Evaluation of the Lipophilicity of Angularly Condensed Diquino- and Quinonaphthothiazines as Potential Candidates for New Drugs
Abstract
1. Introduction
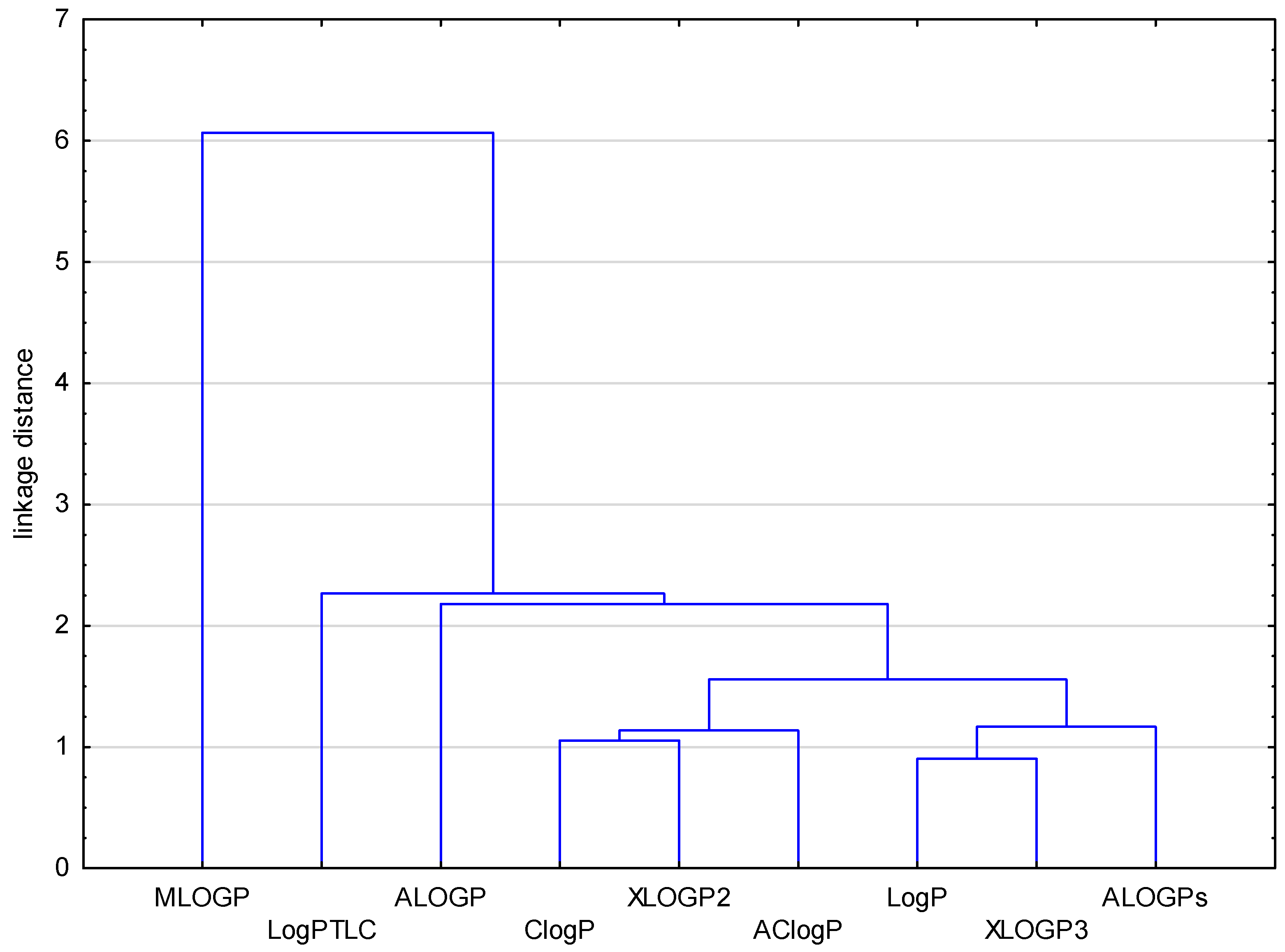
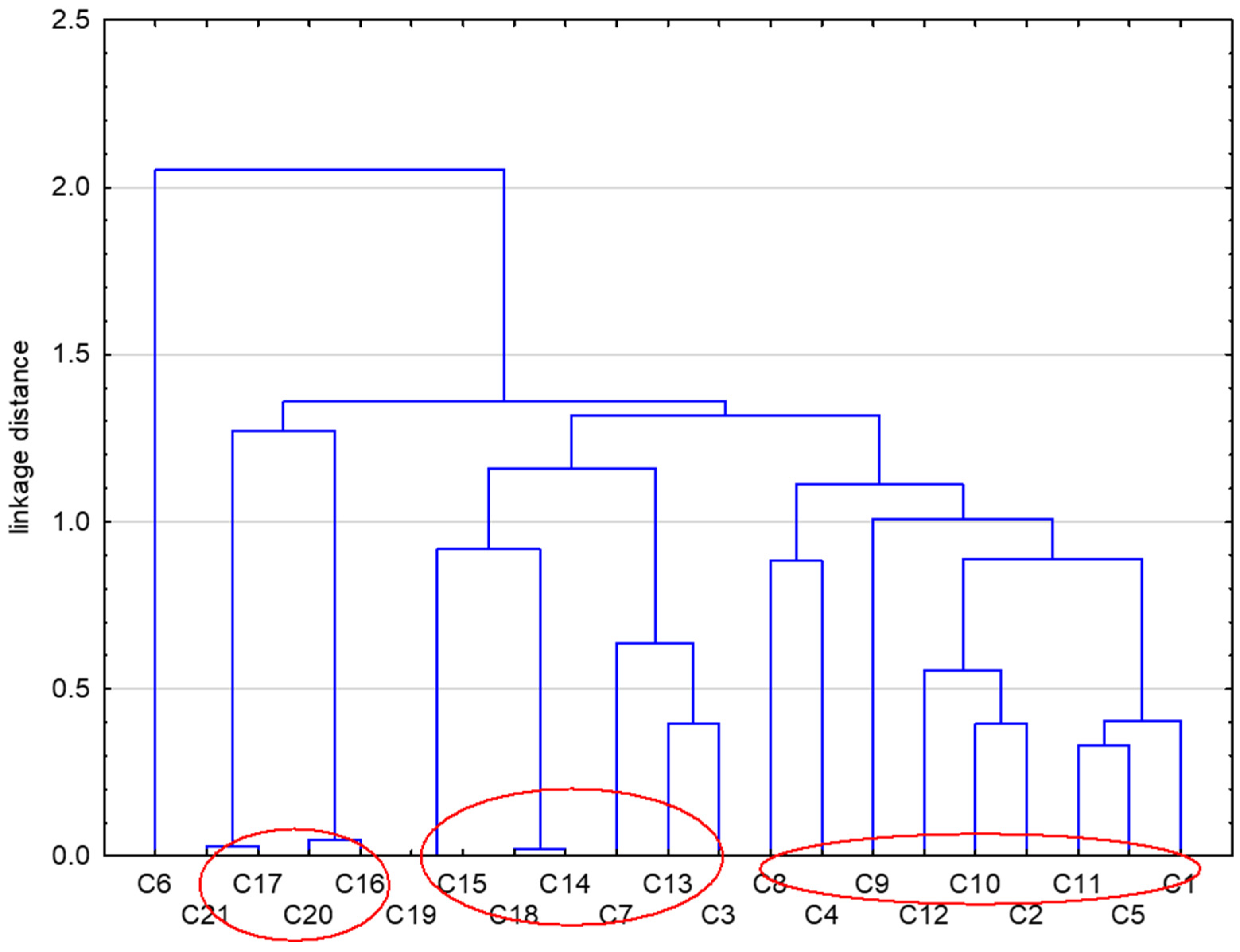
2. Results and Discussion
3. Materials and Methods
3.1. Materials
3.2. Chromatographic Procedure
3.3. Computational Programs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kempińska, D.; Chmiel, T.; Kot-Wasik, A.; Mróz, A.; Mazerska, Z.; Namieśnik, J. State of the art and prospects of methods for determination of lipophilicity of chemical compounds. TrAC Trends Anal. Chem. 2019, 113, 54–73. [Google Scholar] [CrossRef]
- Ferreira, L.L.G.; Andricopulo, A.D. ADMET modeling approaches in drug discovery. Drug Discov. Today 2019, 24, 1157–1165. [Google Scholar] [CrossRef] [PubMed]
- Tsopelas, F.; Giaginis, C.; Tsantili-Kakoulidou, A. Lipophilicity and biomimetic properties to support drug discovery. Expert Opin. Drug Discov. 2017, 12, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Arnott, J.A.; Planey, S.L. The influence of lipophilicity in drug discovery and design. Expert Opin. Drug Discov. 2012, 7, 863–875. [Google Scholar] [CrossRef] [PubMed]
- Jia, C.Y.; Li, J.Y.; Hao, G.F.; Yang, G.F. A drug-likeness toolbox facilitates ADMET study in drug discovery. Drug Discov. Today 2020, 25, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Chmiel, T.; Mieszkowska, A.; Kempińska-Kupczyk, D.; Kot-Wasik, A.; Namieśnik, J.; Mazerska, Z. The impact of lipophilicity on environmental processes, drug delivery and bioavailability of food components. Microchem. J. 2019, 146, 393–406. [Google Scholar] [CrossRef]
- Ginex, T.; Vazquez, J.; Gilbert, E.; Herrero, E.; Luque, F.J. Lipophilicity in drug design: An overview of lipophilicity descriptors in 3D-QSAR studies. Future Med. Chem. 2019, 11, 1177–1193. [Google Scholar] [CrossRef] [PubMed]
