Physicochemical Characterization, Antioxidant and Anticancer Activity Evaluation of an Acidic Polysaccharide from Alpinia officinarum Hance
Abstract
1. Introduction
2. Results
2.1. Isolation and Purification of AHP-3a
2.2. UV–Vis Spectroscopy
2.3. Chemical and Monosaccharide Compositions of AHP-3a
2.4. Molecular Weight Analysis of AHP-3a
2.5. SEM Analysis of AHP-3a
2.6. Congo Red Analysis of AHP-3a
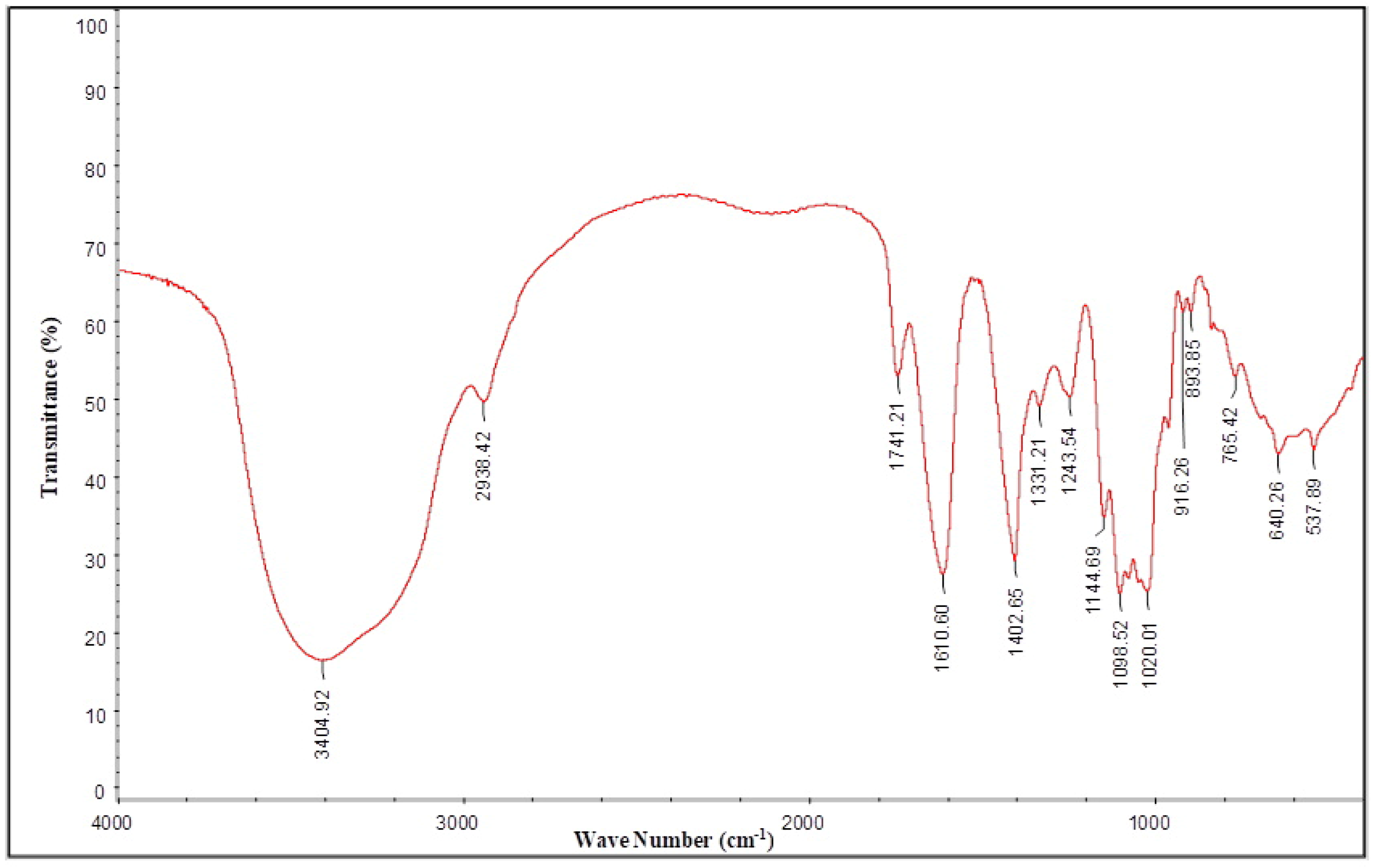
2.7. FT-IR Spectra Analysis of AHP-3a
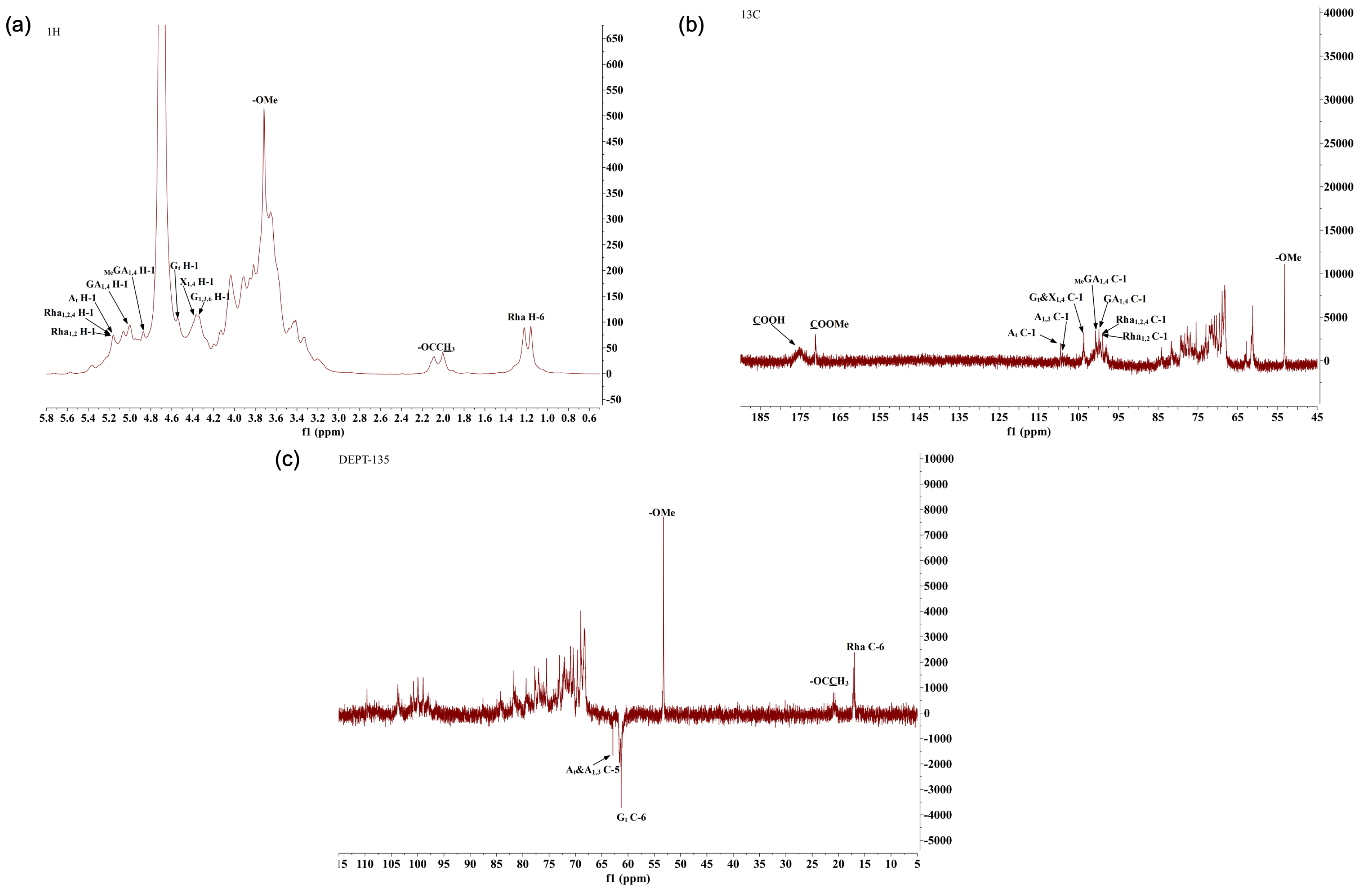
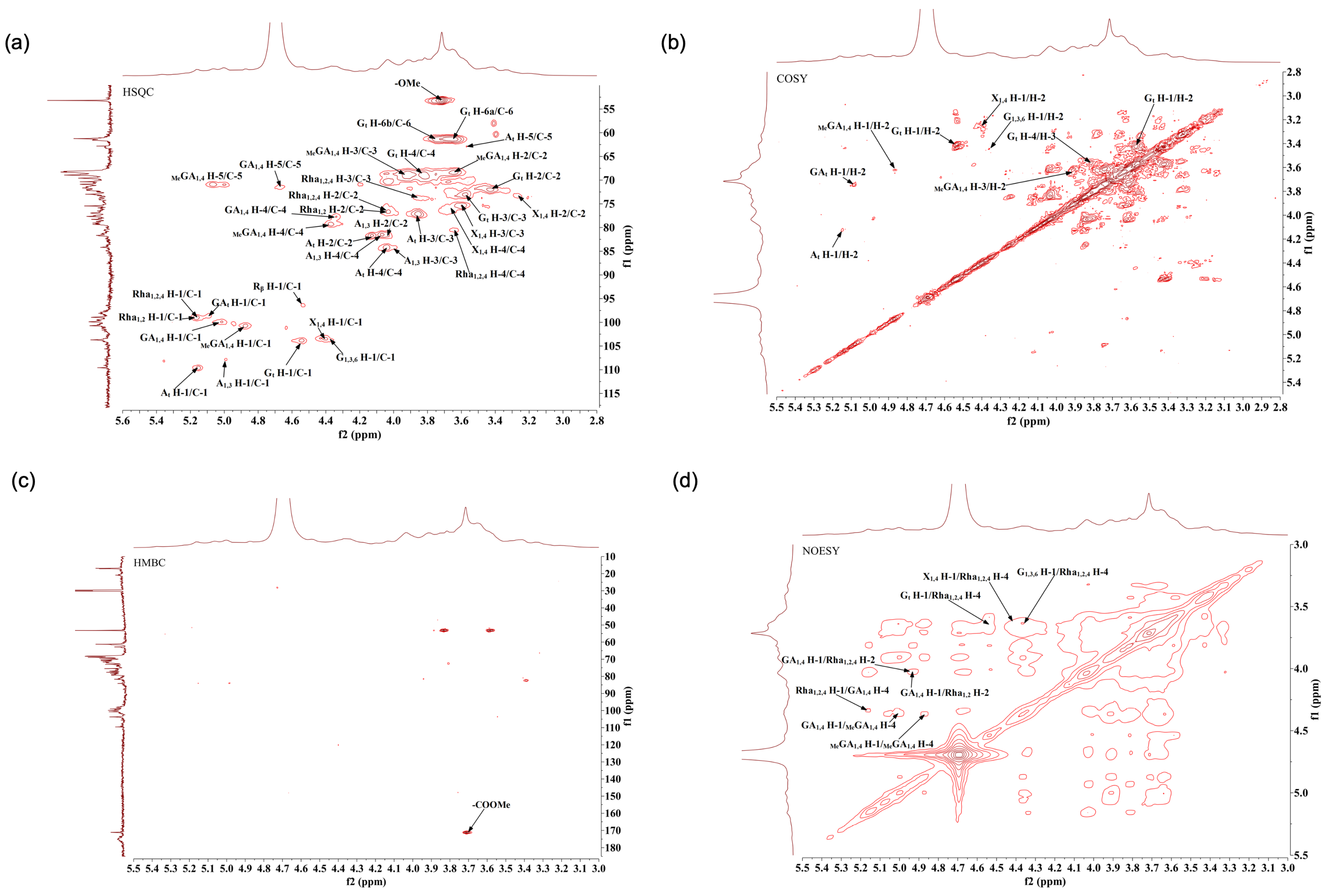
2.8. Methylation Analysis of AHP-3a
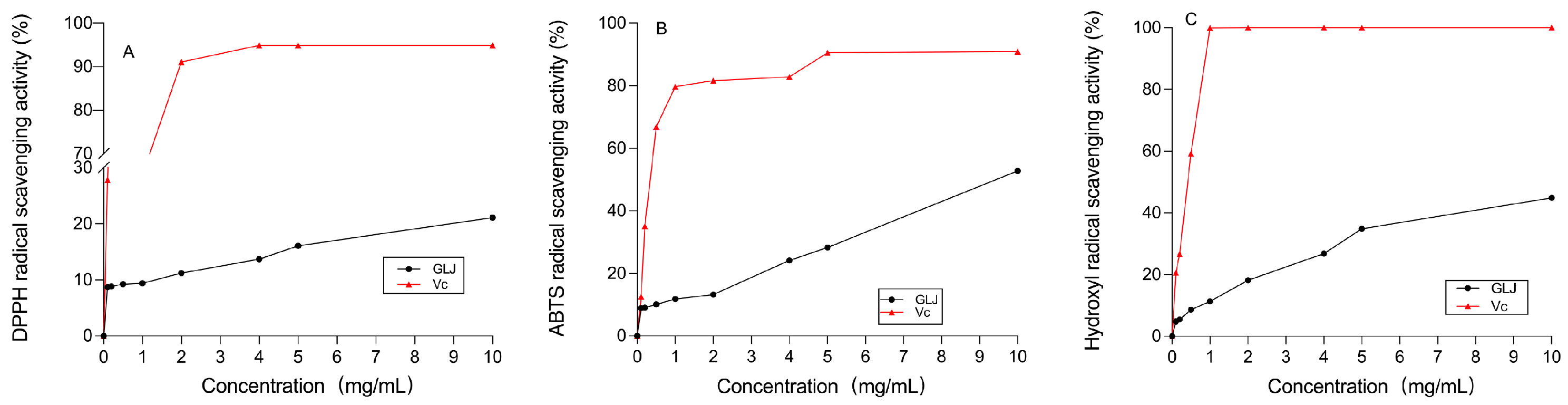
