Evaluation of Urtica dioica Phytochemicals against Therapeutic Targets of Allergic Rhinitis Using Computational Studies
Abstract
1. Introduction
2. Results and Discussion
2.1. Molecular Docking of UD Database against AR Targets
2.2. Stability and Equilibrium of Receptor–Ligand Complexes
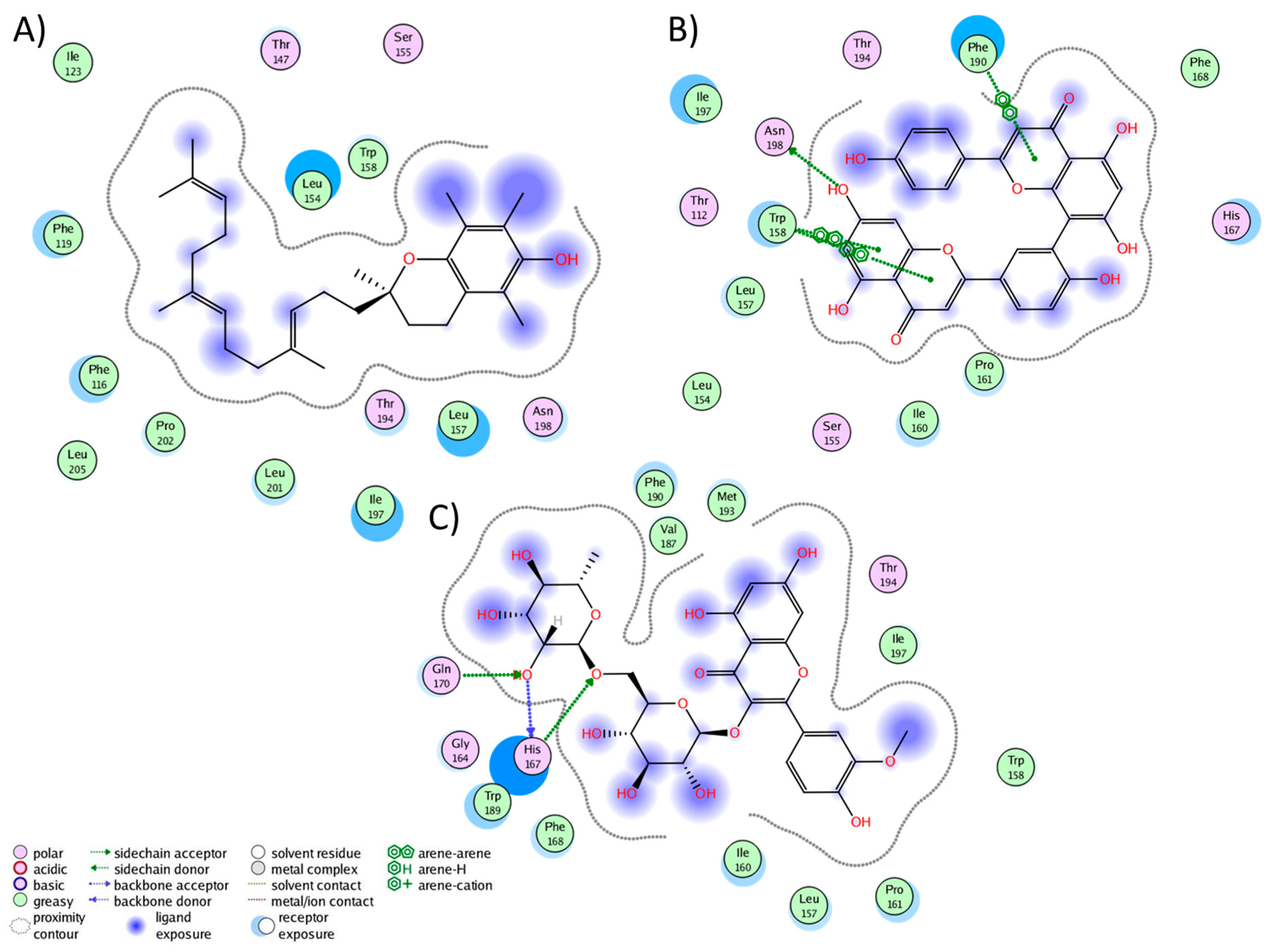
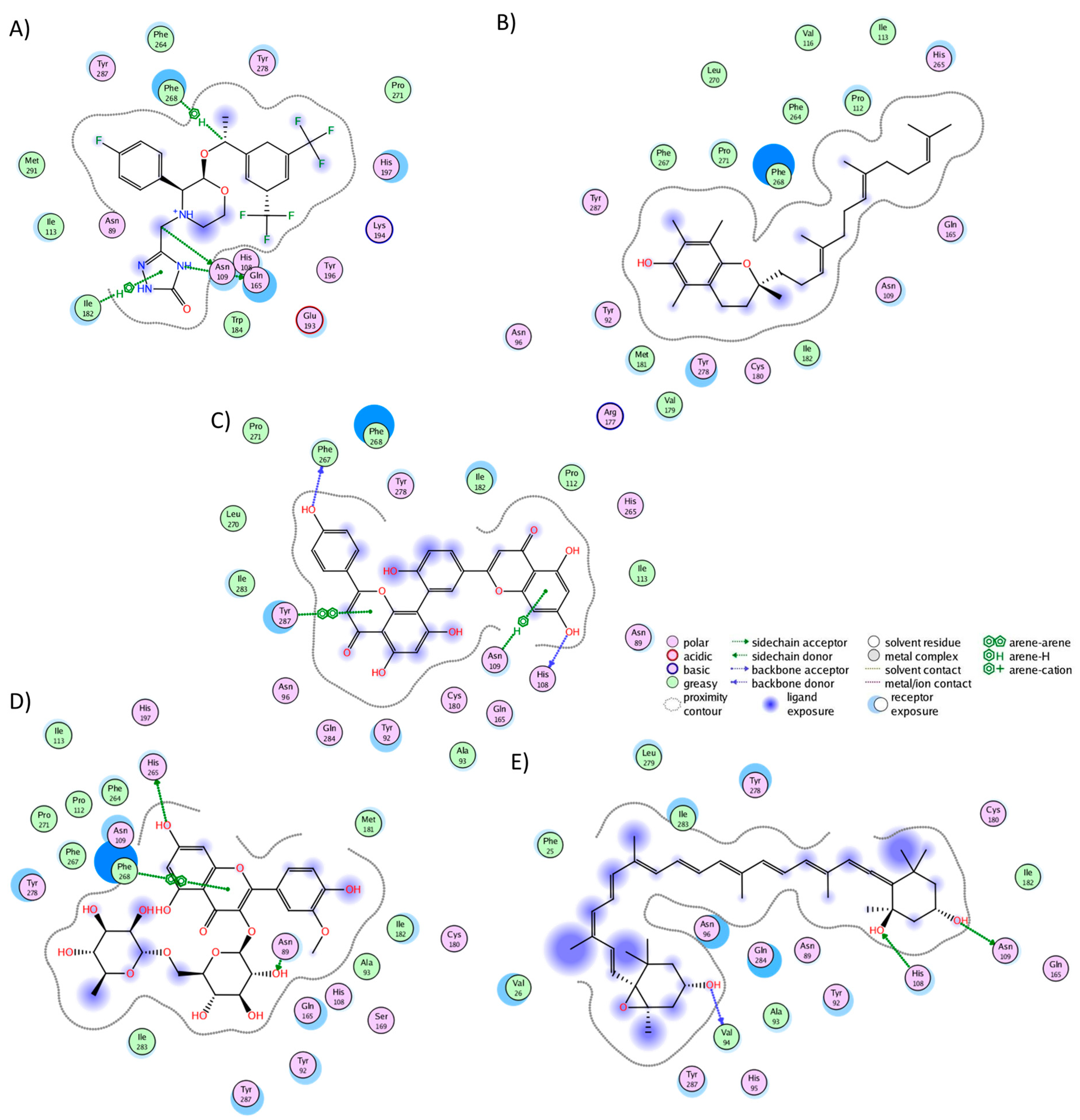
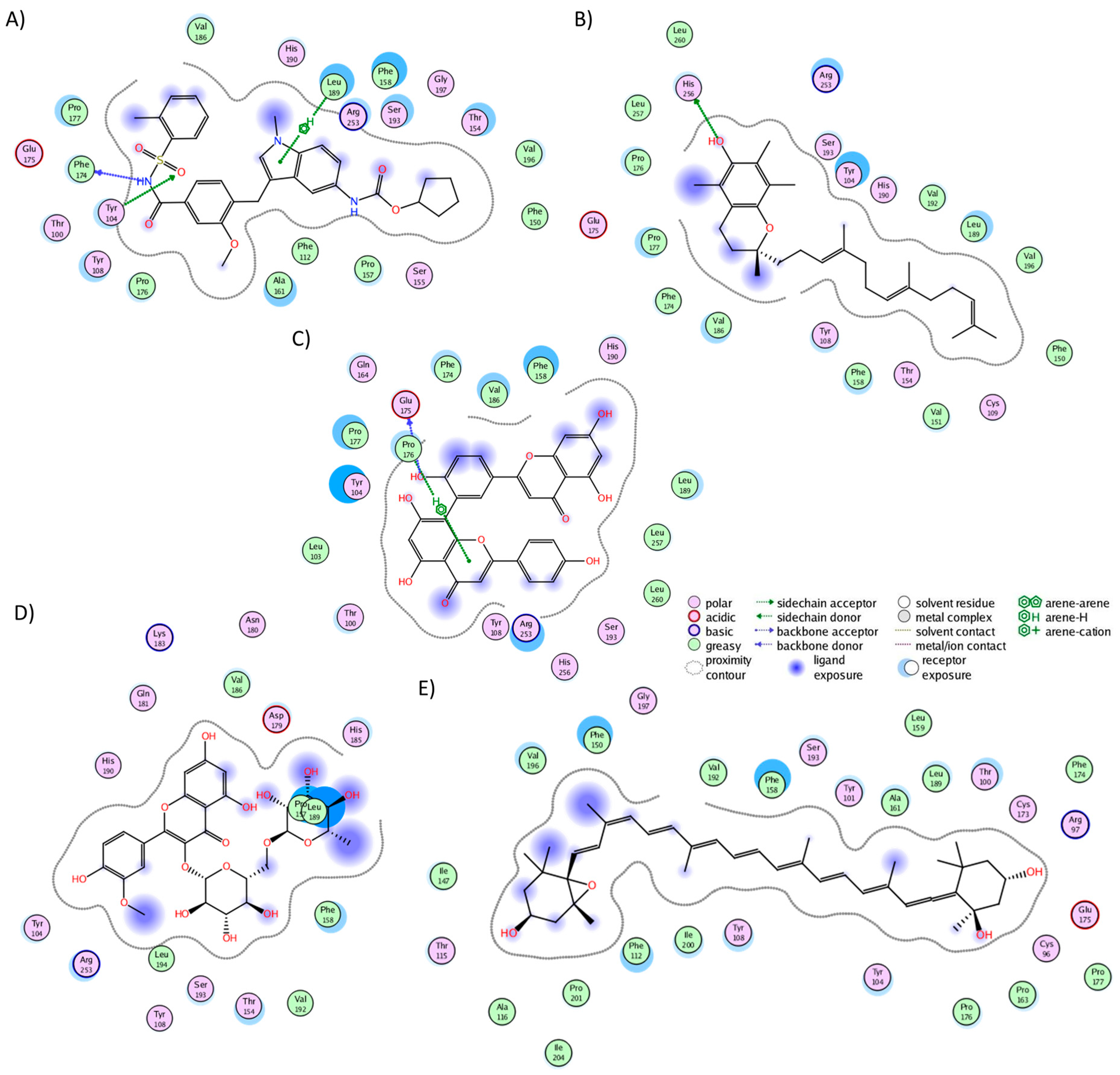
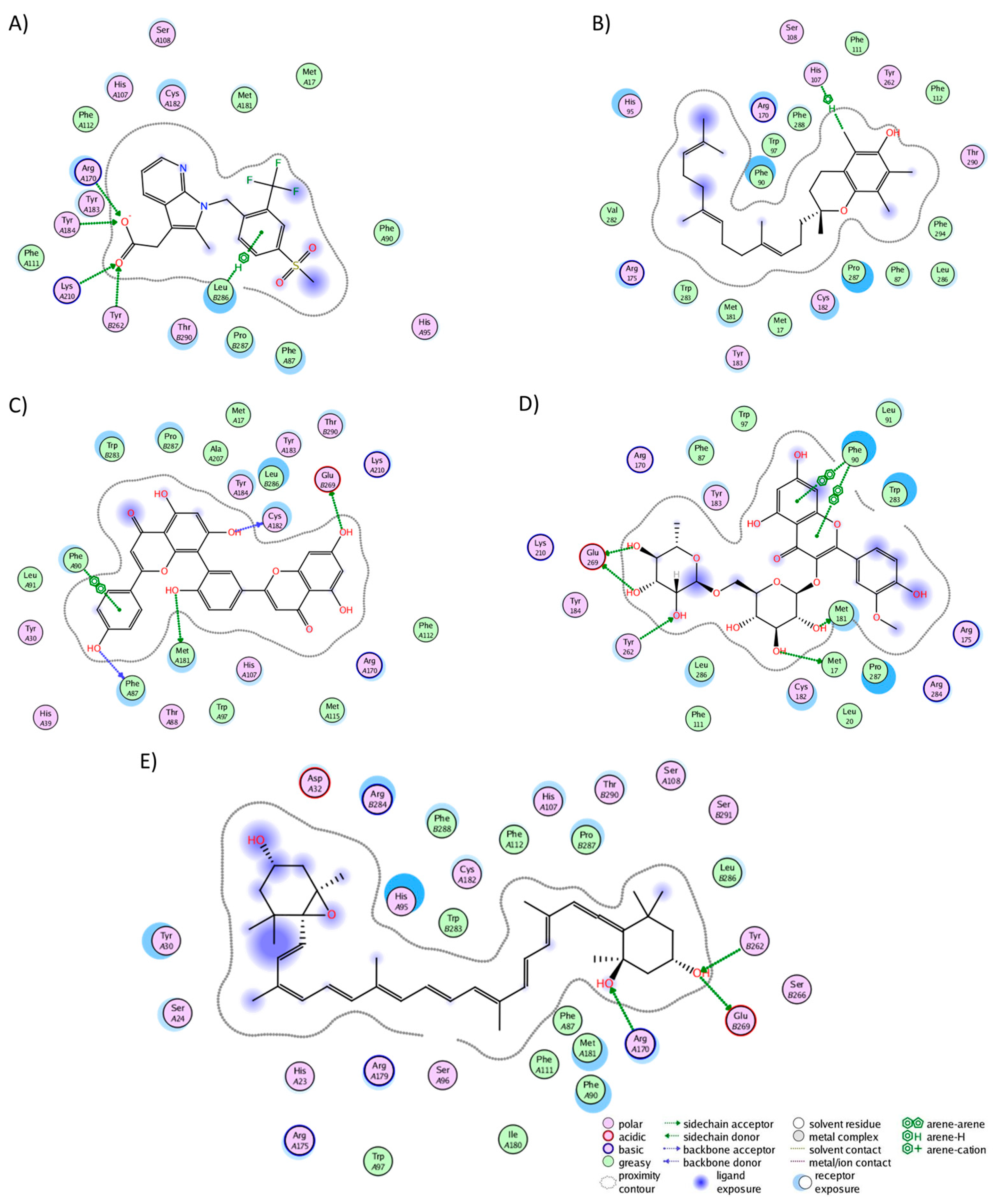
2.3. Analysis of Protein–Ligand Complexes
2.4. Binding Free Energy of UD Phytochemicals
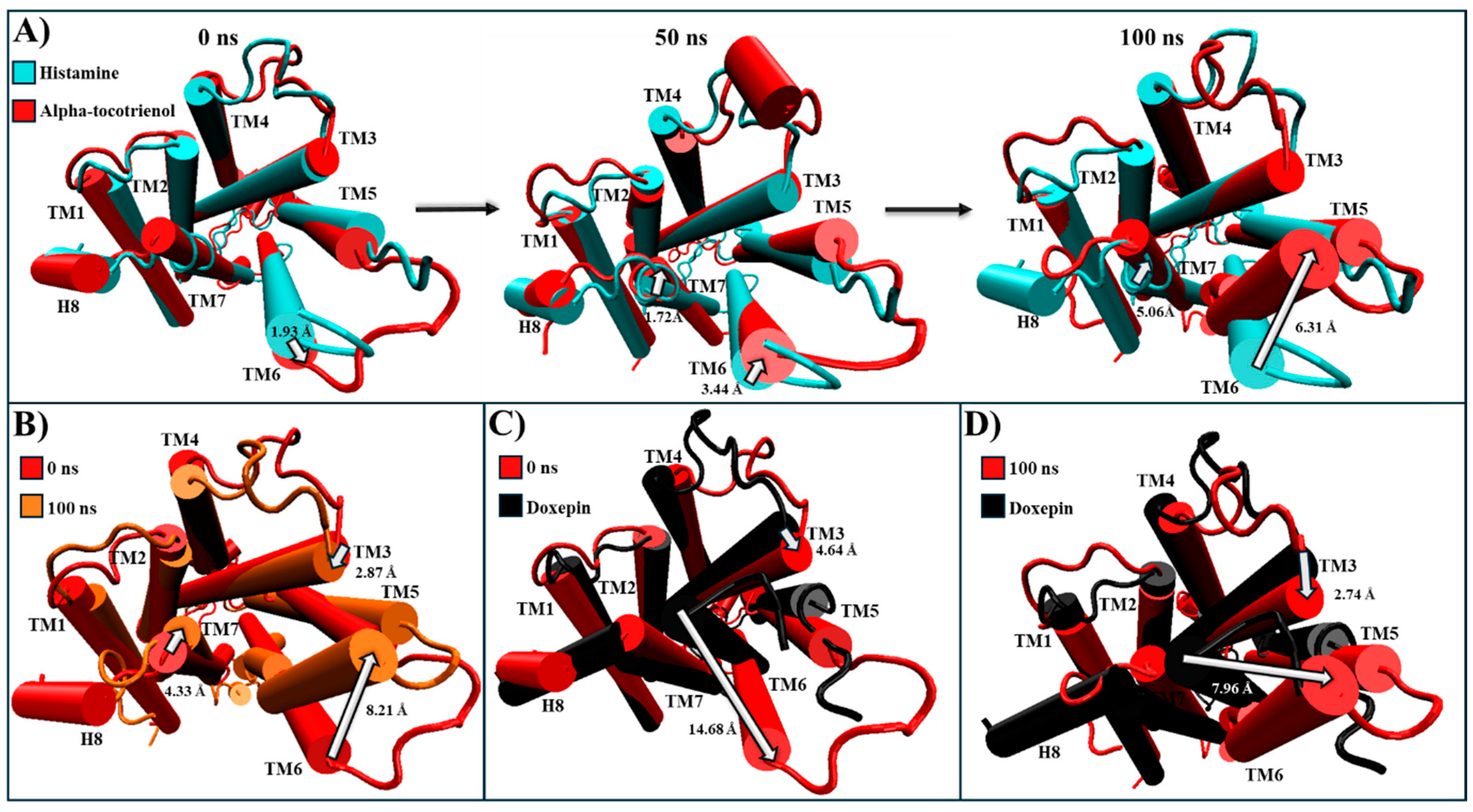
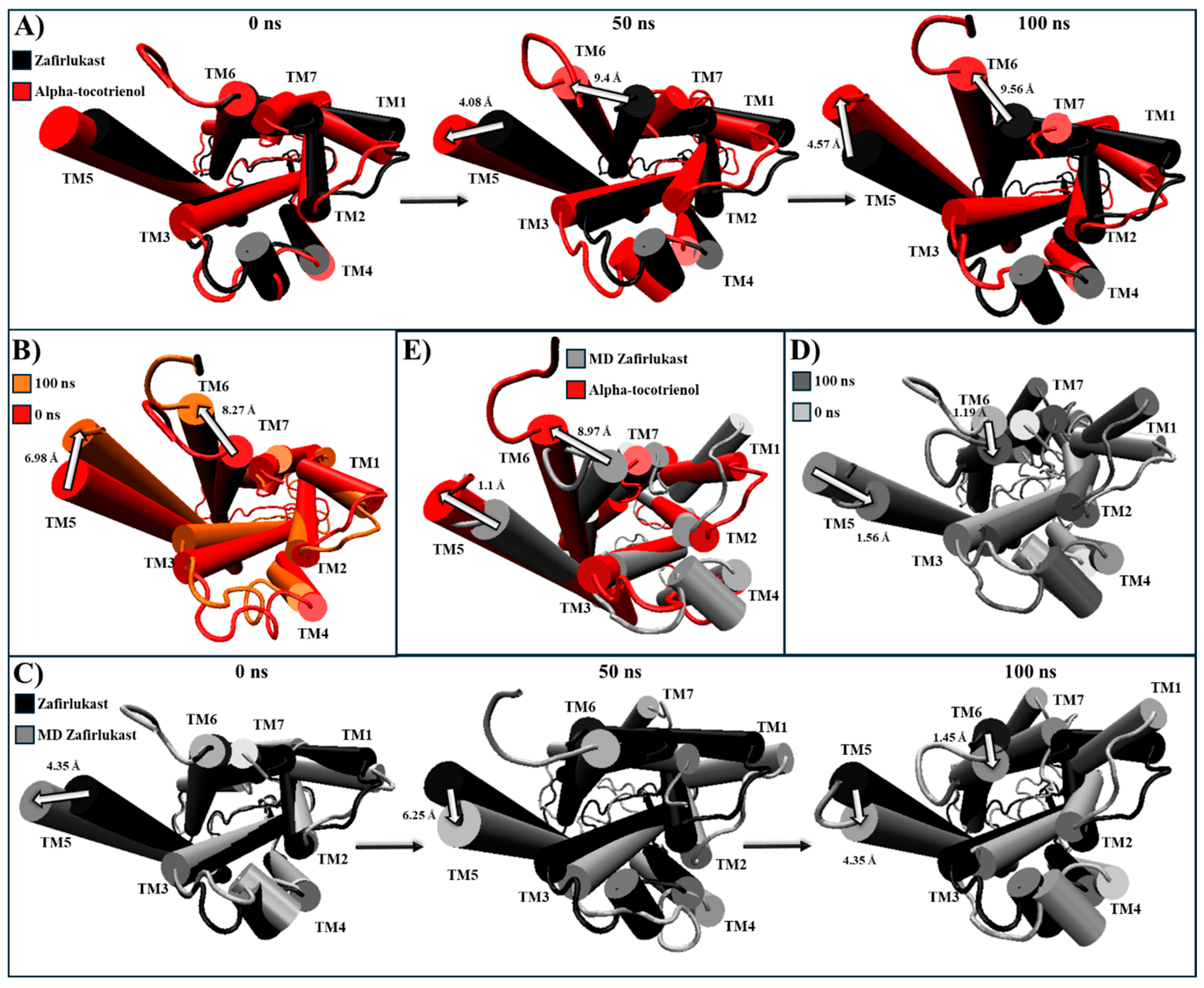
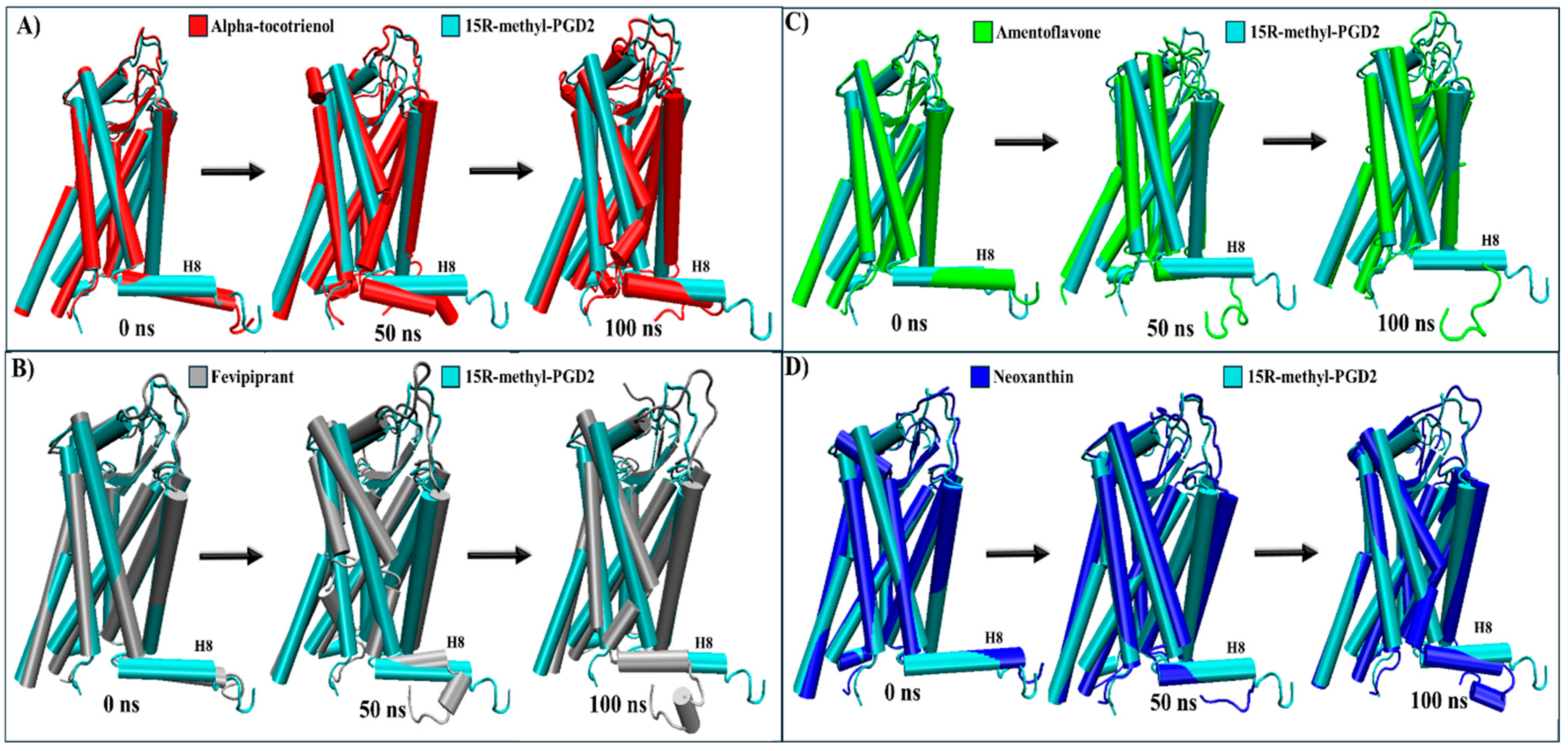
2.5. Evaluation of Structural Inactivation of Target Receptors
2.6. UD Phytochemicals’ Effects on Sensorial Nerves and Immune Cells
2.7. Perspective
3. Materials and Methods
3.1. Urtica dioica Phytochemicals
3.2. Molecular Docking
3.3. MD Positive and Negative Controls
3.4. MD Simulation
3.5. Binding Free Energy Studies with MMGBSA and MMPBSA
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tohidinik, H.R.; Mallah, N.; Takkouche, B. History of Allergic Rhinitis and Risk of Asthma; a Systematic Review and Meta-Analysis. World Allergy Organ. J. 2019, 12, 100069. [Google Scholar] [CrossRef] [PubMed]
- Greiner, A.N.; Hellings, P.W.; Rotiroti, G.; Scadding, G.K. Allergic Rhinitis. Lancet 2011, 378, 2112–2122. [Google Scholar] [CrossRef] [PubMed]
- Rahim, N.A.; Jantan, I.; Said, M.M.; Jalil, J.; Abd Razak, A.F.; Husain, K. Anti-Allergic Rhinitis Effects of Medicinal Plants and Their Bioactive Metabolites via Suppression of the Immune System: A Mechanistic Review. Front. Pharmacol. 2021, 12, 660083. [Google Scholar] [CrossRef] [PubMed]
- Kalmarzi, R.N.; Khazaei, Z.; Shahsavar, J.; Gharibi, F.; Tavakol, M.; Khazaei, S.; Shariat, M. The Impact of Allergic Rhinitis on Quality of Life: A Study in Western Iran. Biomed. Res. Ther. 2017, 4, 1629. [Google Scholar] [CrossRef]
- Bousquet, P.J.; Demoly, P.; Devillier, P.; Mesbah, K.; Bousquet, J. Impact of Allergic Rhinitis Symptoms on Quality of Life in Primary Care. Int. Arch. Allergy Immunol. 2013, 160, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Roger, A.; Arcalá Campillo, E.; Torres, M.C.; Millan, C.; Jáuregui, I.; Mohedano, E.; Liñan, S.; Verdu, P.; Rubira, N.; Santaolalla, M.; et al. Reduced Work/Academic Performance and Quality of Life in Patients with Allergic Rhinitis and Impact of Allergen Immunotherapy. Allergy Asthma Clin. Immunol. 2016, 12, 40. [Google Scholar] [CrossRef] [PubMed]
- Zuberbier, T.; Lötvall, J.; Simoens, S.; Subramanian, S.V.; Church, M.K. Economic Burden of Inadequate Management of Allergic Diseases in the European Union: A GA 2 LEN Review. Allergy 2014, 69, 1275–1279. [Google Scholar] [CrossRef] [PubMed]
- Wheatley, L.M.; Togias, A. Allergic Rhinitis. N. Engl. J. Med. 2015, 372, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Oray, M.; Abu Samra, K.; Ebrahimiadib, N.; Meese, H.; Foster, C.S. Long-Term Side Effects of Glucocorticoids. Expert. Opin. Drug Saf. 2016, 15, 457–465. [Google Scholar] [CrossRef]
- Cordell, G.A. Phytochemistry and Traditional Medicine—A Revolution in Process. Phytochem. Lett. 2011, 4, 391–398. [Google Scholar] [CrossRef]
- Yamprasert, R.; Chanvimalueng, W.; Mukkasombut, N.; Itharat, A. Ginger Extract versus Loratadine in the Treatment of Allergic Rhinitis: A Randomized Controlled Trial. BMC Complement Med. Ther. 2020, 20, 119. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.; Bravo-Clouzet, R. Anti-Inflammatory Herbs for Arthritis. In Bioactive Food as Dietary Interventions for Arthritis and Related Inflammatory Diseases; Elsevier: Amsterdam, The Netherlands, 2013; pp. 619–631. [Google Scholar]
- Bhusal, K.K.; Magar, S.K.; Thapa, R.; Lamsal, A.; Bhandari, S.; Maharjan, R.; Shrestha, S.; Shrestha, J. Nutritional and Pharmacological Importance of Stinging Nettle (Urtica dioica L.): A Review. Heliyon 2022, 8, e09717. [Google Scholar] [CrossRef] [PubMed]
- Mittman, P. Randomized, Double-Blind Study of Freeze-Dried Urtica dioica in the Treatment of Allergic Rhinitis. Planta Med. 1990, 56, 44–47. [Google Scholar] [CrossRef] [PubMed]
- Bakhshaee, M.; Mohammad Pour, A.H.; Esmaeili, M.; Jabbari Azad, F.; Alipour Talesh, G.; Salehi, M.; Noorollahian Mohajer, M. Efficacy of Supportive Therapy of Allergic Rhinitis by Stinging Nettle (Urtica dioica) Root Extract: A Randomized, Double-Blind, Placebo- Controlled, Clinical Trial. Iran J. Pharm. Res. 2017, 16, 112–118. [Google Scholar]