- Alqahtani, M.S.; Kazi, M.; Alsenaidy, M.A.; Ahmad, M.Z. Advances in oral drug delivery. Front. Pharmacol. 2021, 12, 618411. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Testa, B.; Fahr, A. Lipophilicity and its relationship with passive drug permeation. Pharm. Res. 2011, 28, 962–977. [Google Scholar] [CrossRef] [PubMed]
- Young, R.J.; Green, D.V.; Luscombe, C.N.; Hill, A.P. Getting physical in drug discovery II: The impact of chromatographic hydrophobicity measurements and aromaticity. Drug Discov. Today 2011, 16, 822–830. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, R.N. Determination of lipophilicity and its use as a predictor of blood-brain barrier penetration of molecular imaging agents. Mol. Imaging Biol. 2003, 5, 376–389. [Google Scholar] [CrossRef] [PubMed]
- Ciura, K.; Dziomba, S. Application of separation methods for in vitro prediction of blood–brain barrier permeability-The state of the art. J. Pharm. Biomed. Anal. 2020, 177, 112891. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, F.; Cole, S.; Green, C.; van de Waterbeemd, H. Lipophilicity and other parameters affecting brain penetration. Curr. Med. Chem.—Cent. Nerv. Syst. Agents 2002, 2, 229–240. [Google Scholar] [CrossRef]
- Whelan, R.; Hargaden, G.C.; Knox, A.J.S. Modulating the blood-brain barrier: A comprehensive review. Pharmaceutics 2021, 13, 1980. [Google Scholar] [CrossRef] [PubMed]
- Laznicek, M.; Laznickova, A. The effect of lipophilicity on the protein binding and blood cell uptake of some acidic drugs. J. Pharm. Biomed. Anal. 1995, 13, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Soares, J.; Santos, Á.; Fernandes, C.; Pinto, M.M. Liquid chromatography on the different methods for the determination of lipophilicity: An essential analytical tool in medicinal chemistry. Chemosensors 2022, 10, 340. [Google Scholar] [CrossRef]
- Rutkowska, E.; Pajak, K.; Jozwiak, K. Lipophilicity—Methods of determination and its role in medicinal chemistry. Acta Pol. Pharm. 2013, 70, 3–18. [Google Scholar]