2.9. Evaluation of In Vitro Antioxidant Activity
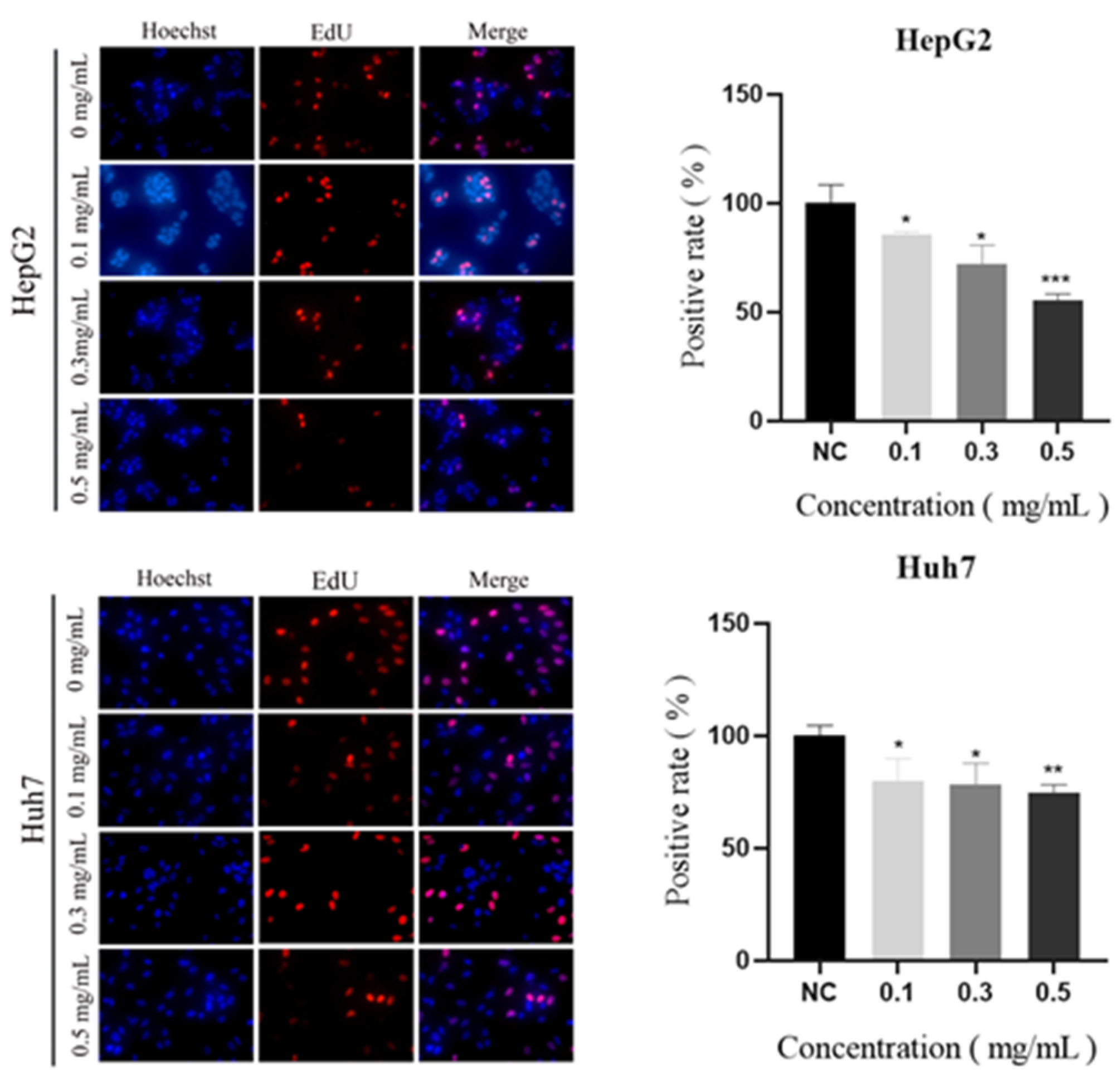
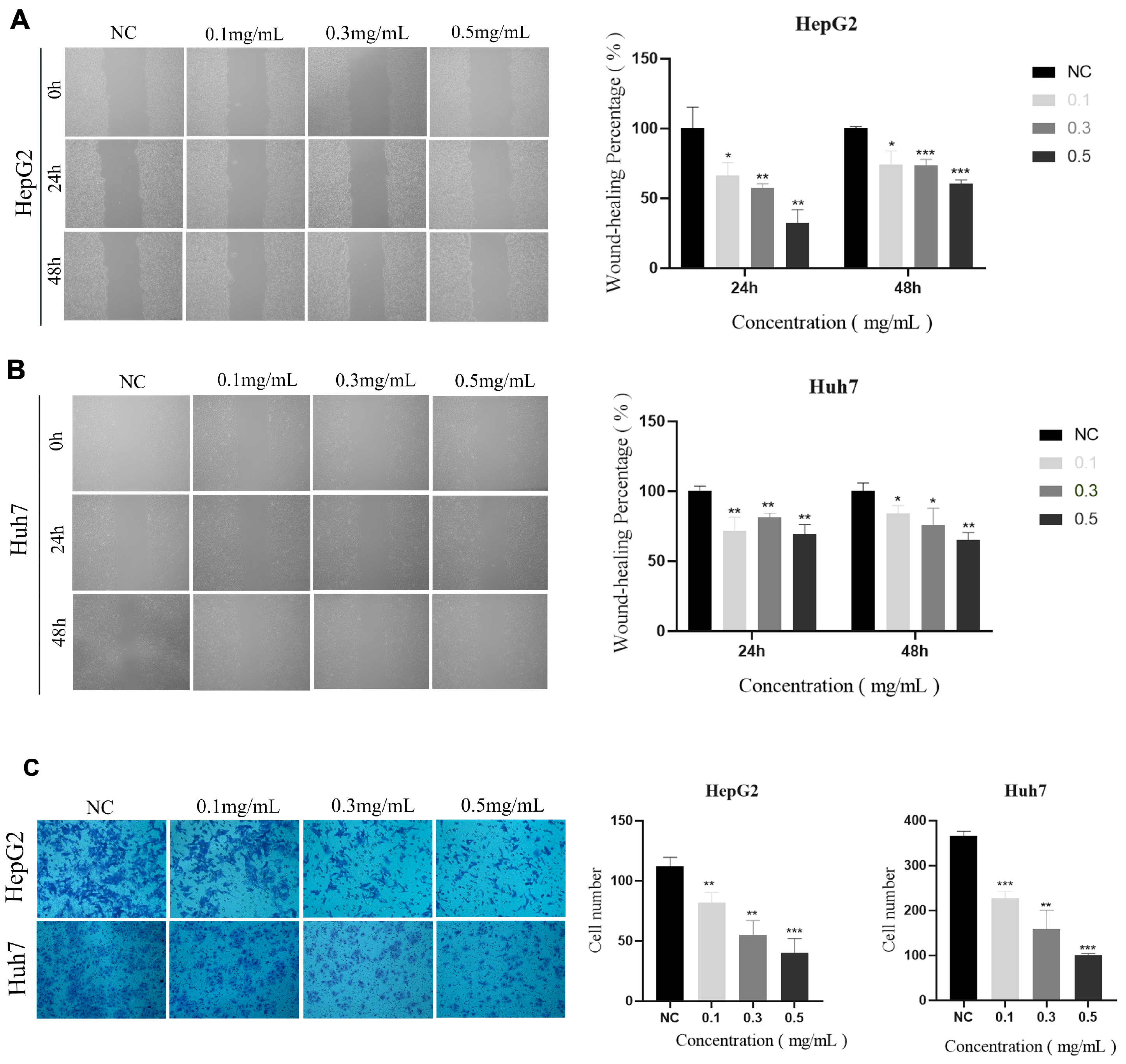
2.10. Vitro Anticancer Activity
3. Materials and Methods
3.1. Materials and Chemicals
3.2. Preparation, Isolation, and Purification of AHP
3.3. Chemical and Monosaccharide Composition Analyses
3.4. Congo Red Analysis
3.5. UV–Vis Spectroscopy
3.6. Molecular Weight Determination
3.7. FT-IR Analysis
3.8. SEM Analysis
3.9. Methylation Analysis
3.10. NMR Analysis
3.11. Vitro Antioxidant Activity
3.12. Evaluation of In Vitro Anticancer Activity
3.12.1. Cell Culture
3.12.2. Cell Viability Determination
3.12.3. Cell Proliferation Measurement
3.12.4. Wound-Healing Assay
3.12.5. Transwell Assay
3.13. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Zhao, H.; Luo, Q.H.; Yin, M.; Zhao, W.J. Oxidative stress and Alzheimer’s disease. Chin. J. Gerontol. 2013, 33, 4090–4093. [Google Scholar]
- Yuan, Y. The Preliminary Research on the Relationship between Oxidative Stress and Allergic Asthma Which Induced by Di-(2-exylhexyl) Phthalate in Ovalbumin-Immunized Mouse Models. Central China Normal University. 2013. Available online: https://kns.cnki.net/kns8/Detail?sfield=fn&QueryID=3&CurRec=1&FileName=1013278159.nh&DbName=CMFD201502&DbCode=CMFD (accessed on 1 April 2013).