- Roschek, B., Jr.; Fink, R.C.; McMichael, M.; Alberte, R.S. Nettle Extract (Urtica dioica) Affects Key Receptors and Enzymes Associated with Allergic Rhinitis. Phytother. Res. Int. J. Devoted Pharmacol. Toxicol. Eval. Nat. Prod. Deriv. 2009, 23, 920–926. [Google Scholar] [CrossRef] [PubMed]
- Zemmouri, H.; Sekiou, O.; Ammar, S.; El Feki, A.; Bouaziz, M.; Messarah, M.; Boumendjel, A. Urtica dioica Attenuates Ovalbumin-Induced Inflammation and Lipid Peroxidation of Lung Tissues in Rat Asthma Model. Pharm. Biol. 2017, 55, 1561–1568. [Google Scholar] [CrossRef] [PubMed]
- Mwamatope, B.; Tembo, D.; Kampira, E.; Maliwichi-Nyirenda, C.; Ndolo, V. Seasonal Variation of Phytochemicals in Four Selected Medicinal Plants. Pharmacogn. Res. 2021, 13, 218–226. [Google Scholar] [CrossRef]
- Culhuac, E.B.; Maggiolino, A.; Elghandour, M.M.M.Y.; De Palo, P.; Salem, A.Z.M. Antioxidant and Anti-Inflammatory Properties of Phytochemicals Found in the Yucca Genus. Antioxidants 2023, 12, 574. [Google Scholar] [CrossRef]
- Raya, K.B.; Ahmad, S.H.; Farhana, S.F.; Mohammad, M.; Tajidin, N.E.; Parvez, A. Changes in Phytochemical Contents in Different Parts of Clinacanthus Nutans (Burm. f.) Lindau Due to Storage Duration. Bragantia 2015, 74, 445–452. [Google Scholar] [CrossRef]
- Aremu, A.O.; Masondo, N.A.; Molnár, Z.; Stirk, W.A.; Ördög, V.; Van Staden, J. Changes in Phytochemical Content and Pharmacological Activities of Three Chlorella Strains Grown in Different Nitrogen Conditions. J. Appl. Phycol. 2016, 28, 149–159. [Google Scholar] [CrossRef]
- Nobossé, P.; Fombang, E.N.; Mbofung, C.M.F. Effects of Age and Extraction Solvent on Phytochemical Content and Antioxidant Activity of Fresh Moringa Oleifera L. Leaves. Food Sci. Nutr. 2018, 6, 2188–2198. [Google Scholar] [CrossRef] [PubMed]
- Henning, T.; Quandt, D.; Grosse-Veldmann, B.; Monro, A.; Weigend, M. Weeding the Nettles II: A Delimitation of “Urtica dioica L.” (Urticaceae) Based on Morphological and Molecular Data, Including a Rehabilitation of Urtica Gracilis Ait. Phytotaxa 2014, 162, 61. [Google Scholar] [CrossRef]
- Grosse-Veldmann, B.; Nürk, N.M.; Smissen, R.; Breitwieser, I.; Quandt, D.; Weigend, M. Pulling the Sting out of Nettle Systematics—A Comprehensive Phylogeny of the Genus Urtica L. (Urticaceae). Mol. Phylogenet Evol. 2016, 102, 9–19. [Google Scholar] [CrossRef]
- Semalty, A.; Semalty, M.; Rawat, M.S.M.; Franceschi, F. Supramolecular Phospholipids–Polyphenolics Interactions: The PHYTOSOME® Strategy to Improve the Bioavailability of Phytochemicals. Fitoterapia 2010, 81, 306–314. [Google Scholar] [CrossRef]
- Thangam, E.B.; Jemima, E.A.; Singh, H.; Baig, M.S.; Khan, M.; Mathias, C.B.; Church, M.K.; Saluja, R. The Role of Histamine and Histamine Receptors in Mast Cell-Mediated Allergy and Inflammation: The Hunt for New Therapeutic Targets. Front. Immunol. 2018, 9, 1873. [Google Scholar] [CrossRef]
- Jutel, M.; Akdis, M.; Akdis, C.A. Histamine, Histamine Receptors and Their Role in Immune Pathology. Clin. Exp. Allergy 2009, 39, 1786–1800. [Google Scholar] [CrossRef]
- Hoyte, F.C.L.; Katial, R.K. Antihistamine Therapy in Allergic Rhinitis. Immunol. Allergy Clin. N. Am. 2011, 31, 509–543. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, R.; Wu, J.; Hu, H. Knockdown of Neurokinin-1 Receptor Expression by Small Interfering RNA Prevents the Development of Allergic Rhinitis in Rats. Inflamm. Res. 2013, 62, 903–910. [Google Scholar] [CrossRef] [PubMed]
- Joachim, R.A.; Sagach, V.; Quarcoo, D.; Dinh, Q.T.; Arck, P.C.; Klapp, B.F. Neurokinin-1 Receptor Mediates Stress-Exacerbated Allergic Airway Inflammation and Airway Hyperresponsiveness in Mice. Psychosom. Med. 2004, 66, 564–571. [Google Scholar] [CrossRef]
- Peters-Golden, M.; Henderson, W.R. The Role of Leukotrienes in Allergic Rhinitis. Ann. Allergy Asthma Immunol. 2005, 94, 609–618. [Google Scholar] [CrossRef]