- Giaginis, C.; Tsantili-Kakoulidou, A. Alternative measures of lipophilicity: From octanol-water partitioning to IAM retention. J. Pharm. Sci. 2008, 97, 2984–3004. [Google Scholar] [CrossRef]
- Hartmann, T.; Schmitt, J. Lipophilicity—Beyond octanol/water: A short comparison of modern technologies. Drug Discov. Today Technol. 2004, 1, 431–439. [Google Scholar] [CrossRef]
- Ciura, K.; Dziomba, S.; Nowakowska, J.; Markuszewski, M.J. Thin layer chromatography in drug discovery process. J. Chromatogr. A 2017, 1520, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Komsta, Ł.; Skibiński, R.; Berecka, A.; Gumieniczek, A.; Radkiewicz, B.; Radoń, M. Revisiting thin-layer chromatography as a lipophilicity determination tool-a comparative study on several techniques with a model solute set. J. Pharm. Biomed. Anal. 2010, 53, 911–918. [Google Scholar] [CrossRef]
- Valkó, K. Application of high-performance liquid chromatography based measurements of lipophilicity to model biological distribution. J. Chromatogr. A. 2004, 1037, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Mannhold, R.; Poda, G.I.; Ostermann, C.; Tetko, I.V. Calculation of molecular lipophilicity: State-of-the-art and comparison of log P methods on more than 96,000 compounds. J. Pharm. Sci. 2009, 98, 861–893. [Google Scholar] [CrossRef] [PubMed]
- Więckowski, K.; Czaja, A.; Woźniak, A.; Musiał, A.; Malawska, B. A study of the lipophilicity of amide derivatives of α-(1,2,3,4-tetrahydroisoquinolin-2-yl)-γ-hydroxybutyric acid by use of RP-TLC and calculation. JPC-J. Planar Chromat 2007, 20, 101–106. [Google Scholar] [CrossRef]
- Dobričić, V.; Turković, N.; Ivković, B.; Csuvik, O.; Vujić, Z. Evaluation of the lipophilicity of chalcones by RP-TLC and computational methods. JPC-J. Planar Chromat. 2020, 33, 245–253. [Google Scholar] [CrossRef]
- Wardecki, D.; Dołowy, M.; Bober-Majnusz, K.; Jampilek, J. Comparative study of the lipophilicity of selected anti-androgenic and blood uric acid lowering compounds. Molecules 2023, 28, 166. [Google Scholar] [CrossRef] [PubMed]
