsipeptide dolastatin 15. J. Org. Chem. 1989, 54, 6005–6006. [Google Scholar] [CrossRef]
- Pettit, G.R.; Yoshiaki, K.; Claude, D.; Cerny, R.L.; Herald, C.L.; Schmidt, J.M. Malyngamide A, a novel chlorinated metabolite of the marine cyanophyte Lyngbya majuscule. J. Am. Chem. Soc. 1979, 101, 240–242. [Google Scholar]
- Kenney, G.E.; Rosenzweig, A.C. Chalkophores. Annu. Rev. Biochem. 2018, 87, 645–676. [Google Scholar] [CrossRef] [PubMed]
- Kenney, G.E.; Goering, A.W.; Ross, M.O.; DeHart, C.J.; Thomas, P.M.; Hoffman, B.M.; Kelleher, N.L.; Rosenzweig, A.C. Characterization of methanobactin from Methylosinus sp. LW4. J. Am. Chem. Soc. 2016, 138, 11124–11127. [Google Scholar] [CrossRef] [PubMed]
- Kenney, G.E.; Dassama, L.M.K.; Pandelia, M.E.; Gizzi, A.S.; Martinie, R.J.; Gao, P.; DeHart, C.J.; Schachner, L.F.; Skinner, O.S.; Ro, S.Y.; et al. The biosynthesis of methanobactin. Science 2018, 359, 1411–1416. [Google Scholar] [CrossRef]
- Kenney, G.E.; Rosenzweig, A.C. Methanobactins: Maintaining copper homeostasis in methanotrophs and beyond. J. Biol. Chem. 2018, 293, 4606–4615. [Google Scholar] [CrossRef]
- Dershwitz, P.; Gu, W.; Roche, J.; Kang-Yun, C.S.; Semrau, J.D.; Bobik, T.A.; Fulton, B.; Zischka, H.; DiSpirito, A.A. MbnC is not required for the formation of the N-terminal oxazolone in the methanobactin from Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 2022, 88, e0184121. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhu, M.; Zhang, Q.; Zhang, X.; Yang, P.; Liu, Z.; Deng, Y.; Zhu, Y.; Huang, X.; Han, L.; et al. Diisonitrile natural product SF2768 functions as a chalkophore that mediates copper acquisition in Streptomyces thioluteus. ACS Chem. Biol. 2017, 12, 3067–3075. [Google Scholar] [CrossRef] [PubMed]
- DiSpirito, A.A.; Semrau, J.D.; Murrell, J.C.; Gallagher, W.H.; Dennison, C.; Vuilleumier, S. Methanobactin and the link between copper and bacterial methane oxidation. Microbiol. Mol. Biol. Rev. 2016, 80, 387–409. [Google Scholar] [CrossRef]
- Buglino, J.A.; Ozakman, Y.; Xu, Y.; Chowdhury, F.; Tan, D.S.; Glickman, M.S. Diisonitrile lipopeptides mediate resistance to copper starvation in pathogenic mycobacteria. mBio 2022, 13, e0251322. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, K.; Maruyama, H.; Imachi, K.; Ikeda, H.; Wakimoto, T. Actinobacterial chalkophores: The biosynthesis of hazimycins. J. Antibiot. 2024, 77, 228–237. [Google Scholar] [CrossRef]
- Park, Y.J.; Jodts, R.J.; Slater, J.W.; Reyes, R.M.; Winton, V.J.; Montaser, R.A.; Thomas, P.M.; Dowdle, W.B.; Ruiz, A.; Kelleher, N.L.; et al. A mixed-valent Fe(II)Fe(III) species converts cysteine to an oxazolone/thioamide pair in methanobactin biosynthesis. Proc. Natl. Acad. Sci. USA 2022, 119, e2123566119. [Google Scholar] [CrossRef]
- Katumba, G.L.; Tran, H.; Henderson, J.P. The yersinia high-pathogenicity island encodes a siderophore-dependent copper response system in uropathogenic Escherichia coli. mBio 2022, 13, e0239121. [Google Scholar] [CrossRef]