- Okano, M.; Fujiwara, T.; Sugata, Y.; Gotoh, D.; Masaoka, Y.; Sogo, M.; Tanimoto, W.; Yamamoto, M.; Matsumoto, R.; Eguchi, N.; et al. Presence and Characterization of Prostaglandin D2–Related Molecules in Nasal Mucosa of Patients with Allergic Rhinitis. Am. J. Rhinol. 2006, 20, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Turner, P.; Dear, J.; Scadding, G.; Foreman, J.C. Role of Kinins in Seasonal Allergic Rhinitis: Icatibant, a Bradykinin B2 Receptor Antagonist, Abolishes the Hyperresponsiveness and Nasal Eosinophilia Induced by Antigen. J. Allergy Clin. Immunol. 2001, 107, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Abraham, W.M.; Scuri, M.; Farmer, S.G. Peptide and Non-Peptide Bradykinin Receptor Antagonists: Role in Allergic Airway Disease. Eur. J. Pharmacol. 2006, 533, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Ohta, K.; Hayashi, H.; Mizuguchi, H.; Kagamiyama, H.; Fujimoto, K.; Fukui, H. Site-Directed Mutagenesis of the Histamine H1 Receptor: Roles of Aspartic Acid107, Asparagine198 and Threonine194. Biochem. Biophys Res. Commun. 1994, 203, 1096–1101. [Google Scholar] [CrossRef] [PubMed]
- Kiss, R.; Kovári, Z.; Keserű, G.M. Homology Modelling and Binding Site Mapping of the Human Histamine H1 Receptor. Eur. J. Med. Chem. 2004, 39, 959–967. [Google Scholar] [CrossRef] [PubMed]
- Shimamura, T.; Shiroishi, M.; Weyand, S.; Tsujimoto, H.; Winter, G.; Katritch, V.; Abagyan, R.; Cherezov, V.; Liu, W.; Han, G.W.; et al. Structure of the Human Histamine H1 Receptor Complex with Doxepin. Nature 2011, 475, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, Y.; Pan, Y.; Wang, J.; Lin, F.; Wang, C.; Zhang, S.; Yang, L. Computational Analysis of Structure-Based Interactions for Novel H1-Antihistamines. Int. J. Mol. Sci. 2016, 17, 129. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Lu, M.; Liu, D.; Yang, L.; Yi, C.; Ma, L.; Zhang, H.; Liu, Q.; Frimurer, T.M.; Wang, M.-W.; et al. Human Substance P Receptor Binding Mode of the Antagonist Drug Aprepitant by NMR and Crystallography. Nat. Commun. 2019, 10, 638. [Google Scholar] [CrossRef]
- Luginina, A.; Gusach, A.; Marin, E.; Mishin, A.; Brouillette, R.; Popov, P.; Shiriaeva, A.; Besserer-Offroy, É.; Longpré, J.-M.; Lyapina, E.; et al. Structure-Based Mechanism of Cysteinyl Leukotriene Receptor Inhibition by Antiasthmatic Drugs. Sci. Adv. 2019, 5, eaax2518. [Google Scholar] [CrossRef]
- Liu, H.; Deepak, R.N.V.K.; Shiriaeva, A.; Gati, C.; Batyuk, A.; Hu, H.; Weierstall, U.; Liu, W.; Wang, L.; Cherezov, V.; et al. Molecular Basis for Lipid Recognition by the Prostaglandin D 2 Receptor CRTH2. Proc. Natl. Acad. Sci. USA 2021, 118, e2102813118. [Google Scholar] [CrossRef]
- Gibson, C.; Schnatbaum, K.; Pfeifer, J.R.; Locardi, E.; Paschke, M.; Reimer, U.; Richter, U.; Scharn, D.; Faussner, A.; Tradler, T. Novel Small Molecule Bradykinin B 2 Receptor Antagonists. J. Med. Chem. 2009, 52, 4370–4379. [Google Scholar] [CrossRef] [PubMed]
- Faussner, A.; Schüssler, S.; Feierler, J.; Bermudez, M.; Pfeifer, J.; Schnatbaum, K.; Tradler, T.; Jochum, M.; Wolber, G.; Gibson, C. Binding Characteristics of [3 H]-JSM10292: A New Cell Membrane-permeant Non-peptide Bradykinin B 2 Receptor Antagonist. Br. J. Pharmacol. 2012, 167, 839–853. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, S.F. Combination Therapy in the Treatment of Allergic Rhinitis. Allergy Asthma Proc. 2002, 23, 1–3. [Google Scholar]
- Kudlacz, E.; Shatzer, S.; Logan, D.; Olsen, K.; Knippenberg, R.; Hsieh, L.; Esteve, H.; Maynard, G. A Role for Histamine and Substance P in Immediate Allergic Responses in Guinea Pig Airways: Characterization of MDL 108,207DA, a Dual H1/NK-1 Receptor Antagonist. Int. Arch. Allergy Immunol. 1998, 115, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-K.; Lee, S.Y.; Park, H.-S.; Yoon, H.J.; Kim, S.-H.; Cho, Y.J.; Yoo, K.-H.; Lee, S.-K.; Kim, H.-K.; Park, J.-W.; et al. A Randomized, Multicenter, Double-Blind, Phase III Study to Evaluate the Efficacy on Allergic Rhinitis and Safety of a Combination Therapy of Montelukast and Levocetirizine in Patients With Asthma and Allergic Rhinitis. Clin. Ther. 2018, 40, 1096–1107.e1. [Google Scholar] [CrossRef] [PubMed]
- Repajić, M.; Cegledi, E.; Zorić, Z.; Pedisić, S.; Elez Garofulić, I.; Radman, S.; Palčić, I.; Dragović-Uzelac, V. Bioactive Compounds in Wild Nettle (Urtica dioica L.) Leaves and Stalks: Polyphenols and Pigments upon Seasonal and Habitat Variations. Foods 2021, 10, 190. [Google Scholar] [CrossRef] [PubMed]
- Pinelli, P.; Ieri, F.; Vignolini, P.; Bacci, L.; Baronti, S.; Romani, A. Extraction and HPLC Analysis of Phenolic Compounds in Leaves, Stalks, and Textile Fibers of Urtica dioica L. J. Agric. Food Chem. 2008, 56, 9127–9132. [Google Scholar] [CrossRef] [PubMed]
- Garcìa, L.M.; Ceccanti, C.; Negro, C.; De Bellis, L.; Incrocci, L.; Pardossi, A.; Guidi, L. Effect of Drying Methods on Phenolic Compounds and Antioxidant Activity of Urtica dioica L. Leaves. Horticulturae 2021, 7, 10. [Google Scholar] [CrossRef]