- Ciura, K.; Fedorowicz, J.; Andrić, F.; Greber, K.E.; Gurgielewicz, A.; Sawicki, W.; Sączewski, J. Lipophilicity Determination of Quaternary (fluoro)quinolones by chromatographic and theoretical approaches. Int. J. Mol. Sci. 2019, 20, 5288. [Google Scholar] [CrossRef] [PubMed]
- Jeleń, M.; Morak-Młodawska, B.; Korlacki, R. Anticancer activities of tetra-, penta-, and hexacyclic phenothiazines modified with quinoline moiety. J. Mol. Struct. 2023, 1287, 135700. [Google Scholar] [CrossRef]
- Jeleń, M.; Pluta, K.; Latocha, M.; Morak-Młodawska, B.; Suwińska, K.; Kuśmierz, D. Evaluation of angularly condensed diquinothiazines as potential anticancer agents. Bioorg. Chem. 2019, 87, 810–820. [Google Scholar] [CrossRef] [PubMed]
- Jeleń, M.; Pluta, K.; Suwinska, K.; Morak-Młodawska, B.; Latocha, M.; Shkurenko, A. Quinonaphthothiazines, syntheses, structures and anticancer activities. J. Mol. Struct. 2015, 1099, 10–15. [Google Scholar] [CrossRef]
- Jeleń, M.; Bavavea, E.I.; Pappa, M.; Kourounakis, A.P.; Morak-Młodawska, B.; Pluta, K. Synthesis of quinoline/naphthalene-containing azaphenothiazines and their potent in vitro antioxidant properties. Med. Chem. Res. 2015, 24, 1725–1732. [Google Scholar] [CrossRef] [PubMed]
- Jeleń, M.; Ying, P.T.C.; Hao, Y.J.; Balachandran, A.; Anamalay, K.; Morak-Młodawska, B.; Gaurav, A.; Lavilla, C.A., Jr.; Uy, M.M.; Billacura, M.P.; et al. In vitro study of antioxidant, antigylycation, sugar hydrolysis enzyme inhibitory effect and molecular in silico docking study of angularly condensed diquinothiazines. J. Mol. Struct. 2024, 1296, 136856. [Google Scholar] [CrossRef]
- Available online: http://www.vcclab.org (accessed on 10 January 2023).
- ACD labs. Release (Build 2726. Nov 2014); Advanced Chemistry Development, Inc.: Toronto, ON, Canada, 2015. [Google Scholar]
- ChemDraw: ChemDraw Ultra, version 12. PerkinElmer Informatics. CambridgeSoft: Cambridge, MA, USA, 2007.