- Koh, E.I.; Henderson, J.P. Microbial copper-binding siderophores at the host-pathogen interface. J. Biol. Chem. 2015, 290, 18967–18974. [Google Scholar] [CrossRef]
- Pham, V.N.; Chang, C.J. Metalloallostery and transition metal signaling: Bioinorganic copper chemistry beyond active sites. Angew. Chem. Int. Ed. Engl. 2023, 62, e202213644. [Google Scholar] [CrossRef]
- Tsvetkov, P.; Coy, S.; Petrova, B.; Dreishpoon, M.; Verma, A.; Abdusamad, M.; Rossen, J.; Joesch-Cohen, L.; Humeidi, R.; Spangler, R.D.; et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022, 375, 1254–1261. [Google Scholar] [CrossRef]
- Einer, C.; Munk, D.E.; Park, E.; Akdogan, B.; Nagel, J.; Lichtmannegger, J.; Eberhagen, C.; Rieder, T.; Vendelbo, M.H.; Michalke, B.; et al. ARBM101 (methanobactin SB2) drains excess liver copper via biliary excretion in Wilson’s disease rats. Gastroenterology 2023, 165, 187–200.e7. [Google Scholar] [CrossRef] [PubMed]
- Zulkifli, M.; Spelbring, A.N.; Zhang, Y.; Soma, S.; Chen, S.; Li, L.; Le, T.; Shanbhag, V.; Petris, M.J.; Chen, T.Y.; et al. FDX1-dependent and independent mechanisms of elesclomol-mediated intracellular copper delivery. Proc. Natl. Acad. Sci. USA 2023, 120, e2216722120. [Google Scholar] [CrossRef] [PubMed]
- He, H.Y.; Niikura, H.; Du, Y.L.; Ryan, K.S. Synthetic and biosynthetic routes to nitrogen-nitrogen bonds. Chem. Soc. Rev. 2022, 51, 2991–3046. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.L.; Rohac, R.; Mitchell, A.J.; Boal, A.K.; Balskus, E.P. An N-nitrosating metalloenzyme constructs the pharmacophore of streptozotocin. Nature 2019, 566, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Ryan, K.S. Reductases produce nitric oxide in an alternative pathway to form the diazeniumdiolate group of l-alanosine. J. Am. Chem. Soc. 2023, 145, 16718–16725. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Sadar, M.D.; Williams, D.E.; Mawji, N.R.; Patrick, B.O.; Wikanta, T.; Chasanah, E.; Irianto, H.E.; Soest, R.V.; Andersen, R.J. Sintokamides A to E, chlorinated peptides from the sponge Dysidea sp. that inhibit transactivation of the N-terminus of the androgen receptor in prostate cancer cells. Org. Lett. 2008, 10, 4947–4950. [Google Scholar] [CrossRef] [PubMed]
- Seunguk, P.; Shmuel, C.; Jean, C.; Moore, R.E.; Patterson, G.M.L.; Tius, M.A. Mirabimide E, an unusual N-acylpyrrolinone from the blue-green alga Scytonema mirabile: Structure determination and synthesis. J. Am. Chem. Soc. 1994, 116, 8116–8125. [Google Scholar]
- Guo, X.; Crnovcic, I.; Chang, C.Y.; Luo, J.; Lohman, J.R.; Papinski, M.; Bechthold, A.; Horsman, G.P.; Shen, B. PokMT1 from the polyketomycin biosynthetic machinery of Streptomyces diastatochromogenes tü6028 belongs to the emerging family of C-methyltransferases that act on CoA-activated aromatic substrates. Biochemistry 2018, 57, 1003–1011. [Google Scholar] [CrossRef]
- Crooke, A.M.; Chand, A.K.; Balskus, E.P. Elucidation of chalkophomycin biosynthesis reveals N-hydroxypyrrole-forming enzymes. bioRxiv 2024. [Google Scholar] [CrossRef]