- Grauso, L.; de Falco, B.; Lanzotti, V.; Motti, R. Stinging Nettle, Urtica dioica L.: Botanical, Phytochemical and Pharmacological Overview. Phytochem. Rev. 2020, 19, 1341–1377. [Google Scholar] [CrossRef]
- Orčić, D.; Francišković, M.; Bekvalac, K.; Svirčev, E.; Beara, I.; Lesjak, M.; Mimica-Dukić, N. Quantitative Determination of Plant Phenolics in Urtica dioica Extracts by High-Performance Liquid Chromatography Coupled with Tandem Mass Spectrometric Detection. Food Chem. 2014, 143, 48–53. [Google Scholar] [CrossRef]
- Guil-Guerrero, J.L.; Rebolloso-Fuentes, M.M.; Isasa, M.E.T. Fatty Acids and Carotenoids from Stinging Nettle (Urtica dioica L.). J. Food Compos. Anal. 2003, 16, 111–119. [Google Scholar] [CrossRef]
- Farag, M.A.; Weigend, M.; Luebert, F.; Brokamp, G.; Wessjohann, L.A. Phytochemical, Phylogenetic, and Anti-Inflammatory Evaluation of 43 Urtica Accessions (Stinging Nettle) Based on UPLC–Q-TOF-MS Metabolomic Profiles. Phytochemistry 2013, 96, 170–183. [Google Scholar] [CrossRef] [PubMed]
- Chaurasia, N.; Wichtl, M. Flavonolglykoside Aus Urtica dioica1,2. Planta Med. 1987, 53, 432–434. [Google Scholar] [CrossRef] [PubMed]
- Sader, S.; Cai, J.; Muller, A.C.G.; Wu, C. Can Human Allergy Drug Fexofenadine, an Antagonist of Histamine (H1) Receptor, Be Used to Treat Dog and Cat? Homology Modeling, Docking and Molecular Dynamic Simulation of Three H1 Receptors in Complex with Fexofenadine. J. Mol. Graph. Model. 2017, 75, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Xia, R.; Wang, N.; Xu, Z.; Lu, Y.; Song, J.; Zhang, A.; Guo, C.; He, Y. Cryo-EM Structure of the Human Histamine H1 Receptor/Gq Complex. Nat. Commun. 2021, 12, 2086. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.A.; Faust, B.; Gondin, A.B.; Dämgen, M.A.; Suomivuori, C.-M.; Veldhuis, N.A.; Cheng, Y.; Dror, R.O.; Thal, D.M.; Manglik, A. Selective G Protein Signaling Driven by Substance P–Neurokinin Receptor Dynamics. Nat. Chem. Biol. 2022, 18, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yao, D.; Deepak, R.N.V.K.; Liu, H.; Xiao, Q.; Fan, H.; Gong, W.; Wei, Z.; Zhang, C. Structures of the Human PGD2 Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition. Mol. Cell 2018, 72, 48–59.e4. [Google Scholar] [CrossRef]
- Sun, H.; Li, Y.; Tian, S.; Xu, L.; Hou, T. Assessing the Performance of MM/PBSA and MM/GBSA Methods. 4. Accuracies of MM/PBSA and MM/GBSA Methodologies Evaluated by Various Simulation Protocols Using PDBbind Data Set. Phys. Chem. Chem. Phys. 2014, 16, 16719–16729. [Google Scholar] [CrossRef]
- Weis, W.I.; Kobilka, B.K. The Molecular Basis of G Protein–Coupled Receptor Activation. Annu. Rev. Biochem. 2018, 87, 897–919. [Google Scholar] [CrossRef]
- Schröder, R.; Merten, N.; Mathiesen, J.M.; Martini, L.; Kruljac-Letunic, A.; Krop, F.; Blaukat, A.; Fang, Y.; Tran, E.; Ulven, T.; et al. The C-Terminal Tail of CRTH2 Is a Key Molecular Determinant That Constrains Gαi and Downstream Signaling Cascade Activation. J. Biol. Chem. 2009, 284, 1324–1336. [Google Scholar] [CrossRef]
- Issahaku, A.R.; Agoni, C.; Kumi, R.O.; Olotu, F.A.; Soliman, M.E.S. Lipid-Embedded Molecular Dynamics Simulation Model for Exploring the Reverse Prostaglandin D2 Agonism of CT-133 towards CRTH2 in the Treatment of Type-2 Inflammation Dependent Diseases. Chem. Biodivers 2020, 17, e1900548. [Google Scholar] [CrossRef] [PubMed]
- Thom, C.; Ehrenmann, J.; Vacca, S.; Waltenspühl, Y.; Schöppe, J.; Medalia, O.; Plückthun, A. Structures of Neurokinin 1 Receptor in Complex with Gq and Gs Proteins Reveal Substance P Binding Mode and Unique Activation Features. Sci. Adv. 2021, 7, eabk2872. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhang, D.; Fu, Y.; Chen, A.; Yang, X.; Zhang, H. Cryo-EM Structures of Human Bradykinin Receptor-Gq Proteins Complexes. Nat. Commun. 2022, 13, 714. [Google Scholar] [CrossRef] [PubMed]
- Pawankar, R.; Mori, S.; Ozu, C.; Kimura, S. Overview on the Pathomechanisms of Allergic Rhinitis. Asia Pac. Allergy 2011, 1, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Al-Shamlan, F.; El-Hashim, A.Z. Bradykinin Sensitizes the Cough Reflex via a B2 Receptor Dependent Activation of TRPV1 and TRPA1 Channels through Metabolites of Cyclooxygenase and 12-Lipoxygenase. Respir. Res. 2019, 20, 110. [Google Scholar] [CrossRef] [PubMed]