- Bodor, N.; Gabanyi, Z.; Wong, C.K. A new method for the estimation of partition coefficient. J. Am. Chem. Soc. 1989, 111, 3783–3786. [Google Scholar] [CrossRef]
- Mannhold, R.; Cruciani, G.; Dross, K.; Rekker, R. Multivariate analysis of experimental and computational descriptors of molecular lipophilicity. J. Comput. Mol. Des. 1998, 12, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Brooke, D.; Dobbs, J.; Williams, N. Octanol:water partition coefficients (P): Measurement, estimation, and interpretation, particularly for chemicals with P > 105. Ecotoxic. Environ. Saf. 1986, 11, 251–260. [Google Scholar] [CrossRef]
- Castillo-Garit, J.A.; Marrero-Ponc, Y.; Torrens, F.; Garcia-Domenech, R. Estimation of ADME properties in drug discovery: Predicting Caco-2 cell premeability using atom-based stochastic and non-stochastic linear indices. J. Pharm. Sci. 2008, 97, 1946–1976. [Google Scholar] [CrossRef] [PubMed]
- Lund, M.; Petersen, T.S.; Dalhoff, K.P. Clinical implications of P-glycoprotein modulation in drug-drug interactions. Drugs 2017, 77, 859–883. [Google Scholar] [CrossRef] [PubMed]
- Karthika, C.; Sureshkumar, R.; Zehravi, M.; Akter, R.; Ali, F.; Ramproshad, S.; Mondal, B.; Tagde, P.; Ahmed, Z.; Khan, F.S.; et al. Multidrug resistance of cancer cells and the vital role of P-Glycoprotein. Life 2022, 12, 897. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P. Inhibition of cytochrome p450 enzymes by drugs-molecular basis and practical applications. Biomol. Ther. 2022, 30, 1–18. [Google Scholar] [CrossRef]
- Zhao, T.; He, Y.-Q.; Wang, J.; Ding, K.-M.; Wang, C.-H.; Wang, Z.-T. Inhibition of human cytochrome P450 enzymes 3A4 and 2D6 by β-carboline alkaloids, harmine derivatives. Phythother. Res. 2011, 25, 1671–1677. [Google Scholar] [CrossRef] [PubMed]
- Manikandan, P.; Nagini, S. Cytochrome P450 structure, function and clinical significance: A review. Curr. Drug Targets 2018, 19, 38–54. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.F.; Chan, E.; Zhou, Z.W.; Xue, C.C.; Lai, X.; Duan, W. Insights into the structure, function, and regulation of human cytochrome P450 1A2. Curr. Drug Metab. 2009, 10, 713–729. [Google Scholar] [CrossRef] [PubMed]
- Bansal, A.; Kaushik, V.; Sharma, N.R. Synthesis and in silico anti-metastatic evaluation of carvacrol derivative, 2-hydroxy-6-isopropyl-3-methylbenzalehyde. Mater. Today Proc. 2022, 57, 739–747. [Google Scholar] [CrossRef]
- Martinez-Quintana, E.; Rodriguez-Gonzalez, F.; Medina-Gil, J.M.; Garay-Sanchez, P.; Tugores, A. CYP2C19 activity and cardiovascular risk factors in patients with an acute coronary syndrome. Med. Clin. 2017, 149, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J. Clinical application of CYP2C19 pharmacogenetics toward more personalized medicine. Front. Genet. 2013, 3, 318. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.; Crosby, I.; Yip, V.; Maguire, P.; Pirmohamed, M.; Turner, R.M. A review of the important role of CYP2D6 in pharmacogenomics. Genes 2020, 30, 1295. [Google Scholar] [CrossRef]
- Luo, G.; Guenthner, T.; Gan, L.S.; Humphreys, W.G. CYP3A4 induction by xenobiotics: Biochemistry, experimental methods and impact on drug discovery and development. Curr. Drug Metab. 2004, 5, 483–505. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://swissadme.ch (accessed on 12 March 2024).