- Cuzzocrea, S.; Riley, D.P.; Caputi, A.P.; Salvemini, D. Antioxidant therapy: A new pharmacological approach in shock, inflation, and ischemia/reperfusion injury. Pharmacol. Rev. 2001, 53, 135–159. [Google Scholar]
- Tang, J. Effect of Different Doses of Cyathula Officinalis Polysaccharide on Antioxidant Activity in Mice. Master’s Thesis, Sichuan Agricultural University, Yaan, China, 2013. [Google Scholar]
- Wang, Y.C. Study of Clinical and Metabolomics on the Treatment of Non-Small Cell Lung Cancer Based on Pathogenesis Theory of Cancerous Toxin. Ph.D. Thesis, Nanjing University of Chinese Medicine, Nanjing, China, 2022. [Google Scholar] [CrossRef]
- Chen, S.; Huang, S.W. Research progress in anti-tumor drug formulations at home and abroad. Med. J. 2009, 28, 215–217. [Google Scholar] [CrossRef]
- Ding, X.Q. Clinical Observation of Fufang Zhebei Granule in Reducing the Toxic Reaction of Chemotherapy. Ph.D. Thesis, Beijing University of Chinese Medicine, Beijing, China, 2011. [Google Scholar]
- Lv, S.S.; Li, S.G. Research on Plant Derived Natural Food Antioxidants and Their Applications. Grain Oil Food Tech. 2013, 21, 60–65. [Google Scholar] [CrossRef]
- Yu, Y.; Shen, M.; Song, Q.; Xie, J. Biological activities and pharmaceutical applications of polysaccharide from natural resources: A review. Carbohydr. Polym. 2018, 183, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.H.; Jin, M.L.; Morris, G.A.; Zha, X.Q.; Chen, H.Q.; Yi, Y.; Li, J.E.; Wang, Z.J.; Gao, J.; Nie, S.P.; et al. Advances on Bioactive Polysaccharides from Medicinal Plants. Crit. Rev. Food Sci. Nutr. 2016, 56 (Suppl. 1), S60–S84. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Huang, G.; Huang, H. Extraction, derivatization and antioxidant activities of onion polysaccharide. Food. Chem. 2022, 338, 133000. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.; Date, A.; Chawda, H.; Patel, K. Polysaccharides as potential anticancer agents—A review of their progress. Carbohydr. Polym. 2019, 210, 412–428. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; He, C.; Fan, Y.; Si, H.; Wang, Y.; Shi, Z.; Zhao, X.; Zheng, Y.; Liu, Q.; Zhang, H. Immune-enhancing activity of polysaccharides from Cyrtomium macrophyllum. Int. J. Biol. Macromol. 2014, 70, 590–595. [Google Scholar] [CrossRef]
- Ma, F.W.; Kong, S.Y.; Tan, H.S.; Wu, R.; Xia, B.; Zhou, Y.; Xu, H.X. Structural characterization and antiviral effect of a novel polysaccharide PSP-2B from Prunellae Spica. Carbohydr. Polym. 2016, 152, 699–709. [Google Scholar] [CrossRef]
- Jiang, H.; Sun, T.; Yao, H.Y.; Li, X.K.; Wang, Y.L.; Lei, P.; Wang, R.; Xu, H.; Li, S.; Sun, D.F. Research Progress of Edible Fungal Polysaccharides. Food Ind. Sci. Technol. 2022, 43, 447–456. [Google Scholar] [CrossRef]
- Zhao, S.; Xue, H.; Tao, Y.; Chen, K.; Li, X.; Wang, M. An Acidic Heteropolysaccharide Isolated from Pueraria lobata and Its Bioactivities. Int. J. Mol. Sci. 2023, 24, 6247. [Google Scholar] [CrossRef] [PubMed]
- Geng, C.Y. Preparation of a Galangal Polysaccharide and Its Application in Emulsion Delivery System Loaded with Galangal Flavonoids. Master’s Thesis, South China University of Technology, Guangzhou, China, 2021. [Google Scholar] [CrossRef]
- Alasmary, F.A.; Assirey, E.A.; El-Meligy, R.M.; Awaad, A.S.; El-Sawaf, L.A.; Allah, M.M.; Alqasoumi, S.I. Analysis of Alpina officinarum Hance, chemically and biologically. Saudi Pharm. J. 2019, 27, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, S.S.; Zhang, T.H.; Huang, R.Q. Optimization of Extraction Process of Alpinia officinarum Hance Polysaccharides and Study on Antioxidant Activity. Farm Products Process. 2023, 569, 34–38. [Google Scholar] [CrossRef]
- Hailiwu, R. Antitumor Activity and Mechanism of Alpinia Officinarum Hance against Stomach Cancer. Master’s Thesis, Xinjiang Medical University, Urumqi, China, 2019. [Google Scholar]
- Ni, J.; Chen, H.; Zhang, C.; Luo, Q.; Qin, Y.; Yang, Y.; Chen, Y. Characterization of Alpinia officinarum Hance polysaccharide and its immune modulatory activity in mice. Food Funct. 2022, 13, 2228–2237. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.Y.; Huang, S.J.; Mau, J.L. Antioxidant properties of hot water extracts from Agrocybe cylindracea. Food Chem. 2006, 98, 670–677. [Google Scholar] [CrossRef]