- Kopřiva, F.; Sobolová, L.; Szotkowská, J.; Zápalka, M. Treatment of Chronic Cough in Children with Montelukast, a Leukotriene Receptor Antagonist. J. Asthma 2004, 41, 715–720. [Google Scholar] [CrossRef] [PubMed]
- Ikoma, A.; Steinhoff, M.; Ständer, S.; Yosipovitch, G.; Schmelz, M. The Neurobiology of Itch. Nat. Rev. Neurosci. 2006, 7, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Broadhead, A.; Stebbins, K.J.; Stock, N.S.; Coate, H.; Evans, J.F.; Lorrain, D.S. AM206, A Novel CRTH2 Selective Antagonist, Inhibits Sneezing And Nasal Rubs In A Mouse Allergic Rhinitis Model. In Proceedings of the C21. Mechanisms of Th2 Inflammation in the Lung, San Diego, CA, USA, 4 May 2010; American Thoracic Society: New York, NY, USA, 2010; p. A4047. [Google Scholar]
- Driessen, A.K.; McGovern, A.E.; Behrens, R.; Moe, A.A.K.; Farrell, M.J.; Mazzone, S.B. A Role for Neurokinin 1 Receptor Expressing Neurons in the Paratrigeminal Nucleus in Bradykinin-evoked Cough in Guinea-pigs. J. Physiol. 2020, 598, 2257–2275. [Google Scholar] [CrossRef] [PubMed]
- Veldhuis, N.A.; Poole, D.P.; Grace, M.; McIntyre, P.; Bunnett, N.W. The G Protein–Coupled Receptor–Transient Receptor Potential Channel Axis: Molecular Insights for Targeting Disorders of Sensation and Inflammation. Pharmacol. Rev. 2015, 67, 36–73. [Google Scholar] [CrossRef]
- Geppetti, P.; Veldhuis, N.A.; Lieu, T.; Bunnett, N.W. G Protein-Coupled Receptors: Dynamic Machines for Signaling Pain and Itch. Neuron 2015, 88, 635–649. [Google Scholar] [CrossRef]
- Backaert, W.; Steelant, B.; Hellings, P.W.; Talavera, K.; Van Gerven, L. A TRiP Through the Roles of Transient Receptor Potential Cation Channels in Type 2 Upper Airway Inflammation. Curr. Allergy Asthma Rep. 2021, 21, 20. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-Set Enrichment Tool for Animals and Plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Wilzopolski, J.; Kietzmann, M.; Mishra, S.K.; Stark, H.; Bäumer, W.; Rossbach, K. TRPV1 and TRPA1 Channels Are Both Involved Downstream of Histamine-Induced Itch. Biomolecules 2021, 11, 1166. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cang, C.-L.; Kawasaki, Y.; Liang, L.-L.; Zhang, Y.-Q.; Ji, R.-R.; Zhao, Z.-Q. Neurokinin-1 Receptor Enhances TRPV1 Activity in Primary Sensory Neurons via PKCε: A Novel Pathway for Heat Hyperalgesia. J. Neurosci. 2007, 27, 12067–12077. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, S.; Devesa, I.; Changeux, J.-P.; Ferrer-Montiel, A. Bradykinin Induces TRPV1 Exocytotic Recruitment in Peptidergic Nociceptors. Front. Pharmacol. 2016, 7, 178. [Google Scholar] [CrossRef]
- Alenmyr, L.; Högestätt, E.D.; Zygmunt, P.M.; Greiff, L. TRPV1-mediated Itch in Seasonal Allergic Rhinitis. Allergy 2009, 64, 807–810. [Google Scholar] [CrossRef] [PubMed]
- Samivel, R.; Kim, D.W.; Son, H.R.; Rhee, Y.-H.; Kim, E.H.; Kim, J.H.; Bae, J.-S.; Chung, Y.-J.; Chung, P.-S.; Raz, E.; et al. The Role of TRPV1 in the CD4+ T Cell-Mediated Inflammatory Response of Allergic Rhinitis. Oncotarget 2016, 7, 148–160. [Google Scholar] [CrossRef] [PubMed]
- Tourangeau, L.M.; Christiansen, S.C.; Herschbach, J.; Brooks, S.M.; Eddleston, J.; Zuraw, B. Nasal Mucosal TRPA1 and TRPV1 Levels in Human Rhinitis. J. Allergy Clin. Immunol. 2011, 127, AB52. [Google Scholar] [CrossRef]
- Bao, C.; Chen, O.; Sheng, H.; Zhang, J.; Luo, Y.; Hayes, B.W.; Liang, H.; Liedtke, W.; Ji, R.-R.; Abraham, S.N. A Mast Cell–Thermoregulatory Neuron Circuit Axis Regulates Hypothermia in Anaphylaxis. Sci. Immunol. 2023, 8, eadc9417. [Google Scholar] [CrossRef]
- Church, D.S.; Church, M.K. Pharmacology of Antihistamines. World Allergy Organ. J. 2011, 4, S22–S27. [Google Scholar] [CrossRef]
- Han, E.D.; MacFarlane, R.C.; Mulligan, A.N.; Scafidi, J.; Davis, A.E. Increased Vascular Permeability in C1 Inhibitor–Deficient Mice Mediated by the Bradykinin Type 2 Receptor. J. Clin. Investig. 2002, 109, 1057–1063. [Google Scholar] [CrossRef] [PubMed]
- Ashina, K.; Tsubosaka, Y.; Nakamura, T.; Omori, K.; Kobayashi, K.; Hori, M.; Ozaki, H.; Murata, T. Histamine Induces Vascular Hyperpermeability by Increasing Blood Flow and Endothelial Barrier Disruption In Vivo. PLoS ONE 2015, 10, e0132367. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wu, J.; Zhang, R. Effect of Neurokinin-1 Receptor Knockdown on the Expression of RANTES in Allergic Rhinitis. Am. J. Rhinol. Allergy 2023, 37, 730–738. [Google Scholar] [CrossRef]