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and premeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef] [PubMed]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. Knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Egan, W.J.; Merz, K.M., Jr.; Baldwin, J.J. Prediction of drug absorption using multivariate statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef] [PubMed]
- Stanisz, A. Przystępny Kurs Statystyki z Zastosowaniem STATISTICA PL na Przykładach Medycyny; Analizy Wielowymiarowe; StatSoft Polska: Kraków, Poland, 2007. [Google Scholar]
| No. of Compound | Chemical Structure | No. of Compound | Chemical Structure | No. of Compound | Chemical Structure |
|---|---|---|---|---|---|
| 1 | 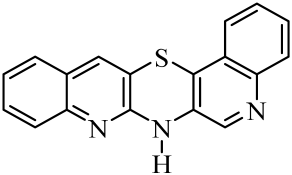 | 8 | 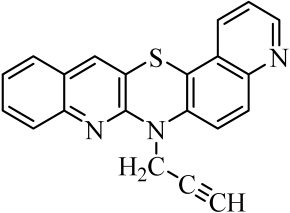 | 15 |  |
| 2 |  | 9 | 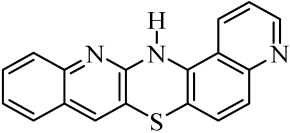 | 16 | 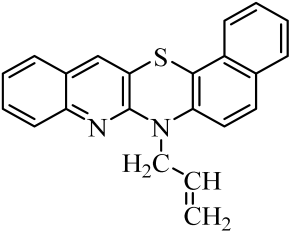 |
| 3 |  | 10 | 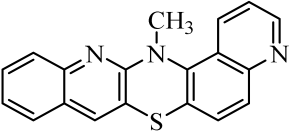 | 17 |  |
| 4 | 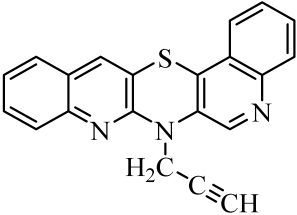 | 11 |  | 18 |  |
| 5 | 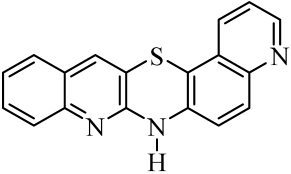 | 12 | 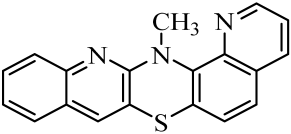 | 19 | 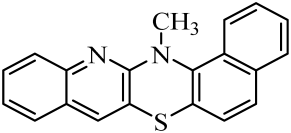 |
| 6 | 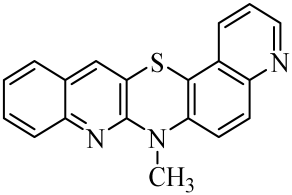 | 13 | 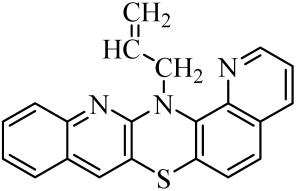 | 20 | 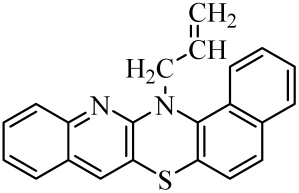 |
| 7 |  | 14 | 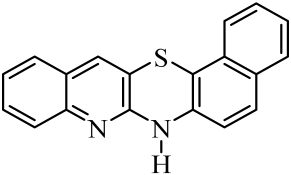 | 21 |  |
| No. of Compound | ALOGPs | AClogP | ALOGP | MLOGP | XLOGP2 | XLOGP3 | LogP * | ClogP ** |
|---|---|---|---|---|---|---|---|---|
| 1 | 4.90 | 4.66 | 4.58 | 2.78 | 4.72 | 4.48 | 4.54 | 5.23 |
| 2 | 4.61 | 5.37 | 4.79 | 3.01 | 5.02 | 4.62 | 4.92 | 5.09 |
| 3 | 5.06 | 5.97 | 5.40 | 3.39 | 5.64 | 5.26 | 5.35 | 5.86 |
| 4 | 4.87 | 5.26 | 5.98 | 3.39 | 5.29 | 4.73 | 4.77 | 5.51 |
| 5 | 4.93 | 4.66 | 4.58 | 3.17 | 4.72 | 4.48 | 4.54 | 5.13 |
| 6 | 4.64 | 5.37 | 4.79 | 3.40 | 5.02 | 4.62 | 4.92 | 5.03 |
| 7 | 5.06 | 5.97 | 5.40 | 3.78 | 5.64 | 5.26 | 5.35 | 5.80 |
| 8 | 4.91 | 5.26 | 5.98 | 3.78 | 5.29 | 4.73 | 4.77 | 5.48 |
| 9 | 4.92 | 4.66 | 5.58 | 3.17 | 4.72 | 4.48 | 4.41 | 5.13 |
| 10 | 4.64 | 5.37 | 4.79 | 3.40 | 5.02 | 4.64 | 4.92 | 5.03 |
| 11 | 4.93 | 4.66 | 4.58 | 3.17 | 4.72 | 4.48 | 4.87 | 5.13 |
| 12 | 4.59 | 5.37 | 4.79 | 3.40 | 5.02 | 4.62 | 4.92 | 5.03 |
| 13 | 5.04 | 5.97 | 5.40 | 3.78 | 5.64 | 5.26 | 5.35 | 5.80 |