- Wu, Y.T.; Huo, Y.F.; Xu, L.; Xu, Y.Y.; Wang, X.L.; Zhou, T. Purification, characterization and antioxidant activity of polysaccharides from Porphyra haitanensis. Int. J. Biol. Macromol. 2020, 165, 2116–2125. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.S.; Ma, Y.F.; Yang, J.X.; Zhang, R.J.; Zhang, D.S.; Zheng, Y.G.; Wu, L.F. Research Progress in Analytical Methods for Structures of Phytogenic Polysaccharides. Sci. Technol. Food Ind. 2022, 43, 411–421. [Google Scholar] [CrossRef]
- Yu, S.; Yu, J.; Dong, X.; Li, S.; Liu, A. Structural characteristics and anti-tumor/-oxidant activity in vitro of an acidic polysaccharide from Gynostemma pentaphyllum. Int. J. Biol. Macromol. 2020, 161, 721–728. [Google Scholar] [CrossRef]
- Wang, H.; Chen, J.; Ren, P.; Zhang, Y.; Onyango, S.O. Ultrasound irradiation alters the spatial structure and improves the antioxidant activity of the yellow tea polysaccharide. Ultrason. Sonochem. 2021, 70, 105355. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, S.; Song, R.; Cai, J.; Xu, J.; Tang, X.; Li, N. Ginger polysaccharides induced cell cycle arrest and apoptosis in human hepatocellular carcinoma HepG2 cells. Int. J. Biol. Macromol. 2019, 123, 81–90. [Google Scholar] [CrossRef]
- Shang, F.F.; Zhu, R.G.; Zhang, X.Y.; Wang, Y.; Wang, C. Extraction, Isolation and Purification of Haw Polysaccharide and Its Antioxidant and Antiglycation Activities In Vitro. Mod. Food Sci. Technol. 2019, 35, 96–101. [Google Scholar] [CrossRef]
- Xu, L.; Yu, J.Q.; Wang, X.Y.; Xu, N.; Liu, J.L. Microwave extraction optimization using the response surface methodology of Fructus Meliae Toosendan polysaccharides and its antioxidant activity. Int. J. Biol. Macromol. 2018, 118 Pt B, 1501–1510. [Google Scholar] [CrossRef]
- Zheng, W.; Zhao, T.; Feng, W.; Wang, W.; Zou, Y.; Zheng, D.; Takase, M.; Li, Q.; Wu, H.; Yang, L.; et al. Purification, characterization and immunomodulating activity of a polysaccharide from flowers of Abelmoschus esculentus. Carbohydr. Polym. 2014, 106, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Li, Y.; Wang, C.; Zhang, N.; Zhu, X.; Wu, R.; Wu, J. Purification, characterization and antitumor activity of an exopolysaccharide produced by Bacillus velezensis SN-1. Int. J. Biol. Macromol. 2020, 156, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, J.; Liu, Y.; Wu, D.; Cai, P.; Pan, Y. Structural characterization and immunomodulatory activity of a water soluble polysaccharide isolated from Botrychium ternatum. Carbohydr. Polym. 2017, 171, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Qiu, Z.; Dong, H.; Ma, C.; Qiao, Y.; Zheng, Z. Structural characterization and antioxidant activities of one neutral polysaccharide and three acid polysaccharides from the roots of Arctium lappa L.: A comparison. Int. J. Biol. Macromol. 2021, 182, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, X.; Huo, D.; Cao, C.; Li, Y.; Liang, Y.; Li, B.; Li, L. Preliminary characterization, antioxidant and α-glucosidase inhibitory activities of polysaccharides from Mallotus furetianus. Carbohydr. Polym. 2019, 215, 307–315. [Google Scholar] [CrossRef]
- Li, Y.M.; Zhong, R.F.; Chen, J.; Luo, Z.G. Structural characterization, anticancer, hypoglycemia and immune activities of polysaccharides from Russula virescens. Int. J. Biol. Macromol. 2021, 184, 380–392. [Google Scholar] [CrossRef]
- Makarova, E.N.; Shakhmatov, E.G.; Belyy, V.A. Structural characteristics of oxalate-soluble polysaccharides of Sosnowsky’s hogweed (Heracleum sosnowskyi Manden). Carbohydr. Polym. 2016, 153, 66–77. [Google Scholar] [CrossRef]
- Hamed, M.; Coelho, E.; Bastos, R.; Evtuguin, D.V.; Ferreira, S.S.; Lima, T.; Vilanova, M.; Sila, A.; Coimbra, M.A.; Bougatef, A. Isolation and identification of an arabinogalactan extracted from pistachio external hull: Assessment of immunostimulatory activity. Food. Chem. 2022, 373, 131416. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.H.; Cui, S.W.; Goff, H.D.; Chen, J.; Wang, Q.; Han, N.F. Arabinan-rich rhamnogalacturonan-I from flaxseed kernel cell wall. Food Hydrocoll. 2015, 47, 158–167. [Google Scholar] [CrossRef]