- Wang, S.; Liu, L.; Blanco, T.; Zhu, S.; Pang, K.; Ge, H.; Chen, Y.; Dana, R. The Effect of Blocking Substance P in Reducing Ocular Redness. Investig. Ophthalmol. Vis. Sci. 2021, 62, 399. [Google Scholar]
- Doherty, T.A.; Khorram, N.; Lund, S.; Mehta, A.K.; Croft, M.; Broide, D.H. Lung Type 2 Innate Lymphoid Cells Express Cysteinyl Leukotriene Receptor 1, Which Regulates TH2 Cytokine Production. J. Allergy Clin. Immunol. 2013, 132, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Stevens, W.W.; Kato, A. Group 2 Innate Lymphoid Cells in Nasal Polyposis. Ann. Allergy Asthma Immunol. 2021, 126, 110–117. [Google Scholar] [CrossRef]
- Chen, W.; He, S.; Xie, X.; Yang, X.; Duan, C.; Ye, P.; Li, X.; Lawrence, M.G.; Borish, L.; Feng, X. Over-Expression of CRTH2 Indicates Eosinophilic Inflammation and Poor Prognosis in Recurrent Nasal Polyps. Front. Immunol. 2022, 13, 1046426. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, B. The Role of Nutrition in Age-Related Eye Diseases. In Molecular Basis of Nutrition and Aging; Elsevier: Amsterdam, The Netherlands, 2016; pp. 433–446. [Google Scholar]
- Wang, S.; Wang, Y.; Pan, C.; Sun, J. Serum Level and Clinical Significance of Vitamin E in Children with Allergic Rhinitis. BMC Pediatr. 2020, 20, 362. [Google Scholar] [CrossRef]
- Wu, S.; Wang, A. Serum Level and Clinical Significance of Vitamin E in Pregnant Women with Allergic Rhinitis. J. Chin. Med. Assoc. 2022, 85, 597–602. [Google Scholar] [CrossRef]
- Jiang, J.; Mehrabi Nasab, E.; Athari, S.M.; Athari, S.S. Effects of Vitamin E and Selenium on Allergic Rhinitis and Asthma Pathophysiology. Respir. Physiol. Neurobiol. 2021, 286, 103614. [Google Scholar] [CrossRef]
- Shahar, E.; Hassoun, G.; Pollack, S. Effect of Vitamin E Supplementation on the Regular Treatment of Seasonal Allergic Rhinitis. Ann. Allergy Asthma Immunol. 2004, 92, 654–658. [Google Scholar] [CrossRef]
- Cerqua, I.; Neukirch, K.; Terlizzi, M.; Granato, E.; Caiazzo, E.; Cicala, C.; Ialenti, A.; Capasso, R.; Werz, O.; Sorrentino, R.; et al. A Vitamin E Long-Chain Metabolite and the Inspired Drug Candidate α-Amplexichromanol Relieve Asthma Features in an Experimental Model of Allergen Sensitization. Pharmacol. Res. 2022, 181, 106250. [Google Scholar] [CrossRef] [PubMed]
- Teo, C.W.L.; Png, S.J.Y.; Ung, Y.W.; Yap, W.N. Therapeutic Effects of Intranasal Tocotrienol-Rich Fraction on Rhinitis Symptoms in Platelet-Activating Factor Induced Allergic Rhinitis. Allergy Asthma Clin. Immunol. 2022, 18, 52. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Cai, W.; Zhang, H.-H.; Zhong, Y.-S.; Fang, J.; Zhang, W.-Y.; Mo, L.; Wang, L.-C.; Yu, C.-H. Selaginella Uncinata Flavonoids Ameliorated Ovalbumin-Induced Airway Inflammation in a Rat Model of Asthma. J. Ethnopharmacol. 2017, 195, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, M.-S.; Kim, H.-Y.; Kim, S.; Kam, H.; Choi, H.; Shin, D.H. Amentoflavone, a Potent Natural Matrix Metalloproteinase 2 Inhibitor. Nat. Prod. Res. 2023, 1–8. [Google Scholar] [CrossRef]
- Rong, S.; Yang, C.; Wang, F.; Wu, Y.; Sun, K.; Sun, T.; Wu, Z. Amentoflavone Exerts Anti-Neuroinflammatory Effects by Inhibiting TLR4/MyD88/NF-ΚB and Activating Nrf2/HO-1 Pathway in Lipopolysaccharide-Induced BV2 Microglia. Mediat. Inflamm. 2022, 2022, 5184721. [Google Scholar] [CrossRef]
- Ilies, D.C.; Tudor, I.; Radulescu, V. Chemical Composition of the Essential Oil of Urtica dioica. Chem. Nat. Compd. 2012, 48, 506–507. [Google Scholar] [CrossRef]
- Gül, S.; Demirci, B.; Başer, K.H.C.; Akpulat, H.A.; Aksu, P. Chemical Composition and In Vitro Cytotoxic, Genotoxic Effects of Essential Oil from Urtica dioica L. Bull. Environ. Contam. Toxicol. 2012, 88, 666–671. [Google Scholar] [CrossRef]
- Ayers, S.; Roschek, B., Jr.; Williams, J.M.; Alberte, R.S. Pharmacokinetic Analysis of Anti-Allergy and Anti-Inflammation Bioactives in a Nettle (Urtica dioica) Extract. Online J. Pharmacol. Pharm. Kinet. 2008, 5, 6–21. [Google Scholar]
- Grauso, L.; Emrick, S.; Bonanomi, G.; Lanzotti, V. Metabolomics of the Alimurgic Plants Taraxacum Officinale, Papaver Rhoeas and Urtica dioica by Combined NMR and GC–MS Analysis. Phytochem. Anal. 2019, 30, 535–546. [Google Scholar] [CrossRef]
- Majedi, S.; Abdulsattar Faraj, T.; Jalal Ahmed, H.; HS Hussain, F. A Review of Biochemical Structures of Urtica dioica Metabolites and Their Pharmaceutical Effects. Chem. Rev. Lett. 2021, 4, 206–212. [Google Scholar] [CrossRef]