| 14 | 5.87 | 5.53 | 5.30 | 4.24 | 5.88 | 5.46 | 5.63 | 6.12 |
| 15 | 5.54 | 6.24 | 5.51 | 4.47 | 6.19 | 5.60 | 5.80 | 6.12 |
| 16 | 6.10 | 6.84 | 6.13 | 4.83 | 6.80 | 6.24 | 6.28 | 6.90 |
| 17 | 5.63 | 6.13 | 6.70 | 4.83 | 6.45 | 5.72 | 5.98 | 6.54 |
| 18 | 5.85 | 5.53 | 5.30 | 4.24 | 5.88 | 5.46 | 5.63 | 6.11 |
| 19 | 5.54 | 6.24 | 5.51 | 4.47 | 6.19 | 5.60 | 5.80 | 6.12 |
| 20 | 6.05 | 6.84 | 6.13 | 4.83 | 6.80 | 6.24 | 6.28 | 6.90 |
| 21 | 5.60 | 6.13 | 6.70 | 4.83 | 6.45 | 5.72 | 5.98 | 6.54 |
| No. of Compound | RM0 | r | b | LogPTLC |
|---|---|---|---|---|
| 1 | 3.15 | 0.9953 | −0.0383 | 4.26 |
| 2 | 4.14 | 0.9954 | −0.0503 | 5.57 |
| 3 | 4.30 | 0.9944 | −0.058 | 5.77 |
| 4 | 3.35 | 0.9953 | −0.0383 | 4.56 |
| 5 | 3.72 | 0.9927 | −0.0459 | 5.03 |
| 6 | 4.24 | 0.9982 | −0.0582 | 5.69 |
| 7 | 4.46 | 0.9988 | −0.0534 | 5.97 |
| 8 | 3.96 | 0.9987 | −0.0539 | 5.34 |
| 9 | 3.69 | 0.9969 | −0.0498 | 4.99 |
| 10 | 4.37 | 0.9977 | −0.0540 | 5.86 |
| 11 | 3.48 | 0.9960 | −0.0462 | 4.72 |
| 12 | 4.21 | 0.9989 | −0.0576 | 5.65 |
| 13 | 3.58 | 0.9928 | −0.0478 | 4.85 |
| 14 | 4.24 | 0.9967 | −0.0573 | 5.69 |
| 15 | 4.49 | 0.9965 | −0.0527 | 6.01 |
| 16 | 4.60 | 0.9979 | −0.0571 | 6.15 |
| 17 | 4.29 | 0.9985 | −0.0529 | 5.76 |
| 18 | 3.81 | 0.9934 | −0.0484 | 5.14 |
| 19 | 4.18 | 0.9931 | −0.0538 | 5.58 |
| 20 | 4.34 | 0.9973 | −0.0548 | 5.82 |
| 21 | 4.09 | 0.9977 | −0.0507 | 5.50 |
| Lipophilicity Parameter | Standard | ||||
|---|---|---|---|---|---|
| I | II | III | IV | V | |
| logPlit | 1.21 [36] | 1.87 [37] | 3.18 [37] | 4.45 [37] | 6.38 [38] |
| RM0 | 0.78 | 1.16 | 2.51 | 3.33 | 4.69 |
| -b | 0.0162 | 0.0247 | 0.0328 | 0.0412 | 0.0564 |
| r | 0.992 | 0.994 | 0.997 | 0.998 | 0.998 |
| logPTLC | 1.21 | 1.70 | 3.43 | 4.49 | 6.24 |
| No. of Compound | Caco-2 [10−6 cm/s] | PPB [%] | CNS | HIA [%] | P-gp Substrates | CYP1A2 Inhibitor | CYP2C9 Inhibitor | CYP2C19 Inhibitor | CYP2D6 Inhibitor | CYP3A4 Inhibitor |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 226 | 99 | −3.30 | 100 | 0.30 | 0.61 | 0.34 | 0.49 | 0.48 | 0.53 |
| 2 | 231 | 99 | −3.23 | 100 | 0.38 | 0.57 | 0.26 | 0.49 | 0.46 | 0.50 |
| 3 | 207 | 100 | −3.44 | 100 | 0.35 | 0.62 | 0.33 | 0.52 | 0.48 | 0.53 |
| 4 | 235 | 100 | −3.96 | 100 | 0.35 | 0.68 | 0.40 | 0.51 | 0.42 | 0.54 |
| 5 | 226 | 99 | −3.30 | 100 | 0.30 | 0.58 | 0.34 | 0.50 | 0.48 | 0.58 |