- Patova, O.A.; Smirnov, V.V.; Golovchenko, V.V.; Vityazev, F.V.; Shashkov, A.S.; Popov, S.V. Structural, rheological and antioxidant properties of pectins from Equisetum arvense L. and Equisetum sylvaticum L. Carbohydr. Polym. 2019, 209, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.Q.; Huang, R.M.; Wen, P.; Song, Y.; He, B.L.; Tan, J.L.; Hao, H.L.; Wang, H. Structural characterization and immunological activity of pectin polysaccharide from kiwano (Cucumis metuliferus) peels. Carbohydr. Polym. 2021, 254, 117371. [Google Scholar] [CrossRef] [PubMed]
- Perrone, P.; Hewage, C.M.; Thomson, A.R.; Bailey, K.; Sadler, I.H.; Fry, S.C. Patterns of methyl and O-acetyl esterification in spinach pectins: New complexity. Phytochemistry 2002, 60, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wen, L.; Zhao, Y.; Jiang, Y.; Tian, M.; Liu, H.; Liu, J.; Yang, B. Structure identification of an arabinogalacturonan in Citrus reticulata Blanco ‘Chachiensis’ peel. Food Hydrocoll. 2018, 84, 481–488. [Google Scholar] [CrossRef]
- Shakhmatov, E.G.; Toukach, P.V.; Makarova, E.N. Structural studies of the pectic polysaccharide from fruits of Punica granatum. Carbohydr. Polym. 2020, 235, 115978. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.W.; Synytsya, A.; Capek, P.; Bleha, R.; Pohl, R.; Park, Y.I. Structural analysis and anti-obesity effect of a pectic polysaccharide isolated from Korean mulberry fruit Oddi (Morus alba L.). Carbohydr. Polym. 2016, 146, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Dias, I.P.; Barbieri, S.F.; Fetzer, D.E.L.; Corazza, M.L.; Silveira, J.L.M. Effects of pressurized hot water extraction on the yield and chemical characterization of pectins from Campomanesia xanthocarpa Berg fruits. Int. J. Biol. Macromol. 2020, 146, 431–443. [Google Scholar] [CrossRef]
- Chen, G.; Bai, Y.; Zeng, Z.; Peng, Y.; Zhou, W.; Shen, W.; Zeng, X.; Liu, Z. Structural Characterization and Immunostimulatory Activity of Heteropolysaccharides from Fuzhuan Brick Tea. J. Agric. Food Chem. 2021, 69, 1368–1378. [Google Scholar] [CrossRef]
- Petersen, B.O.; Meier, S.; Duus, J.Ø.; Clausen, M.H. Structural characterization of homogalacturonan by NMR spectroscopy—Assignment of reference compounds. Carbohydr. Res. 2008, 343, 2830–2833. [Google Scholar] [CrossRef] [PubMed]
- Coelho, M.N.; Soares, P.A.G.; Frattani, F.S.; Camargo, L.M.M.; Tovar, A.M.F.; de Aguiar, P.F.; Zingali, R.B.; Mourão, P.A.S.; Costa, S.S. Polysaccharide composition of an anticoagulant fraction from the aqueous extract of Marsypianthes chamaedrys (Lamiaceae). Int. J. Biol. Macromol. 2020, 145, 668–681. [Google Scholar] [CrossRef] [PubMed]
- Shakhmatov, E.G.; Udoratina, E.V.; Atukmaev, K.V.; Makarova, E.N. Extraction and structural characteristics of pectic polysaccharides from Abies sibirica L. Carbohydr. Polym. 2015, 128, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Shakhmatov, E.G.; Belyy, V.A.; Makarova, E.N. Structure of acid-extractable polysaccharides of tree greenery of Picea abies. Carbohydr. Polym. 2018, 199, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Tang, W.; Huang, X.J.; Hu, J.L.; Wang, J.Q.; Yin, J.Y.; Nie, S.P.; Xie, M.Y. Structural characteristic of pectin-glucuronoxylan complex from Dolichos lablab L. hull. Carbohydr. Polym. 2022, 298, 120023. [Google Scholar] [CrossRef] [PubMed]
- Makarova, E.N.; Shakhmatov, E.G. Characterization of pectin-xylan-glucan-arabinogalactan proteins complex from Siberian fir Abies sibirica Ledeb. Carbohydr. Polym. 2021, 260, 117825. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Liu, D.; Wang, J.Q.; Huang, X.J.; Yin, J.Y.; Geng, F.; Nie, S.P. Isolation and structure characterization of a low methyl-esterified pectin from the tuber of Dioscorea opposita Thunb. Food Chem. 2021, 359, 129899. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Chen, Y.; Chen, Z.; Gao, X.; Wang, C.; Panichayupakaranant, P.; Chen, H. Ultrafiltration isolation, physicochemical characterization, and antidiabetic activities analysis of polysaccharides from green tea, oolong tea, and black tea. J. Food Sci. 2020, 85, 4025–4032. [Google Scholar] [CrossRef]
- Chen, R.Z.; Li, Y.; Dong, H.; Liu, Z.Q.; Li, S.Z.; Yang, S.M.; Li, X.L. Optimization of ultrasonic extraction process of polysaccharides from ornithogalum caudatum ait and evaluation of its biological activities. Ultrason. Sonochem. 2012, 19, 1160–1168. [Google Scholar] [CrossRef]