- Yi, L.; Kong, J.; Xiong, Y.; Yi, S.; Gan, T.; Huang, C.; Duan, Y.; Zhu, X. Genome mining of Streptomyces sp. CB00271 as a natural high-producer of β-rubromycin and the resulting discovery of β-rubromycin acid. Biotechnol. Bioeng. 2021, 118, 2243–2254. [Google Scholar] [CrossRef]
- Walsh, C.T.; Garneau-Tsodikova, S.; Howard-Jones, A.R. Biological formation of pyrroles: Nature’s logic and enzymatic machinery. Nat. Prod. Rep. 2006, 23, 517–531. [Google Scholar] [CrossRef]
- Garneau, S.; Dorrestein, P.C.; Kelleher, N.L.; Walsh, C.T. Characterization of the formation of the pyrrole moiety during clorobiocin and coumermycin A1 biosynthesis. Biochemistry 2005, 44, 2770–2780. [Google Scholar] [CrossRef]
- Thomas, M.G.; Burkart, M.D.; Walsh, C.T. Conversion of l-proline to pyrrolyl-2-carboxyl-S-PCP during undecylprodigiosin and pyoluteorin biosynthesis. Chem. Biol. 2002, 9, 171–184. [Google Scholar] [CrossRef]
- Du, L.; Sánchez, C.; Shen, B. Hybrid peptide–polyketide natural products: Biosynthesis and prospects toward engineering novel molecules. Metab. Eng. 2001, 3, 78–95. [Google Scholar] [CrossRef]
- Tang, G.L.; Cheng, Y.Q.; Shen, B. Leinamycin biosynthesis revealing unprecedented architectural complexity for a hybrid polyketide synthase and nonribosomal peptide synthetase. Chem. Biol. 2004, 11, 33–45. [Google Scholar] [CrossRef]
- Huang, Y.; Tang, G.L.; Pan, G.; Chang, C.Y.; Shen, B. Characterization of the ketosynthase and acyl carrier protein domains at the LnmI nonribosomal peptide synthetase-polyketide synthase interface for leinamycin biosynthesis. Org. Lett. 2016, 18, 4288–4291. [Google Scholar] [CrossRef]
- Klau, L.J.; Podell, S.; Creamer, K.E.; Demko, A.M.; Singh, H.W.; Allen, E.E.; Moore, B.S.; Ziemert, N.; Letzel, A.C.; Jensen, P.R. The natural product domain seeker version 2 (NaPDoS2) webtool relates ketosynthase phylogeny to biosynthetic function. J. Biol. Chem. 2022, 298, 102480. [Google Scholar] [CrossRef]
- O’Connor, S.E.; Chen, H.; Walsh, C.T. Enzymatic assembly of epothilones: The EpoC subunit and reconstitution of the EpoA-ACP/B/C polyketide and nonribosomal peptide interfaces. Biochemistry 2002, 41, 5685–5694. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Medvedev, K.E.; Kinch, L.N.; Schaeffer, R.D.; Pei, J.; Grishin, N.V. A fifth of the protein world: Rossmann-like proteins as an evolutionarily successful structural unit. J. Mol. Biol. 2021, 433, 166788. [Google Scholar] [CrossRef]
- Medvedev, K.E.; Kinch, L.N.; Schaeffer, R.D.; Grishin, N.V. Functional analysis of Rossmann-like domains reveals convergent evolution of topology and reaction pathways. PLoS Comput. Biol. 2019, 15, e1007569. [Google Scholar] [CrossRef]
- Jerome, E.; Diogo, S.-M.; Tillack, A.F.; Stefano, F. AutoDock Vina 1.2.0: New docking methods, expanded force field, and python bindings. J. Chem. Inform. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Liu, X.; Walsh, C.T. Cyclopiazonic acid biosynthesis in Aspergillus sp.: Characterization of a reductase-like R* domain in cyclopiazonate synthetase that forms and releases cyclo-acetoacetyl-l-tryptophan. Biochemistry 2009, 48, 8746–8757. [Google Scholar] [CrossRef]