| 6 | 231 | 99 | −3.23 | 100 | 0.38 | 0.61 | 0.26 | 0.49 | 0.46 | 0.54 |
| 7 | 207 | 100 | −3.44 | 100 | 0.35 | 0.55 | 0.33 | 0.51 | 0.48 | 0.53 |
| 8 | 235 | 100 | −3.96 | 100 | 0.35 | 0.67 | 0.40 | 0.50 | 0.37 | 0.54 |
| 9 | 230 | 99 | −3.30 | 100 | 0.30 | 0.58 | 0.34 | 0.50 | 0.48 | 0.58 |
| 10 | 231 | 99 | −3.23 | 100 | 0.38 | 0.61 | 0.26 | 0.49 | 0.46 | 0.54 |
| 11 | 211 | 99 | −3.30 | 100 | 0.30 | 0.58 | 0.34 | 0.50 | 0.48 | 0.58 |
| 12 | 231 | 99 | −3.23 | 100 | 0.38 | 0.61 | 0.26 | 0.49 | 0.46 | 0.54 |
| 13 | 207 | 100 | −3.44 | 100 | 0.35 | 0.55 | 0.33 | 0.51 | 0.48 | 0.53 |
| 14 | 131 | 100 | −3.51 | 100 | 0.28 | 0.66 | 0.33 | 0.51 | 0.49 | 0.53 |
| 15 | 163 | 100 | −3.40 | 100 | 0.38 | 0.68 | 0.26 | 0.53 | 0.47 | 0.50 |
| 16 | 94 | 100 | −3.72 | 100 | 0.38 | 0.63 | 0.32 | 0.57 | 0.50 | 0.51 |
| 17 | 137 | 100 | −4.30 | 100 | 0.34 | 0.71 | 0.39 | 0.53 | 0.43 | 0.56 |
| 18 | 131 | 100 | −3.51 | 100 | 0.28 | 0.66 | 0.33 | 0.51 | 0.49 | 0.53 |
| 19 | 163 | 100 | −3.40 | 100 | 0.38 | 0.68 | 0.26 | 0.53 | 0.47 | 0.50 |
| 20 | 94 | 100 | −3.72 | 100 | 0.38 | 0.63 | 0.32 | 0.57 | 0.50 | 0.51 |
| 21 | 137 | 100 | −4.30 | 100 | 0.34 | 0.71 | 0.39 | 0.53 | 0.43 | 0.56 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klimoszek, D.; Jeleń, M.; Morak-Młodawska, B.; Dołowy, M. Evaluation of the Lipophilicity of Angularly Condensed Diquino- and Quinonaphthothiazines as Potential Candidates for New Drugs. Molecules 2024, 29, 1683. https://doi.org/10.3390/molecules29071683
Klimoszek D, Jeleń M, Morak-Młodawska B, Dołowy M. Evaluation of the Lipophilicity of Angularly Condensed Diquino- and Quinonaphthothiazines as Potential Candidates for New Drugs. Molecules. 2024; 29(7):1683. https://doi.org/10.3390/molecules29071683
Chicago/Turabian StyleKlimoszek, Daria, Małgorzata Jeleń, Beata Morak-Młodawska, and Małgorzata Dołowy. 2024. "Evaluation of the Lipophilicity of Angularly Condensed Diquino- and Quinonaphthothiazines as Potential Candidates for New Drugs" Molecules 29, no. 7: 1683. https://doi.org/10.3390/molecules29071683
APA StyleKlimoszek, D., Jeleń, M., Morak-Młodawska, B., & Dołowy, M. (2024). Evaluation of the Lipophilicity of Angularly Condensed Diquino- and Quinonaphthothiazines as Potential Candidates for New Drugs. Molecules, 29(7), 1683. https://doi.org/10.3390/molecules29071683