- Wang, Y.; Xiao, J.; Suzek, T.O.; Zhang, J.; Wang, J.; Bryant, S.H. PubChem: A Public Information System for Analyzing Bioactivities of Small Molecules. Nucleic Acids. Res. 2009, 37, W623–W633. [Google Scholar] [CrossRef] [PubMed]
- Chemical Computing Group Inc. Molecular Operating Environment (MOE); Chemical Computing Group Inc.: Montrea, QC, Canada, 2016; p. 1010. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System (Version 2.3.4.); DeLano Scientific LLC.: San Carlos, CA, USA, 2022. [Google Scholar]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids. Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- Arcaniolo, D.; Conquy, S.; Tarcan, T. Flavoxate: Present and Future. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 719–731. [Google Scholar]
- Cazzulani, P.; Panzarasa, R.; Luca, C.; Oliva, D.; Graziani, G. Pharmacological Studies on the Mode of Action of Flavoxate. Arch. Int. Pharmacodyn. Ther. 1984, 268, 301–312. [Google Scholar]
- Case, D.A.; Cheatham, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber Biomolecular Simulation Programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and Testing of a General Amber Force Field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, Efficient Generation of High-quality Atomic Charges. AM1-BCC Model: II. Parameterization and Validation. J. Comput. Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef]
- Tian, C.; Kasavajhala, K.; Belfon, K.A.A.; Raguette, L.; Huang, H.; Migues, A.N.; Bickel, J.; Wang, Y.; Pincay, J.; Wu, Q.; et al. Ff19SB: Amino-Acid-Specific Protein Backbone Parameters Trained against Quantum Mechanics Energy Surfaces in Solution. J. Chem. Theory Comput. 2020, 16, 528–552. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald: An N ⋅log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- York, D.M.; Darden, T.A.; Pedersen, L.G. The Effect of Long-Range Electrostatic Interactions in Simulations of Macromolecular Crystals: A Comparison of the Ewald and Truncated List Methods. J. Chem. Phys. 1993, 99, 8345–8348. [Google Scholar] [CrossRef]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J.C. Numerical Integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of n-Alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.Py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Bello, M.; Morales-González, J.A. Molecular Recognition between Potential Natural Inhibitors of the Keap1-Nrf2 Complex. Int. J. Biol. Macromol. 2017, 105, 981–992. [Google Scholar] [CrossRef]
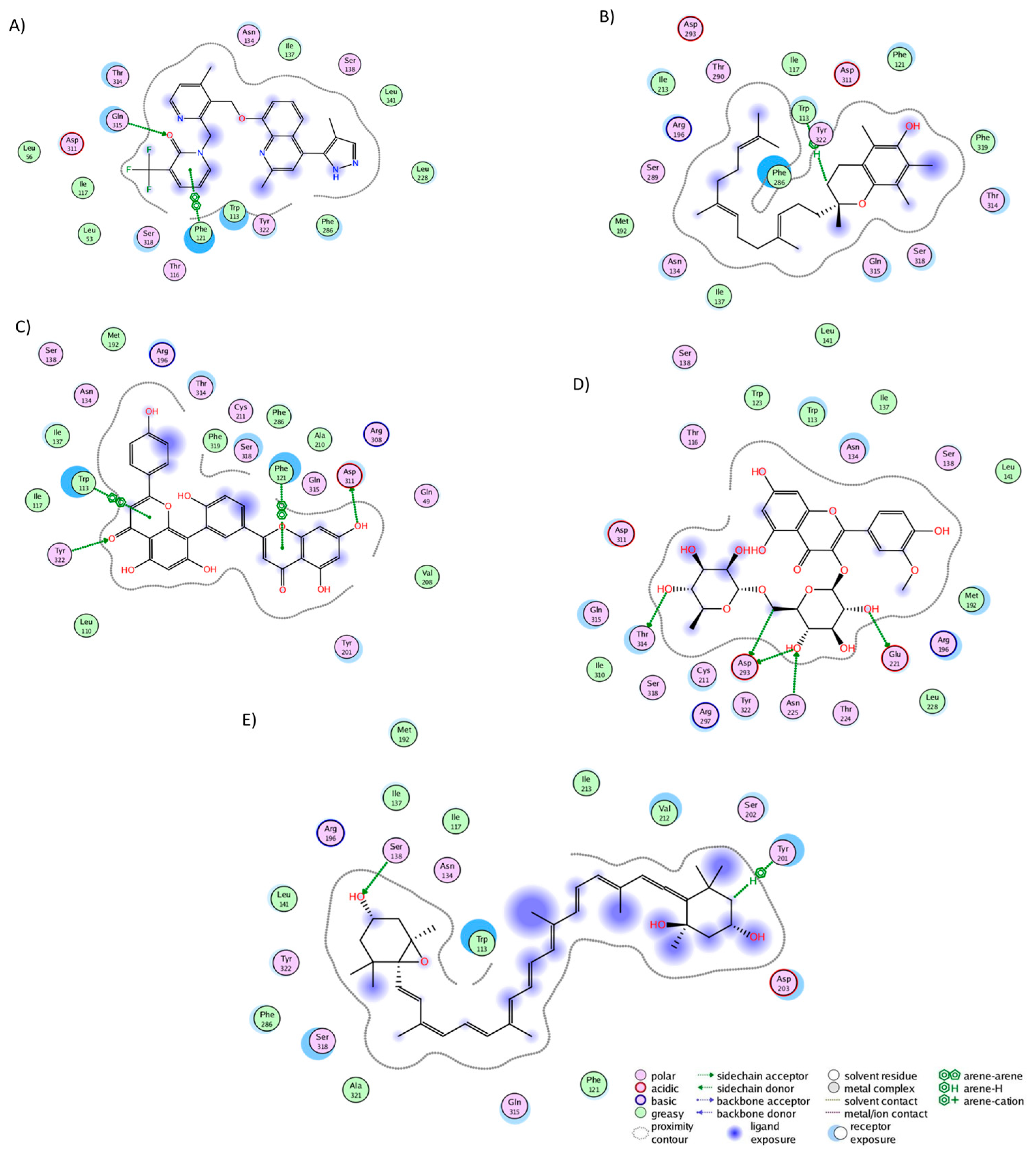


| Phytochemical Name | HR1 | Nkr1 | CLR | CRTH2 | BK2R | Mean Value | Top 15 Appearances |
|---|---|---|---|---|---|---|---|
| Amentoflavone | −6.362 | −9.604 | −9.565 | −8.880 | −8.384 | −8.559 | 4 |
| Alpha-tocotrienol | −7.310 | −8.402 | −9.108 | −9.258 | −6.959 | −8.207 | 4 |
| Neoxanthin | −6.856 | −7.964 | −8.872 | −9.529 | −7.184 | −8.081 | 4 |
| 7α-Hydroxy-sitosterol | −7.465 | −7.963 | −7.939 | −8.445 | −8.569 | −8.076 | 3 |
| Isorhamnetin 3-O-rutinoside | −6.138 | −8.519 | −8.373 | −8.698 | −8.181 | −7.982 | 4 |
| γ-Sitosterol | −7.337 | −7.738 | −7.603 | −8.356 | −8.603 | −7.927 | 2 |
| Cholecalciferol | −7.294 | −7.647 | −8.180 | −8.705 | −7.596 | −7.884 | 3 |
| Kaempferol-3-rutinoside | −5.993 | −8.404 | −8.418 | −8.147 | −8.278 | −7.848 | 3 |
| Isorhamnetin rutinoside | −6.069 | −7.814 | −7.916 | −8.620 | −8.718 | −7.828 | 2 |
| Epigallocatechin gallate | −6.431 | −7.826 | −8.923 | −8.355 | −7.516 | −7.810 | 1 |
| Hecogenin | −7.091 | −7.953 | −8.059 | −8.852 | −6.849 | −7.761 | 3 |
| Solanidine | −7.234 | −7.349 | −7.705 | −8.703 | −7.781 | −7.755 | 3 |
| 7β-Hydroxy-sitosterol | −5.974 | −7.701 | −7.867 | −8.482 | −8.645 | −7.734 | 1 |
| Dicaffeoylquinic acid | −6.519 | −8.128 | −8.253 | −8.511 | −7.205 | −7.723 | 1 |
| Epicatechin gallate | −6.458 | −7.329 | −8.938 | −8.610 | −7.180 | −7.703 | 2 |
| Known inhibitor | −7.311 | −9.039 | −11.292 | −9.207 | −8.375 | −9.045 | - |
| Receptor | Alpha-Tocotrienol | Amentoflavone | Isorhamnetin 3-O-Rutinoside | Neoxanthin | Known Inhibitor |
|---|---|---|---|---|---|
| HR1 | −38.493 (±5.11) | −28.679 (±3.30) | −26.446 (±4.21) | - | - |
| NKR1 | −45.400 (±3.28) | −33.954 (±3.70) | −46.756 (±4.78) | −39.455 (±7.95) | −38.343 (±3.61) |
| CLR1 | −54.785 (±2.89) | −36.620 (±3.39) | −41.335 (±3.13) | −61.436 (±5.20) | −49.684 (±4.43) |
| CRTH2 | −53.334 (±4.96) | −45.005 (±3.37) | −41.325 (±6.32) | −56.836 (±4.22) | −43.221 (±4.58) |