- Du, L.; Lou, L. PKS and NRPS release mechanisms. Nat. Prod. Rep. 2010, 27, 255–278. [Google Scholar] [CrossRef]
- Little, R.F.; Hertweck, C. Chain release mechanisms in polyketide and non-ribosomal peptide biosynthesis. Nat. Prod. Rep. 2022, 39, 163–205. [Google Scholar] [CrossRef]
- Nivina, A.; Yuet, K.P.; Hsu, J.; Khosla, C. Evolution and diversity of assembly-line polyketide synthases. Chem. Rev. 2019, 119, 12524–12547. [Google Scholar] [CrossRef]
- Marahiel, M.A. A structural model for multimodular NRPS assembly lines. Nat. Prod. Rep. 2016, 33, 136–140. [Google Scholar] [CrossRef]
- Guzman, K.M.; Cogan, D.P.; Brodsky, K.L.; Soohoo, A.M.; Li, X.; Sevillano, N.; Mathews, I.I.; Nguyen, K.P.; Craik, C.S.; Khosla, C. Discovery and characterization of antibody probes of module 2 of the 6-deoxyerythronolide B synthase. Biochemistry 2023, 62, 1589–1593. [Google Scholar] [CrossRef]
- Li, H.; Hu, Y.; Zhang, Y.; Ma, Z.; Bechthold, A.; Yu, X. Identification of RimR2 as a positive pathway-specific regulator of rimocidin biosynthesis in Streptomyces rimosus M527. Microb. Cell Fact. 2023, 22, 32. [Google Scholar] [CrossRef]
- Rai, D.; Mehra, S. The mycobacterial efflux pump EfpA can induce high drug tolerance to many antituberculosis drugs, including moxifloxacin, in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 2021, 65, e0026221. [Google Scholar] [CrossRef]
- Gilchrist, C.L.M.; Booth, T.J.; Wersch, B.V.; Grieken, L.V.; Medema, M.H.; Yit-Heng, C. Cblaster: A remote search tool for rapid identification and visualization of homologous gene clusters. Bioinform. Adv. 2021, 1, vbab016. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Kalkreuter, E.; Kautsar, S.A.; Yang, D.; Teijaro, C.N.; Bader, C.D.; Fluegel, L.L.; Davis, C.M.; Simpson, J.R.; Steele, A.D.; Gui, C.; et al. The Natural Products Discovery Center: Release of the first 8490 sequenced strains for exploring Actinobacteria biosynthetic diversity. bioRxiv 2023. [Google Scholar] [CrossRef]
- Navarro-Muñoz, J.C.; Selem-Mojica, N.; Mullowney, M.W.; Kautsar, S.A.; Tryon, J.H.; Parkinson, E.I.; De Los Santos, E.L.C.; Yeong, M.; Cruz-Morales, P.; Abubucker, S.; et al. A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 2020, 16, 60–68. [Google Scholar] [CrossRef]
- Otasek, D.; Morris, J.H.; Bouças, J.; Pico, A.R.; Demchak, B. Cytoscape automation: Empowering workflow-based network analysis. Genome Biol. 2019, 20, 185. [Google Scholar] [CrossRef]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- De Lano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newslett. Pro. Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Gilchrist, C.L.M.; Chooi, Y.H. Clinker & clustermap.js: Automatic generation of gene cluster comparison figures. Bioinformatics 2021, 37, 2473–2475. [Google Scholar] [CrossRef]