- Li, X.L.; Zhou, A.G.; Han, Y. Anti-oxidation and anti-microorganism activities of purification polysaccharide from Lygodium japonicum in vitro. Carbohydr. Polym. 2006, 66, 34–42. [Google Scholar] [CrossRef]
- Chen, H.; Niu, H.; Zhang, H.; Yun, Y.; Chen, W.; Zhong, Q.; Chen, W.; Fu, X. Preparation and properties of ferulic acid-sugar beet pulp pectin ester and its application as a physical and antioxidative stabilizer in a fish oil-water emulsion. Int. J. Biol. Macromol. 2019, 139, 81–90. [Google Scholar] [CrossRef]
- Kiddane, A.T.; Kim, G.D. Anticancer and Immunomodulatory Effects of Polysaccharides. Nutr. Cancer 2021, 73, 2219–2231. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Chen, M.; Ding, Y.; Yang, P.; Wang, M.; Zhang, H.; He, Y.; Ma, H. Polysaccharides as Potential Anti-tumor Biomacromolecules—A Review. Front. Nutr. 2022, 28, 838179. [Google Scholar] [CrossRef]
- Liao, D.W.; Cheng, C.; Liu, J.P.; Zhao, L.Y.; Huang, D.C.; Chen, G.T. Characterization and antitumor activities of polysaccharides obtained from ginger (Zingiber officinale) by different extraction methods. Int. J. Biol. Macromol. 2020, 152, 894–903. [Google Scholar] [CrossRef] [PubMed]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.T.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Bitter, T.; Muir, H.M. A modified uronic acid carbazole reaction. Anal. Biochem. 1962, 4, 330–334. [Google Scholar] [CrossRef]
- Xu, G.Y. Study on Physicochemical Properties and Functionality of Potato Peel Polysaccharide. Master’s Thesis, Hefei University of Technology, Hefei, China, 2020. [Google Scholar] [CrossRef]
- Nie, C.; Zhu, P.; Ma, S.; Wang, M.; Hu, Y. Purification, characterization and immunomodulatory activity of polysaccharides from stem lettuce. Carbohydr. Polym. 2018, 188, 236–242. [Google Scholar] [CrossRef]
- Li, X.; Wei, J.; Lin, L.; Li, J.; Zheng, G. Structural characterization, antioxidant and antimicrobial activities of polysaccharide from Akebia trifoliata (Thunb.) Koidz stem. Coll. Surf. B Biointerfaces 2023, 231, 113573. [Google Scholar] [CrossRef]
- Du, Y.; Zhang, Z.; Liu, X.J.; Guo, W.C. Extraction, characterization and in vitro biological activity of polysaccharide from Actinidia arguta Planch.pollen. Cereals Oils 2023, 36, 85–91. [Google Scholar]
- Xiong, G.; Ma, L.; Zhang, H. Physicochemical properties, antioxidant activities and α-glucosidase inhibitory effects of polysaccharides from Evodiae fructus extracted by different solvents. Int. J. Biol. Macromol. 2022, 194, 484–498. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.L.; Conrad, H.E. Stoichiometric depolymerization of polyuronides and glycosaminoglycuronans to monosaccharides following reduction of their carbodiimide-activated carboxyl groups. Biochemistry 1972, 11, 1383–1388. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.C.; Song, T.Y.; Shih, P.H.; Yen, G.C. Antioxidant properties of watersoluble polysaccharides from Antrodia cinnamomea in submerged culture. J. Food Chem. Food Chem. 2007, 104, 1115–1122. [Google Scholar] [CrossRef]
- Wu, J.; Wang, J.; Su, Q.; Ding, W.; Li, T.; Yu, J.; Cao, B. Traditional Chinese medicine Astragalus polysaccharide enhanced antitumor effects of the angiogenesis inhibitor apatinib in pancreatic cancer cells on proliferation, invasiveness, and apoptosis. OncoTargets Ther. 2018, 11, 2685–2698. [Google Scholar] [CrossRef]
- Wang, X.; Dong, K.; Jin, Q.; Ma, Y.; Yin, S.; Wang, S. Upregulation of lncRNA FER1L4 suppresses the proliferation and migration of the hepatocellular carcinoma via regulating PI3K/AKT signal pathway. J. Cell. Biochem. 2019, 120, 6781–6788. [Google Scholar] [CrossRef]
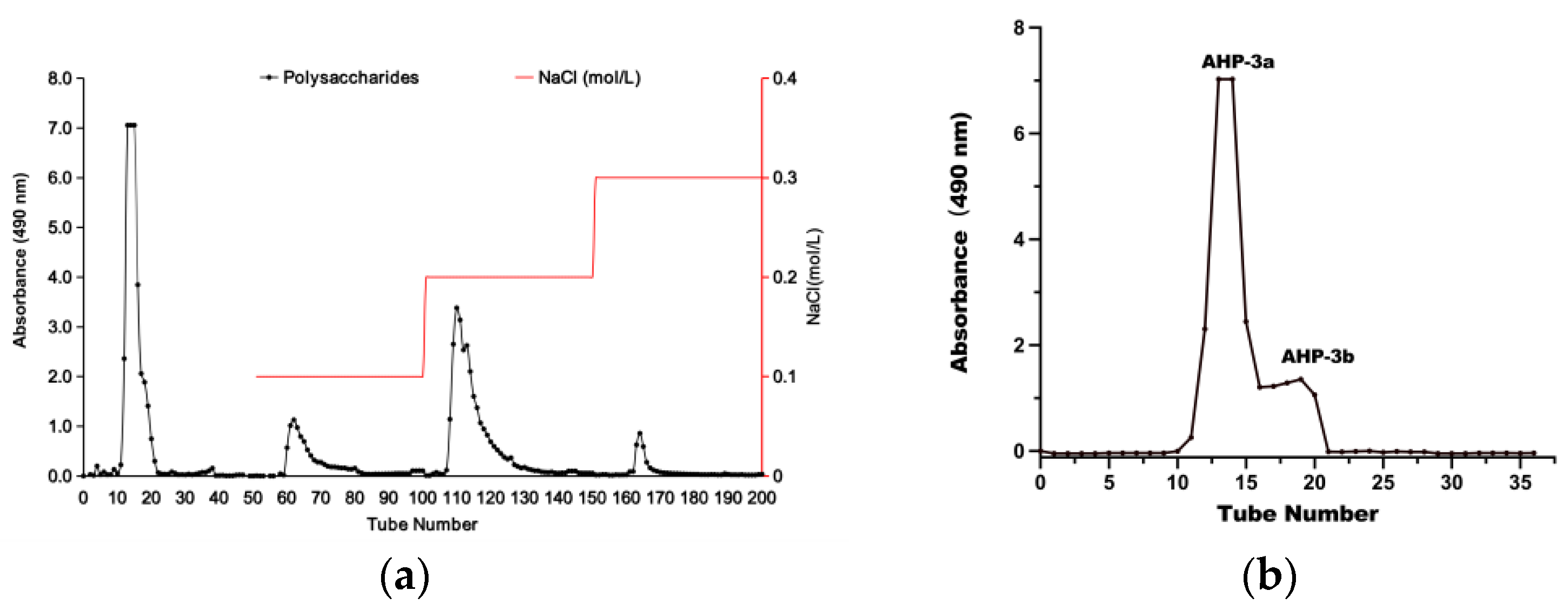


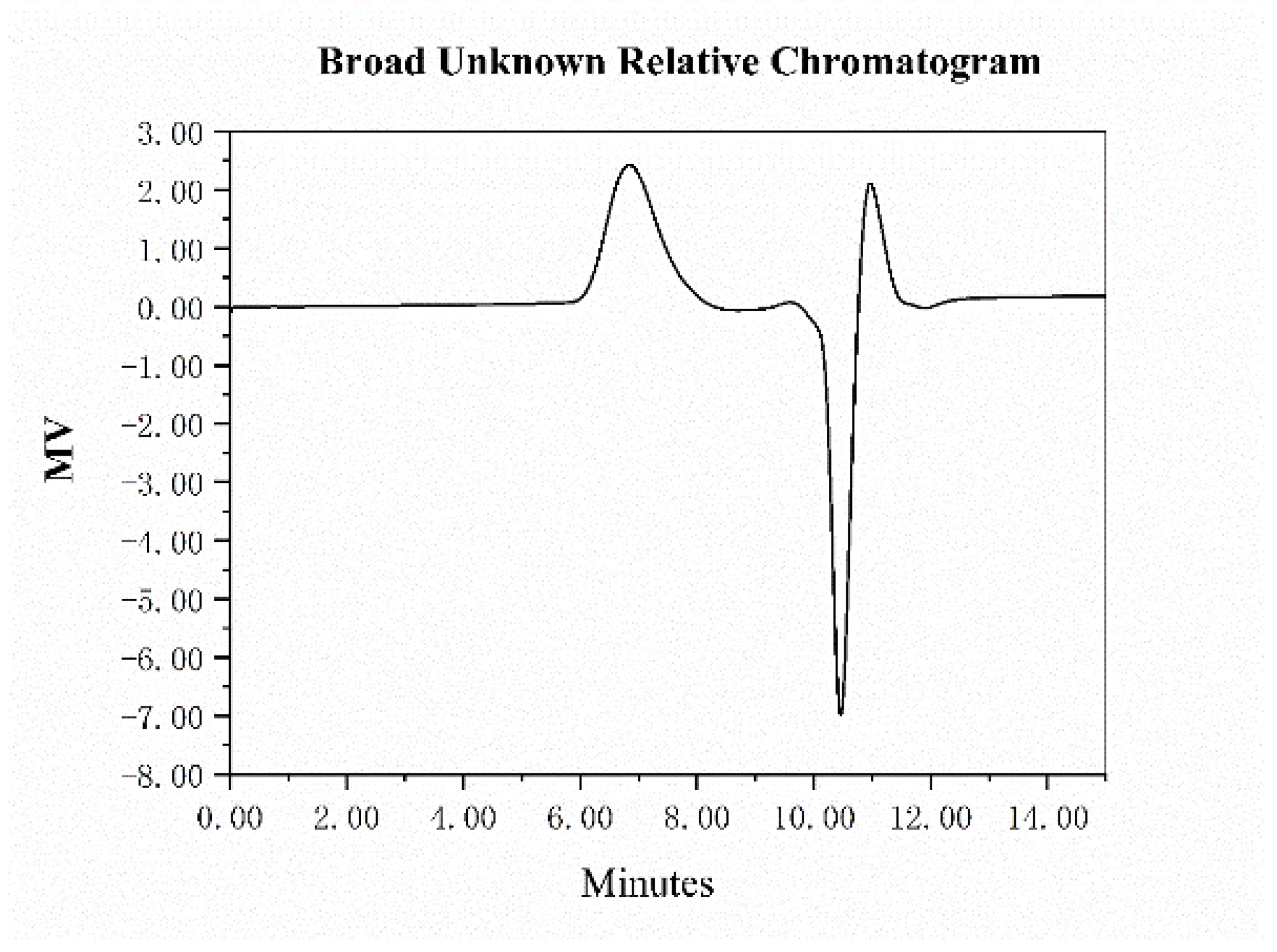
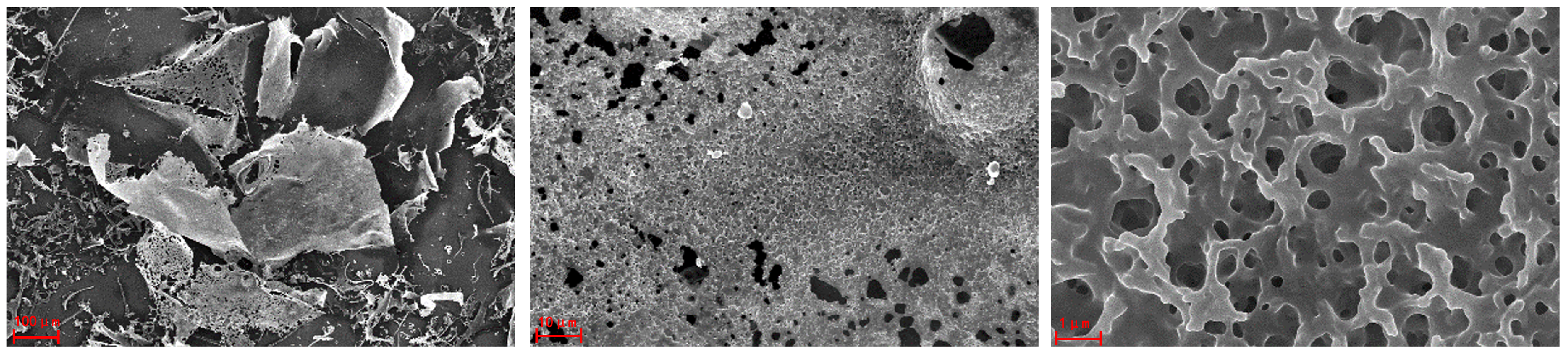
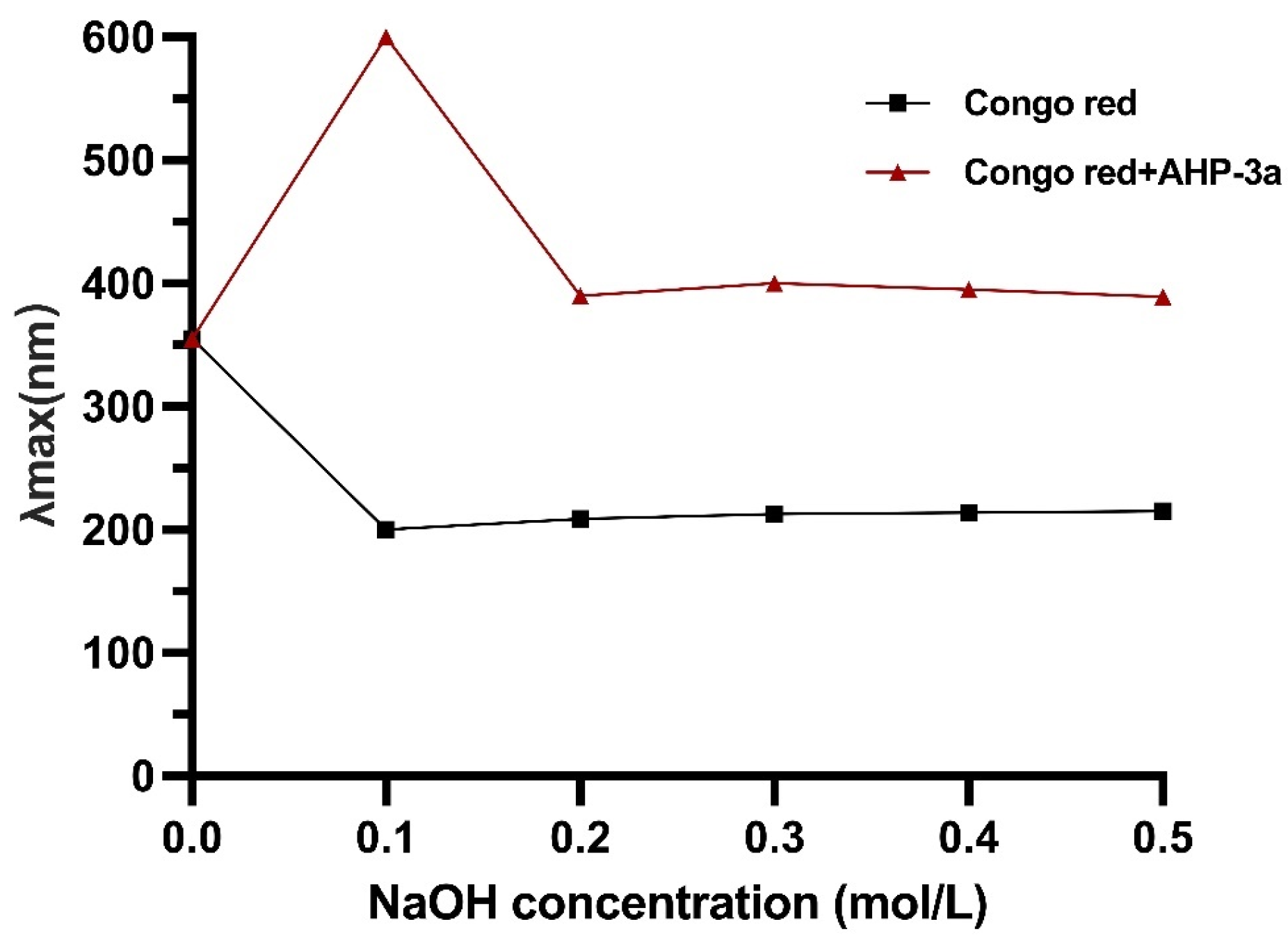


| Neutral Sugar (%) | Uronic Sugar (%) | Protein (%) | Total Phenolics (%) |
|---|---|---|---|
| 28.08 ± 0.13 | 45.93 ± 0.22 | ND | 8.16 ± 0.07 |
| Fuc | Rha | Ara | Gal | Glc | Xyl | Man | GalA | GlcA |
|---|---|---|---|---|---|---|---|---|
| 0.5 | 8.9 | 16.9 | 21.4 | 3.1 | 11.8 | 0.0 | 35.4 | 2.0 |
| Mn (Daltons) | Mw (Daltons) | MP (Daltons) | Polydispersity | |
|---|---|---|---|---|