| BK2R | −42.654 (±3.71) | −38.984 (±5.37) | −37.080 (±4.76) | −47.394 (±4.81) | −32.310 (±4.73) |
| Receptor | Alpha-Tocotrienol | Known Inhibitor |
|---|---|---|
| HR1 | −34.65 (±4.70) | - |
| NKR1 | −35.7 (±4.65) | −29.52 (±5.01) |
| CLR1 | −45.55 (±3.07) | −38.95 (±4.65) |
| CRTH2 | −31.73 (±5.38) | −52.55 (±5.93) |
| BK2R | −31.93 (±3.71) | −32.310 (±4.73) |
| Receptor | Reported Inactivation | Ligand | Conformational Change Observed |
|---|---|---|---|
| HR1 | Extracellular: Antagonist pushes TM3, TM6, and TM7 away from its core [56]. Intracellular: Moves TM6 towards TM2, TM3, and TM7, which closes the G protein intracellular cavity [56]. | Alpha-tocotrienol | Extracellular: Doxepin pushes TM3 and TM6 away to a degree almost comparable to that of doxepin (Figure S3). Intracellular: Bring TM3 and TM6, as well as TM2 and TM7, together, which closed the G protein intracellular cavity (Figure S3). |
| NKR1 | Extracellular: The antagonist prompts TM5, TM6, and TM7 to move away from the center [57,63]. Intracellular: Closure of the G protein cavity between TM6, TM7, and TM5 [57,63]. | Alpha-tocotrienol | Extracellular: TM2 and TM7 become closer to the core’s perimeter, while TM6 moves in the direction of TM7 (Figure S5). Intracellular: Moves TM2 and TM7 inward, while also bringing TM3, TM5, and TM6 remarkably close (Figure S5). |
| Isorhamnetin-3-O-rutinoside | Extracellular: Moves TM6 and TM5 away from the core, while TM7 moves close to TM2 (Figure S5). Intracellular: Moves TM2, TM6, and TM7 inward (Figure S5). | ||
| Neoxanthin | |||
| CLR1 | Extracellular: Zafirlukast induces TM3, TM4, TM5, and TM7 to move away from the center (Figure S7). Intracellular: G proteins bind within TM3, TM5, TM6, and TM7 [60]. Zafirlukast shifts TM6 towards the center, effectively closing the cavity (Figure S7). | Alpha-tocotrienol | Extracellular: Slightly moves MT2, TM4, and TM7 away from the core (Figure S7) Intracellular: Induces an inward movement of TM2 and TM7 while slightly separating TM6 and TM7 (Figure S7). |
| Neoxanthin | Extracellular: Moves TM6 towards the core (Figure S7) Intracellular: Induces an outward movement of TM5, TM6, and TM7, comparable to an agonist-induced movement (Figure S7). | ||
| CRTH2 | Intracellular: The only known structural change is that the agonist causes helix 8 to undergo compaction, inhibiting arrestin recruitment [61,62]. | Alpha-tocotrienol | Intracellular: Leads to the loosening of helix 8 around Leu323 (Figure S9). |
| Amentoflavone | Intracellular: Results in the relaxation of helix 8 due to a loosening of the structure in Leu327, like the inhibitor (Figure S9). | ||
| Neoxanthin | Intracellular: Causes the loosening of helix 8 around Leu323 (Figure S9). | ||
| BK2R | Extracellular: TM6 moves outside the center of the core [64]. Intracellular: TM6 moves inward to avoid the generation of the BK2R-Gq complex within TM2, TM3, TM5, and TM7 [64]. | Alpha-tocotrienol | Extracellular: A slight movement of TM6 away from the core (Figure S10). Intracellular: Generates a movement of TM6 towards TM3, while TM7 moves towards TM6 (Figure S10). |
| Amentoflavone | No significant movement was observed. | ||
| Isorhamnetin-3-O-rutinoside | Extracellular: A slight movement of TM6 towards TM5 (Figure S10). Intracellular: Movement of TM6 and TM5 outside the core (Figure S10). | ||
| Neoxanthin | No significant movement was observed. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Culhuac, E.B.; Bello, M. Evaluation of Urtica dioica Phytochemicals against Therapeutic Targets of Allergic Rhinitis Using Computational Studies. Molecules 2024, 29, 1765. https://doi.org/10.3390/molecules29081765
Culhuac EB, Bello M. Evaluation of Urtica dioica Phytochemicals against Therapeutic Targets of Allergic Rhinitis Using Computational Studies. Molecules. 2024; 29(8):1765. https://doi.org/10.3390/molecules29081765
Chicago/Turabian StyleCulhuac, Erick Bahena, and Martiniano Bello. 2024. "Evaluation of Urtica dioica Phytochemicals against Therapeutic Targets of Allergic Rhinitis Using Computational Studies" Molecules 29, no. 8: 1765. https://doi.org/10.3390/molecules29081765
APA StyleCulhuac, E. B., & Bello, M. (2024). Evaluation of Urtica dioica Phytochemicals against Therapeutic Targets of Allergic Rhinitis Using Computational Studies. Molecules, 29(8), 1765. https://doi.org/10.3390/molecules29081765