| Gene | Size (a.a.) | Putative Function | Protein Homologue | Identity%/Similarity% |
|---|---|---|---|---|
| Orf(-2) | 293 | Diacylglycerol kinase | WP_011029127.1 | 60/67 |
| Orf(-1) | 427 | Adenylosuccinate synthase | 4M0G_A | 53/67 |
| ChmA′ | 484 | MFS transporter | EfpA (ALB20045) | 34/53 |
| ChmA | 119 | Regulatory LuxR family protein | RimR2 (QAS68949) | 35/59 |
| ChmB | 248 | TetR/AcrR-like transcription regulators | SCO1718 (CAB50933) | 32/40 |
| ChmC | 478 | MFS transporter | EfpA (ALB20045) | 32/51 |
| ChmD | 595 | ABC transporter | SCO7689 (CAC17506) | 48/62 |
| ChmE | 582 | ABC transporter | BDD77077 | 46/60 |
| ChmF | 264 | Proline iminopeptidase | AlmF (CCA29204) | 24/37 |
| ChmG | 395 | Acyl-CoA/acyl-ACP dehydrogenase | TdaE (WP_014881725.1) | 25/42 |
| ChmH | 383 | l-prolyl-PCP dehydrogenase | CloN3 (AAN65232) | 31/41 |
| ChmI | 92 | Peptidyl carrier protein | CloN5 (AAN65234) | 26/54 |
| ChmJ | 517 | Adenylation protein | CloN4 (AAN65233) | 31/48 |
| ChmK | 543 | Adenylation protein | EntE (CAD6013920) | 40/56 |
| ChmL | 1189 | Non-ribosomal peptide synthetase | AlmA (CCA29202) | 33/48 |
| ChmM | 469 | l-graminine biosynthesis | GrbE (WP_006051176.1) | 35/48 |
| ChmN | 264 | l-graminine biosynthesis | GrbD (WP_006051175.1) | 37/51 |
| ChmO | 1152 | Non-ribosomal peptide synthetase | AlmA (CCA29202) | 37/53 |
| ChmP | 1414 | Type I polyketide synthase | AlmB (CCA29203) | 42/54 |
| ChmQ | 339 | O-methyltransferase | PokMT3 (ACN648470 | 36/50 |
| ChmR | 191 | Flavin reductase | VlmR (AAC45645) | 32/50 |
| Orf(+1) | 273 | Chitinase | Chitinase C (1WVU_A) | 58/75 |
| Orf(+2) | 294 | Chitinase | Chitinase C (1WVU_A) | 97/98 |
| Orf(+3) | 371 | DNA polymerase III subunit beta | 5AH2_A | 56/58 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, L.; Yi, L.; Gong, B.; Chen, L.; Li, M.; Zhu, X.; Duan, Y.; Huang, Y. Chalkophomycin Biosynthesis Revealing Unique Enzyme Architecture for a Hybrid Nonribosomal Peptide Synthetase and Polyketide Synthase. Molecules 2024, 29, 1982. https://doi.org/10.3390/molecules29091982
Yang L, Yi L, Gong B, Chen L, Li M, Zhu X, Duan Y, Huang Y. Chalkophomycin Biosynthesis Revealing Unique Enzyme Architecture for a Hybrid Nonribosomal Peptide Synthetase and Polyketide Synthase. Molecules. 2024; 29(9):1982. https://doi.org/10.3390/molecules29091982
Chicago/Turabian StyleYang, Long, Liwei Yi, Bang Gong, Lili Chen, Miao Li, Xiangcheng Zhu, Yanwen Duan, and Yong Huang. 2024. "Chalkophomycin Biosynthesis Revealing Unique Enzyme Architecture for a Hybrid Nonribosomal Peptide Synthetase and Polyketide Synthase" Molecules 29, no. 9: 1982. https://doi.org/10.3390/molecules29091982
APA StyleYang, L., Yi, L., Gong, B., Chen, L., Li, M., Zhu, X., Duan, Y., & Huang, Y. (2024). Chalkophomycin Biosynthesis Revealing Unique Enzyme Architecture for a Hybrid Nonribosomal Peptide Synthetase and Polyketide Synthase. Molecules, 29(9), 1982. https://doi.org/10.3390/molecules29091982