| AHP-3a | 273,062 | 484,410 | 582,790 | 1.77 |
| Sugar Derivatives | Glycosidic Linkage | RT (min) | Molar Ratio (%) |
|---|---|---|---|
| 1,4-di-O-acetyl-2,3,5-tri-O-methyl arabinitol | T-Araf | 11.530 | 8.881 |
| 1,5-di-O-acetyl-6-deoxy-2,3,4-tri-O-methyl rhamnitol | T-Rhap | 12.559 | 3.104 |
| 1,3,4-tri-O-acetyl-2,5-di-O-methyl arabinitol | 1,3-Araf | 14.344 | 1.264 |
| 1,2,5-tri-O-acetyl-6-deoxy-3,4-di-O-methyl rhamnitol | 1,2-Rhap | 15.321 | 5.061 |
| 1,4,5-tri-O-acetyl-2,3-di-O-methyl xylitol | 1,4-Xylp | 15.457 | 6.139 |
| 1,5-di-O-acetyl-2,3,4,6-tetra-O-methyl glucitol | T-GlcpA | 16.576 | 6.390 |
| 1,5-di-O-acetyl-2,3,4,6-tetra-O-methyl galactitol | T-Galp | 17.294 | 12.282 |
| 1,5-di-O-acetyl-2,3,4,6-tetra-O-methyl galactitol | T-GalpA | 17.294 | 3.816 |
| 1,2,4,5-tetra-O-acetyl-6-deoxy-3-O-methyl rhamnitol | 1,2,4-Rhap | 18.200 | 9.370 |
| 1,4,5-tri-O-acetyl-2,3,6-tri-O-methyl galactitol | 1,4-GalpA | 19.597 | 32.634 |
| 1,4,5-tri-O-acetyl-2,3,6-tri-O-methyl galactitol | 1,4-Galp | 19.597 | 1.417 |
| 1,4,5-tri-O-acetyl-2,3,6-tri-O-methyl glucitol | 1,4-Glcp | 19.830 | 2.051 |
| 1,3,5-tri-O-acetyl-2,4,6-tri-O-methyl galactitol | 1,3-Galp | 20.108 | 1.835 |
| 1,5,6-tri-O-acetyl-2,3,4-tri-O-methyl galactitol | 1,6-Galp | 21.330 | 1.447 |
| 1,3,4,5-tetra-O-acetyl-2,6-di-O-methyl galactitol | 1,3,4-GalpA | 21.744 | 1.038 |
| 1,2,4,5-tetra-O-acetyl-3,6-di-O-methyl glucitol | 1,2,4-GlcpA | 22.340 | 0.786 |
| 1,3,5,6-tetra-O-acetyl-2,4-di-O-methyl galactitol | 1,3,6-Galp | 24.157 | 2.486 |
| Glycosyl Residues | Chemical Displacement δ () | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5a/5b | 6a/6b | -OMe | |||
| MeGA1,4 | →4)-α-D-GalpA-6-OMe-(1→ | H | 4.87 | 3.63 | 3.92 | 4.37 | 5.06/5.00 | 3.72 | |
| C | 100.76 | 68.26 | 68.75 | 79.25 | 70.92 | 171.16 | 53.24 | ||
| GA1,4 | →4)-α-D-GalpA-(1→ | H | 5.01 | 3.63 | 3.92 | 4.34 | 4.67 | ||
| C | 99.94 | 68.26 | 68.75 | 77.76 | 71.47 | 175.31 | |||
| GAt | α-D-GalpA-(1→ | H | 5.09 | 3.75 | 3.81 | ||||
| C | 98.75 | 68.75 | 69.05 | 175.31 | |||||
| Rβ | →4)-β-D-GalpA | H | 4.53 | ||||||
| C | 96.45 | 175.31 | |||||||
| X1,4 | →4)-β-D-Xylp-(1→ | H | 4.41 | 3.61 | 3.66 | 3.30/4.03 | |||
| C | 103.45 | 73.34 | 75.49 | 76.16 | 63.14 | ||||
| Gt | β-D-Galp-(1→ | H | 4.53 | 3.42 | 3.57 | 3.81 | 3.65 | 3.64/3.74 | |
| C | 103.85 | 71.99 | 73.22 | 69.05 | 75.64 | 61.24 | |||
| G1,3,6 | →3,6)-β-D-Galp-(1→ | H | 4.37 | 3.45 | 3.76 | ||||
| C | 103.71 | 71.99 | 81.56 | ||||||
| At | α-L-Araf-(1→ | H | 5.15 | 4.12 | 3.85 | 4.04 | 3.57 | ||
| C | 109.63 | 81.82 | 77.09 | 84.24 | 62.87 | ||||
| A1,3 | →3)-α-L-Araf-(1→ | H | 4.99 | 4.03 | 3.99 | 4.06 | 3.57 | ||
| C | 108.03 | 81.67 | 84.24 | 81.41 | 62.87 | ||||
| Rha1,2,4 | →2,4)-α-L-Rhap-(1→ | H | 5.16 | 4.03 | 3.86 | 3.64 | 1.22 | ||
| C | 99.02 | 76.72 | 73.89 | 80.59 | 16.99 | ||||
| Rha1,2 | →2)-α-L-Rhap-(1→ | H | 5.16 | 4.03 | 3.86 | 1.16 | |||
| C | 99.02 | 76.72 | 73.89 | 16.99 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, H.; Kuang, Y.; Lian, X.; Li, H.; Zhou, M.; Tan, Y.; Zhang, X.; Pan, Y.; Zhang, J.; Xu, J. Physicochemical Characterization, Antioxidant and Anticancer Activity Evaluation of an Acidic Polysaccharide from Alpinia officinarum Hance. Molecules 2024, 29, 1810. https://doi.org/10.3390/molecules29081810
Wen H, Kuang Y, Lian X, Li H, Zhou M, Tan Y, Zhang X, Pan Y, Zhang J, Xu J. Physicochemical Characterization, Antioxidant and Anticancer Activity Evaluation of an Acidic Polysaccharide from Alpinia officinarum Hance. Molecules. 2024; 29(8):1810. https://doi.org/10.3390/molecules29081810
Chicago/Turabian StyleWen, Huan, Yangjun Kuang, Xiuxia Lian, Hailong Li, Mingyan Zhou, Yinfeng Tan, Xuguang Zhang, Yipeng Pan, Junqing Zhang, and Jian Xu. 2024. "Physicochemical Characterization, Antioxidant and Anticancer Activity Evaluation of an Acidic Polysaccharide from Alpinia officinarum Hance" Molecules 29, no. 8: 1810. https://doi.org/10.3390/molecules29081810
APA StyleWen, H., Kuang, Y., Lian, X., Li, H., Zhou, M., Tan, Y., Zhang, X., Pan, Y., Zhang, J., & Xu, J. (2024). Physicochemical Characterization, Antioxidant and Anticancer Activity Evaluation of an Acidic Polysaccharide from Alpinia officinarum Hance. Molecules, 29(8), 1810. https://doi.org/10.3390/molecules29081810










